Abstract
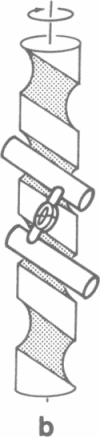
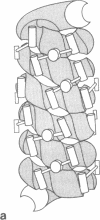
Catabolite activator protein (CAP) is a dimeric molecule (Mr = 2 × 22,500) involved in transcription initiation of several catabolite-sensitive genes of Escherichia coli. The present communication proposes a model for the interaction of CAP with DNA. The model is based upon known geometrical features of the CAP molecule [McKay, D. B. & Steitz, T. A. (1981) Nature (London) 290, 744-749], which allow interaction between dyad-related α-helices of the dimeric protein and major grooves in adjacently aligned sections of right-handed B-DNA. These geometrical features suggest that in vivo CAP binding to closed-circular DNA involves CAP bridging adjacent loops of a DNA solenoidal coil. This interaction pattern is shown to be consistent with the geometrical and stoichiometric properties of nonspecifically bound CAP complexes observed by Chang et al. [Chang, J. J., Dubochet, J., Baudras, A., Blazy, B. & Takahashi, M. (1981) J. Mol. Biol. 150, 435-439]. CAP-induced coil formation is related to in vivo CAP potentiation of RNA polymerase activity in underwound closedcircular DNA. Specifically, it is proposed that CAP binding to the right-interwound form of supercoiled DNA effects a local redistribution of DNA twist-strain energy, thus resulting in the formation of a left-handed solenoidal loop. The production of this localized solenoidal loop, which reflects compensatory alterations in DNA twist and writhe, may provide a conformationally unique site for RNA polymerase binding where the DNA is partially unwound. The proposed interaction pattern is consistent with both recent DNA unwinding experiments and various nuclease protection data. Moreover, features of the model suggest that the repetitive and symmetric character of many promoter sequences may provide the structural basis for a switching mechanism operative in the differential control of gene transcription.
Keywords: gene expression, protein-DNA interactions
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Toroidal elastic supercoiling of DNA. Biopolymers. 1980 Oct;19(10):1705–1713. doi: 10.1002/bip.1980.360191002. [DOI] [PubMed] [Google Scholar]
- Chang J. J., Dubochet J., Baudras A., Blazy B., Takahashi M. Electron microscope observation of a fibre structure formed by non-specific binding of cAMP receptor protein to DNA. J Mol Biol. 1981 Aug 15;150(3):435–439. doi: 10.1016/0022-2836(81)90558-1. [DOI] [PubMed] [Google Scholar]
- De Crombrugghe B., Perlman R. L., Varmus H. E., Pastan I. Regulation of inducible enzyme synthesis in Escherichia coli by cyclic adenosine 3', 5'-monophosphate. J Biol Chem. 1969 Nov 10;244(21):5828–5835. [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Echols H. Developmental pathways for the temperate phage: lysis vs lysogeny,. Annu Rev Genet. 1972;6(0):157–190. doi: 10.1146/annurev.ge.06.120172.001105. [DOI] [PubMed] [Google Scholar]
- Epstein W., Rothman-Denes L. B., Hesse J. Adenosine 3':5'-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2300–2304. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Conformational flexibility of DNA: polymorphism and handedness. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6486–6490. doi: 10.1073/pnas.77.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Control of gene expression in bacteriophage lambda. Annu Rev Genet. 1973;7:289–324. doi: 10.1146/annurev.ge.07.120173.001445. [DOI] [PubMed] [Google Scholar]
- Herskowitz I., Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Kolb A., Buc H. Is DNA unwound by the cyclic AMP receptor protein? Nucleic Acids Res. 1982 Jan 22;10(2):473–485. doi: 10.1093/nar/10.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Musso R. E., Di Lauro R., Adhya S., de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977 Nov;12(3):847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- O'Neill M. C., Amass K., de Crombrugghe B. Molecuar model of the DNA interaction site for the cyclic AMP receptor protein. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2213–2217. doi: 10.1073/pnas.78.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. K. The flexible DNA double helix. II. Superhelix formation. Biopolymers. 1979 May;18(5):1235–1260. doi: 10.1002/bip.1979.360180515. [DOI] [PubMed] [Google Scholar]
- Parker D. E. The vestibular apparatus. Sci Am. 1980 Nov;243(5):118–135. doi: 10.1038/scientificamerican1180-118. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Mazur S. J., Melançon P., Roe J. H., Shaner S. L., Unger L. Double helical DNA: conformations, physical properties, and interactions with ligands. Annu Rev Biochem. 1981;50:997–1024. doi: 10.1146/annurev.bi.50.070181.005025. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Schmitz A. Cyclic AMP receptor proteins interacts with lactose operator DNA. Nucleic Acids Res. 1981 Jan 24;9(2):277–292. doi: 10.1093/nar/9.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B. Interaction of the cAMP receptor protein with the lac promoter. Nucleic Acids Res. 1980 Feb 25;8(4):759–766. [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., O'Neill M., de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hippel P. H., McGhee J. D. DNA-protein interactions. Annu Rev Biochem. 1972;41(10):231–300. doi: 10.1146/annurev.bi.41.070172.001311. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R. B., Berg O. G., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981 Nov 24;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- Zubay G., Schwartz D., Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci U S A. 1970 May;66(1):104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Santis P., Morosetti S., Palleschi A., Savino M. Conformational analysis of double-stranded B-type DNA structures. Biopolymers. 1981 Aug;20(8):1727–1739. doi: 10.1002/bip.1981.360200812. [DOI] [PubMed] [Google Scholar]