Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that negatively regulate gene expression of their targets at the post-transcriptional levels. A single miRNA can target up to several hundred mRNAs, thus capable of significantly altering gene expression regulatory networks. In-depth study and characterization of miRNAs has elucidated their critical functions in development, homeostasis, and disease. A link between miRNAs and longevity has been demonstrated in C. elegans, implicating their role in regulation of lifespan and in the aging process. Recent years have witnessed unprecedented technological advances in studies of miRNAs, including ultra-high throughput sequencing technologies that allow comprehensive discovery of miRNAs and their targets. Here we review the latest experimental approaches from the perspective of understanding miRNA gene expression regulatory networks in aging. We provide a methodological work flow that can be employed to discover aging-related miRNAs and their targets, and to functionally validate their roles in aging. Finally, we review the links between miRNAs known to act in the conserved pathways of aging and major aging-related diseases. Taken together, we hope to provide a focused review to facilitate future endeavor of uncovering the functional role of miRNA in aging.
Keywords: Aging, Longevity, Gene regulatory network, High-throughput sequencing, microRNAs, Target validation.
1. INTRODUCTION
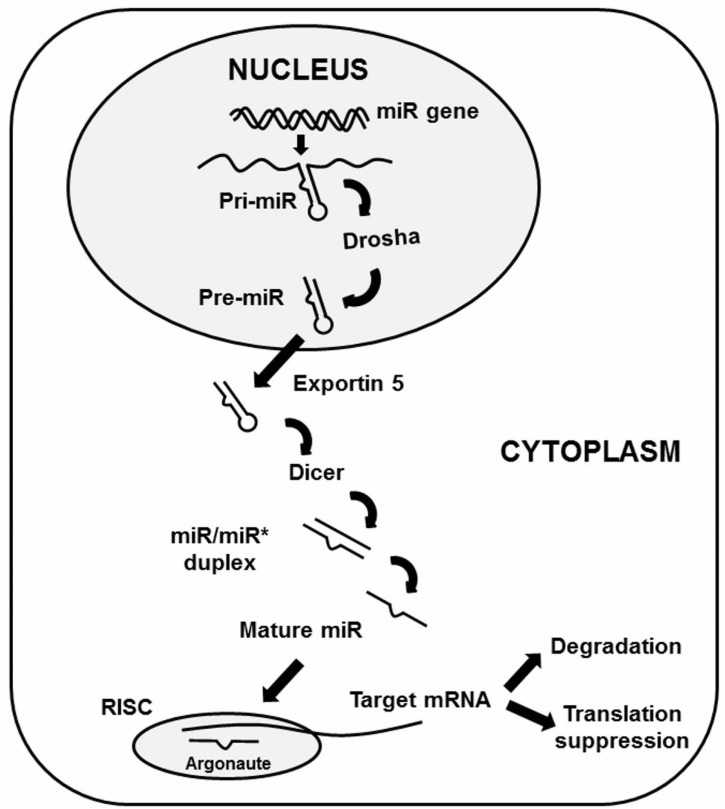
MicroRNAs (miRNAs), first discovered in C. elegans [1], are small non-coding RNA species that regulate gene expression at the post-transcriptional level [2]. Mature miRNA are between 18-25 nucleotides (nt) in length and are initially transcribed as primary-miRNA (pri-miRNA) molecules which contain a characteristic stem loop structure. These non-coding RNAs undergo two processing steps [3] Fig. (1). The first step is the generation of stem-loop precursors (pre-miRNAs) of ~70nt in length by the enzyme Drosha in a micro-processing complex within the nucleus. The pre-miRNAs are then exported into the cytoplasm by exportin 5 and further processed into double stranded RNAs (miRNA-miRNA* duplex) by Dicer. The mature miRNA strand of this duplex is loaded into an Argonaute-containing miRNA-induced silencing complex (miRISC). In contrast, the complementary strand known as miRNA* (miRNA star) were thought to be degraded. However a growing body of work challenges the dogma that miRNA* is simply a non-functional byproduct of miRNA biogenesis, suggesting instead that miRNA* plays a significant role in cellular function and human disease [4, 5]. The mature miRNA within miRISC serves as a guide for recognizing target mRNAs by partial base-pairing.
Fig. (1).
Biogenesis of microRNA. MicroRNA (miRNA) genes are typically transcribed into primary miRNA (pri-miRNA) transcripts that undergo processing by Drosha-containing complexes in animals. The unified hairpin precursor miRNAs (pre-miRNAs) are transported to the cytoplasm by exportin 5 (XPO5). The Dicer complex removes the loop region from pre-miRNAs, and one strand of the resulting duplex is bound by Argonaute to form a miRNA-induced silencing complex (miRISC). After multiple processing, mature miRNAs target complementary transcripts and downregulate gene expression by mRNA degradation or translational repression.
The mature miRNA primarily targets the 3’ UTR of an mRNA strand based on sequence homology [6]. The nucleotides in the 2-7 position of the 5’ end of the mature miRNA comprise a “seed region”. Once an mRNA is targeted by a miRNA, its gene expression is down-regulated either by induction of mRNA degradation or blocking of translation by the miRNA, which occurs through conserved mechanisms [2, 7]. Perfect pairing of a miRNA with its target site supports endonucleolytic cleavage of the mRNA by Argonaute [8, 9]. Binding of the miRISC, which includes GW182 proteins, to 3’UTR target sequences has been shown to induce the recruitment of deadenylation factors that remove the poly(A) tail and make the mRNA susceptible to exonucleolytic degradation [10-15]. Although translation repression by miRNA occurs at the targeted mRNAs through inhibition of translation initiation or elongation [16-22], recent studies suggest that mRNA degradation is the primary mechanism by which miRNAs reduce protein output [23, 24].
One miRNA can target multiple mRNAs, and one mRNA can be targeted by multiple, distinct miRNAs; therefore miRNAs can significantly alter gene expression regulatory networks. The profound impact of miRNA on the gene regulatory networks has led to the in-depth study and characterization of miRNAs, elucidating their critical function in development, homeostasis, and disease such as cardiovascular disease [25] and neurodegenerative disease [26]. Thus far 1048 human miRNA sequences have been identified through cloning, sequencing, or computational analysis [27].
2. HIGH-THROUGHPUT SEQUENCING FOR DISCOVERY OF miRNAs AND TARGETs
The multitude of important roles played by miRNAs indicates that they are a critical genetic component of gene regulatory networks. However, quantification of miRNA has been technically challenging due to small size, low copy number, interference from other small RNAs, and contamination by degradation products of mRNAs or other RNA species. Until recently, the only known and computationally predicted miRNAs have been interrogated using hybridization-based array methods, an assay of limited value due to cross-hybridization, array content, and the inability to discover novel miRNAs. The increased availability and affordability of massively parallel sequencing offers a dramatically improved method to gain high-resolution views of miRNA expression [28, 29]. This technology has recently been utilized to profile expression of miRNAs in several species, including humans. Currently three commercial platforms for high-throughput sequencing are widely employed: Roche’s 454/FLX system, Illumina’s Genome Analyzer (formerly known as Solexa sequencing and succeeded by Illumina’s more recent model the HiSeq 2000) and ABI’s SOLiD. The choice of sequencing method often comes down to cost, read length and sequencing depth. Because miRNAs are in the range of approximately 18 to 30 nt and high sequencing depth is necessary to observe rare species, Illumina and SOLiD are currently the most cost-efficeint platforms for miRNAs sequencing studies.
Illumina uses a four-color, reversible terminator sequencing-by-synthesis technology to sequence one base at a time [28]. SOLiD uses 16 dinucleotide probes, each labeled with one of four fluorophores, to sequence by ligation two nucleotides of each clone at a time [30]. Sequencing cost has been further reduced by multiplex sequencing of indexed libraries, which allows sequencing of two or more samples in a single lane [31-39]. The number of samples for multiplexing varies depending upon the desired sequencing depth. By incorporating a unique sequence called a bar code or index into the 5’- or 3’-adapter of each library, or by adding the bar code during a PCR step after adapter ligation, the identity of each sample can be denoted. Multiplexing library preparation kits are now available for both Illumina and SOLiD.
3. ANALYSIS OF HIGH-THROUGHPUT miRNA SEQUENCING DATA
There exist several analytical tools and databases that allow analysis of unprecedented amounts of miRNA-seq data from the high-throughput sequencing platforms for miRNA discovery and expression profiling as well as comparing miRNA profiles across a broad spectrum of species, tissues and diseases [40]. miRBase (http://www.mirbase.org/) is currently the repository for miRNAs. This database, which is updated regularly, stores information about the mature miRNA sequences, precursor sequences, map locations, and overlapping annotations, as well as predicted targets and a complete list of all publications that support each of the miRNA entries. An increasing number of species are included in every new miRBase version [41]. Other miRNA analysis platforms including miRDeep, miRNAkey, UEAsRNA toolkit, miRanalyzer, SeqBuster, DSAP, mirTools, E-miR and SigTerms are available for miRNA discovery and profiling and the identification of functional miRNA-mRNA pairs from deep sequencing [40]. miRecords is a resource for animal miRNA-target interactions, consisting of two components [42]; the Validated Targets component is a large, high-quality database of experimentally validated miRNA targets resulting from meticulous literature curation and the Predicted Targets component of miRecords is an integration of predicted miRNA targets produced by 11 established miRNA target prediction programs.
Recently GOmir has been developed for the target prediction and ontology clustering, which consists of two different components, JTarget and TAGGO [43]. JTarget combines the data from four different prediction databases (TargetScan, miRanda, RNAhybrid and PicTar) and also from the experimental database TarBase [44]. TAGGO provides detailed assignments from Gene Ontology (GO) resources to gene products. The expression patterns of miRNAs across tissues can be obtained from NCBI Gene Expression Omnibus GEO, miRGator [45], and microRNA.org. Databases such as miRSigDB [46] and miRGator allow high-level integration to understand how miRNAs expressed in a given sample relate to their putative targets in the context of signaling pathways. Furthermore, miRSigDB, miRGator, and recently published microRNA Expression and Sequence Analysis Database (mESAdb) permit miRNA expression and target gene expression to be linked to human diseases [47].
4. IDENTIFICATION OF FUNCTIONAL miRNA TARGETS
One of the challenges in the emerging field of miRNA biology is the identification of functional miRNA targets. A given miRNA may have multiple (up to several hundred) predicted gene targets, and ~60% of mRNAs have predicted binding sites for several miRNAs in their 3’ UTRs. But since miRNA regulation of an mRNA requires only a short (eight-nucleotide or fewer) match in their sequences, it has proved almost impossible to definitively determine which among many predicted mRNA binding sites is the in vivo target for each miRNA. Recently, several methods have been developed to identify molecular targets of miRNAs based on crosslinking approaches.
A. High-Throughput Sequencing of RNA Isolated by Crosslinking Immunoprecipitation (HITS-CLIP)
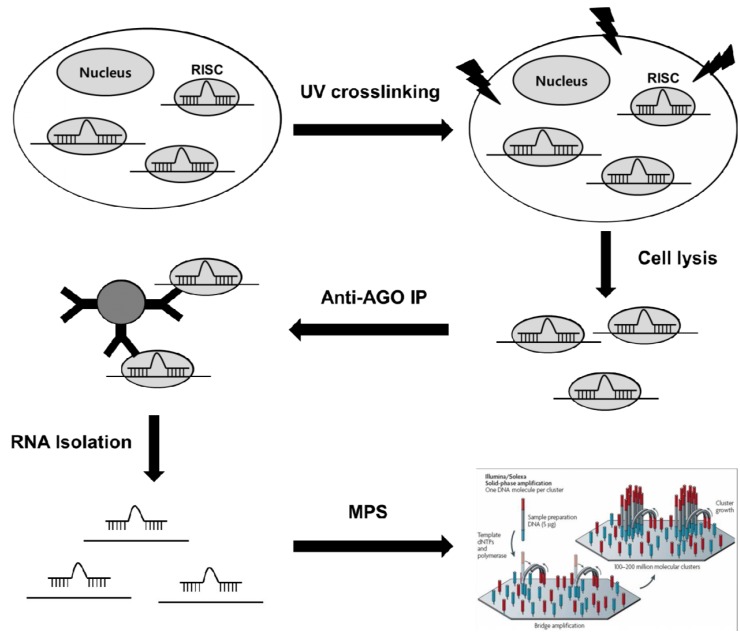
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP), is a genome-wide method to identify functional protein–RNA interaction sites [48]. This method utilizes ultraviolet (UV)-induced covalent crosslinking to stabilize RNA binding to its associated RNA-binding proteins, thereby enhancing the ability to capture more transient miRNA-mRNA interactions, prior to immunoprecipitation with antibodies for the protein. Deep sequencing of bound RNAs then allows comprehensive identification of functional interaction targets. HITS-CLIP can be used to covalently crosslink native Argonaute (Ago) protein-RNA complexes, producing two simultaneous datasets, Ago-miRNA and Ago-mRNA binding sites, that together allow identification of miRNA-target mRNA interaction sites through bioinformatic analysis [48]. Briefly, biological samples are irradiated with UV light (245 nm) and crosslinked samples are then immunoprecipitated with anti-AGO antibodies. Mild RNA digestion releases RNA flanking fragments that are not in direct contact with AGO. After stringent washing, only those miRNA and mRNA fragments directly crosslinked to AGO in RISC are preserved. These RNA tags are then released and deep-sequenced using massively parallel sequencing technology Fig. (2). Approximate miRNA binding sites on captured mRNA fragments are identified through the analysis and have been demonstrated to be enriched in miRNA target sequences [48]. HITS-CLIP of the RNA binding protein AGO has been applied to identify targets of miRNAs in mouse brain [49], and subsequently in C. elegans [50] and embryonic stem cells [51]. Recently, improved bioinformatics was applied to AGO HITS-CLIP, enabling identification of binding sites with single nucleotide resolution [52].
Fig. (2).
High-throughput methods for identification of miRNA target genes. HITS-CLIP and PAR-CLIP utilizes ultraviolet (UV)- induced covalent crosslinking to stabilize RNA-Argonaute (AGO) protein complexes in miRISC, thereby enhancing the ability to capture more transient miRNA-mRNA interactions, prior to immunoprecipitation (IP) with antibodies. Massively parallel sequencing (MPS) of bound RNAs then allows comprehensive identification of functional miRNA-target mRNA interaction sites.
B. Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP)
PAR-CLIP, a modification of HITS-CLIP, utilizes photoactivatable ribonucleosides to enhance crosslinking efficiency and fold-recovery of RNA following wash steps [53]. The method relies on the incorporation of photoreactive ribonucleoside analogs, such as 4-thiouridine (4-SU) and 6-thioguanosine (6-SG), into nascent RNA transcripts by living cells. Irradiation of the cells by UV light (365 nm) induces efficient crosslinking of photoreactive nucleoside-labeled cellular RNAs to interacting RNA binding proteins. Crosslink formation between these modified ribonucleosides and RNA-binding proteins induces a high rate of modified T to C conversions during the reverse transcription process used prior to deep sequencing Fig. (2). Therefore, by analyzing locations of T to C conversions, the exact site of crosslinks can be determined, which greatly aids the process of identifying the miRNA target site on targeted transcripts. Hafner et al. used the epitope tagged Argonaute family members (AGO 1-4) expressed in HEK293T cells [53]. This study demonstrated that the most significantly enriched 7-mer motifs identified in co-immunoprecipitated RNA corresponded to the seed sequences of the most abundant miRNAs, which were generally positioned 1-2 nt downstream of the predominant cross-linking site. This implies that the site of crosslinking lies near the AGO-miRNA-mRNA complex and shows the ability for using T-C transitions to identify more specific sequenced DNA on miRNA:mRNA interaction sites [53].
These crosslinking strategies provide genome-wide data sets of endogenous miRNA targets and, potentially, their direct binding sites [54]. However, the detection of an mRNA bound by miRISC alone does not guarantee that it is actually being regulated, nor does it reveal the potential mechanism of control. HITS-CLIP and PAR-CLIP experiments may also reflect a selection bias for strongly interacting microRNA-ribonucleoprotein complexes [48], suggesting that they likely under-represent all miRNA target sites [48].
5. miRNA IN AGING
The potential of miRNAs to modulate aging in model organisms has recently attracted the interest of the molecular genetics community [55]. The importance of miRNAs in development has been firmly established, and increasingly, studies are linking altered miRNA function to a range of disease mechanisms [56]. Based on this research, there is every reason to believe that miRNAs play a major role in modulating life span and the aging process; indeed, support for this assertion has emerged from studies of model organisms as described below.
A. miRNAs That Modulate Lifespan in Animal Models
Multiple miRNAs have been shown to regulate lifespan of C. elegans both positively and negatively [57-59], adding weight to the hypothesis that this gene class may contribute to robustness required for maintenance of healthy lifespan [60]. For example, the miRNA lin-4 and its target lin-14 control lifespan post-developmentally [57]; loss of function mutation of lin-4 miRNA shortened life span and accelerated tissue aging, whereas knock-down of its target lin-14 extends life span. Conversely, overexpression of lin-4 extended lifespan by suppressing the target gene, lin-14, or lin-14 gain of function mutation, which lacks the lin-4 binding sites in the lin-14 3’UTR leads to decreased longevity. Interestingly, knockdown of lin-14 only during adulthood is sufficient to increase longevity and suppress the lin-4 short-lived phenotype, indicating that these genes function in adulthood to modulate aging processes. In addition to lin-4, several other miRNAs that modulate longevity have recently been characterized and these miRNAs do not affect the developmental progression of C. elegans. mir-71, -238 and -246 mutants display a significantly shorter lifespan than those of wild-type animals, and over expressing miR-71 or miR-246 increases lifespan, indicating that these miRNAs function to promote longevity. Conversely, mir-239 mutants exhibit an increase in lifespan compared with that of wild-type animals, and miR-239 over expression produces the opposite effect, demonstrating that miR-239 antagonizes longevity [61]. Furthermore, expression patterns of these lifespan modulating miRNAs can be a predictor of lifespan [59]. Other miRNAs, including let-7 and the muscle miRNA miR-1, have been described as potential modulators of age-related decline [62]. Recently, Kato et al. have shown that an adult-specific knockdown of alg-1, a C.elegans Argonaute gene, results in a significantly shorter lifespan compared with that of wild-type animals [63]. This indicates that a large-scale perturbation of miRNA maturation and function affects longevity. Since a significant number of miRNAs are evolutionarily conserved [64, 65], regulation of longevity by miRNAs is expected in higher organisms. Indeed, specific miRNAs up regulated during aging differ significantly between the long-lived Ames dwarf mice and their wild-type counterparts [66], implicating the function of miRNAs in delayed aging.
B. Age-Related Changes in miRNA Expression in Animal Models
Recently, in an effort to link miRNAs with processes of aging, global miRNA analyses on various aged tissues or organs have been performed. These studies revealed that more than 50 of the ~200 miRNAs in C.elegans reported in the miRBase are differentially expressed during aging, and more than half of these have conserved sequences in humans [60, 67]. miR-34 stands out in particular, as it was found to be upregulated during aging, as well as during the dauer stage and early dormancy [61, 63, 68].
In mouse models, there exist differences in miRNA expression between young and old organs and tissues. While no significant differences were evident upon comparison of lung tissue from adult and aged lung [69], comparison of the brain and liver tissues of 10-, 18-, 24-, and 33-month-old mice through the miRNA microarrays and global proteomic profiling revealed that deregulated miRNAs were shared between the aging brain and aging liver, as well as between brain- and liver-specific miRNAs during normal aging [70]. Bates et al. have reported specific miRNA profiles in the livers of Ames dwarf mice (which are well known for their remarkable delay in onset of aging) using miRNA microarrays [66]. The results indicate that key enzymes involved in biosynthetic pathways such as ornithine decarboxylase and spermidine synthase are suppressed by miR-27a, and that this feature may contribute to the extended lifespan of the Ames dwarf mouse [66].
In humans, a comparative profiling of genes and miRNAs expressed in newborn, young adult and aged human epididymides was reported [71]. Since tissue is comprised of multiple cell types, the differential gene expression among different types of cells may compromise the ability to compare results. Nonetheless, the authors found that the newborn epididymis expressed the fewest mRNAs but the largest number of miRNAs, whereas the adult and aged epididymides expressed the most mRNAs but the fewest miRNAs, demonstrating a negative correlation between mRNAs and miRNA during aging. More recently, Noren Hooten et al. [72] utilized mononuclear cells from peripheral blood to evaluate miRNA expression in young and old individuals and revealed that the majority of miRNAs decreased in abundance with age. Predicted targets of the age-related down-regulated miRNAs include PI3K, c-Kit and H2AX, which were found to be elevated with advancing age, supporting a possible role for these miRNAs in the aging process [72].
C. miRNAs Acting on the Conserved Pathways of Aging
The rate of aging and lifespan are regulated by multiple conserved pathways of aging. Altering the pathways controlling metabolism, endocrine signaling, nutrient sensing, and stress resistance has been shown to prolong lifespan from yeast to mammals [73, 74]. Interestingly, the longevity-modulating miRNAs discovered in C. elegans are shown to function through the insulin/IGF-1 signaling (IIS) pathway, one of the first and best characterized conserved pathways of aging [57-59]. Mutations that impair the function of the IIS pathway extend lifespan in C. elegans [75], Drosophila [76], and mice [77], and are also implicated in human longevity [78]. This pathway is activated by binding of ligand (IGF-1 or insulin) to its receptor, DAF-2 in C. elegans, leading to an intracellular signaling that antagonizes the activity of Forkhead/FOXO transcription factors. Reduced function mutations in IIS genes activate Forkhead/FOXO proteins, DAF-16 in C. elegans, that regulate expression of several hundred genes implicated in metabolism, stress resistance and antimicrobial defense [79, 80]. Genetic studies have demonstrated that the lin-4 miRNA (a longevity promoting factor) and its target lin-14 (a life span antagonizing factor) function in the same pathway as DAF-2 and DAF-16. It was suggested that DAF-2 and LIN-14 negatively regulate DAF-16 function in parallel, whereas DAF-16 represses lin-4 forming a possible negative feedback regulatory loop. Several studies report that multiple miRNAs regulate the components of the IIS pathway, such as miR-1, miR-320, and miR-206 targeting IGF-1 [25, 81], and miR-216a, miR-217, and miR-21 targeting PTEN [82, 83].
There are number of reports that forge a link between target of rapamycin (TOR) pathway and miRNAs. TOR is a major amino-acid and nutrient sensor that stimulates growth and blocks rescue pathways such as autophagy in response to nutrient and growth factor cues [23, 24, 84]. TOR proteins are highly conserved from yeast to humans. Inhibition of TOR signaling by rapamycin or by chronic dietary restriction decreases translation through activation of the translational repressor eIF4EBP and downregulation of ribosomal S6 kinase (S6K) and increases autophagy [68]. Inhibition of the pathway has also been reported to increase lifespan in many species, from yeast to mice [85-89]. Overexpression of miR-100 inhibits both mTOR mRNA and protein levels, although there is currently no evidence of direct binding [90]. miR-30a targets beclin-1, the mammalian homologue of the yeast Atg6, by binding to its 3’ UTR and inhibits activation of autophagy induced by rapamycin [91]. Still, experimental evidence for a modulation of life span by these miRNAs is lacking, as is analysis of miRNAs in the context of caloric restriction.
Several miRNAs have been reported to regulate the expression of SIRT1, an ortholog of yeast Sir2 implicated in regulation of life span, stress resistance, and metabolism [92]. Sir2 and other related members of the sirtuin family are highly conserved from yeast to mammalian cells. The surtuin proteins are NAD+-dependent protein deacetylases that regulate the activity of many proteins involved in energy metabolism, inflammation, transcription, and cell survival [93]. miR-217 expression is progressively increased during ageing in endothelial cells, and it can modulate SIRT1 expression through binding to the 3’UTR of SIRT1 mRNA [94]. miR-34a, a downstream target of p53, has also been found to target SIRT1 in mouse liver [95], indicating that there is a connection between miR-34a and the ageing signaling pathway. SIRT1 is also a direct target of miR-199a and miR-132 and mediates the regulation of chemokine production [96] or HIF-1α function [97].
D. miRNAs in Age-Associated Diseases
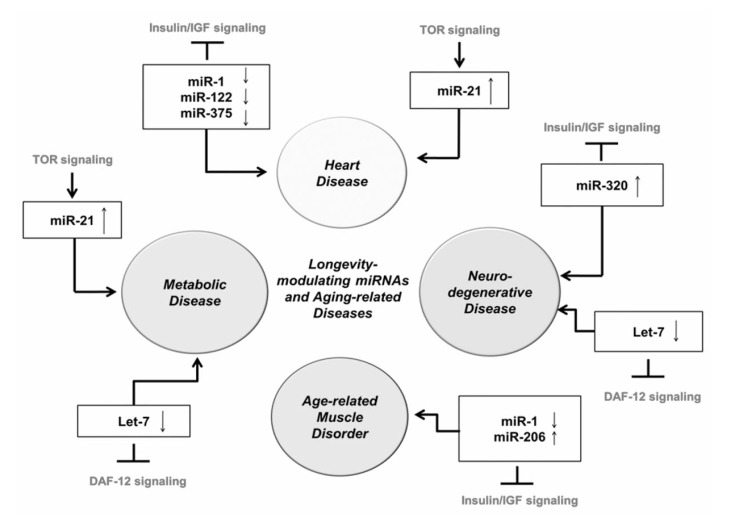
Recently, some miRNAs that target conserved pathways of aging, including Insulin/IGF signaling (IIS), DAF-12 signaling and TOR signaling, have been linked to human aging-related disorders such as heart [98-108], muscle [109, 110], and neurodegenerative disease [111, 112] Fig. (3). miR-1, miR-122 and miR-375 target IIS and have been associated with heart disease. There is evidence that downregulation of miR-1 is correlated with hypertrophic growth of heart in both mice and humans [103]. Plasma levels of miR-122 and miR-375 are decreased in patients that present with myocardial infarction [102]. miR-21, which is activated by TOR and NFκB in hepatocytes, is one of the most highly and consistently upregulated miRNAs during cardiac hypertrophy [99, 106-108]. Reconstitution of miR-21 within an infarct zone reduces cell death and infarct size and ameliorates cardiac dysfunctionalthough the role of miR-21 in heart through the TOR and NFkB pathway is not studied yet.
Fig. (3).
MiRNAs involved in conserved pathways of aging and their role in age-related diseases in humans. A schematic representation of miRNAs known to target genes involved in the conserved pathways of aging and their connections to age-related diseases.
miR-1 and miR-206 regulate IIS [81, 110] and play have a role in skeletal muscle hypertrophy and atrophy [109, 110]. The expression of these miRNAs increased during development of human skeletal muscle, indicating that these miRNAs are also involved in the development of human skeletal muscle [113, 114] and myogenesis by targeting myogenic factors such as MEF2, serum response factor (SRF), and myostatin [115]. miR-1 expression are decreased during skeletal muscle hypertrophy [116]. miRNA profiling in skeletal muscle identified miR-206 as an up-regulated miRNA with age in mice [109]. miR-206 can induce muscle hypertrophy and its increased expression with muscle atrophy in aging may indicate an adaptive, compensatory response to antagonize other catabolic signals [109]. The reason for the differential expression of these miRNAs in muscle disorders is still unclear and needs further investigation.
miRNAs also play key roles in controlling metabolic homeostasis and diseases [117]. miR-21 expression is increased in the livers of rats fed high-fat diets and in human liver biopsies of obese patients with diminished PTEN expression, in line with the findings that miR-21 is activated by an mTOR/NF-κB-dependent mechanism and inhibits PTEN by binding to its 3’UTR [118]. Aberrant up-regulation of miR-21 expression by excessive circulating levels of fatty acids exemplify a novel regulatory mechanism by which fatty acids affect PTEN expression and trigger liver disorders [118]. In contrast, let-7 inhibits adipogenic differentiation through the down-regulation of adipogenic factors [119-124]. The tumor suppressor roles of let-7 are well studied in cancer biology, but let-7 was recently also shown to be involved in the regulation of glucose metabolism [125]. This effect may, at least partially, be mediated by repression of insulin-like growth factor receptor 1 (IGF1R), insulin receptor (INSR) and IRS2 [125].
Many studies have shown the alteration of miRNA expression in neurodegenerative diseases including Alzheimer’s disease and prion-induced neurodegenerative disease [126]. let-7, known to target DAF-12 signaling and regulate lifespan in worms [127], has been implicated in Alzheimer’s disease by its demonstrated genetic interactions with the homolog of amyloid precursor protein (APP), APP-like-1 (apl-1) in worms, suggesting that Aβ peptide formation is under miRNA control in organisms other than mammals [111, 112]. Recently, miR-320 which is known to target IGF-1 and IGF-1R in rats [128] is found to be up-regulated in prion-induced neurodegenerative disease [129]. In summary, de-regulation of miRNAs acting on the conserved pathways of aging in a variety of aging-related disorders strengthens the notion that aging is a root cause of aging-related diseases.
6. CONCLUSION
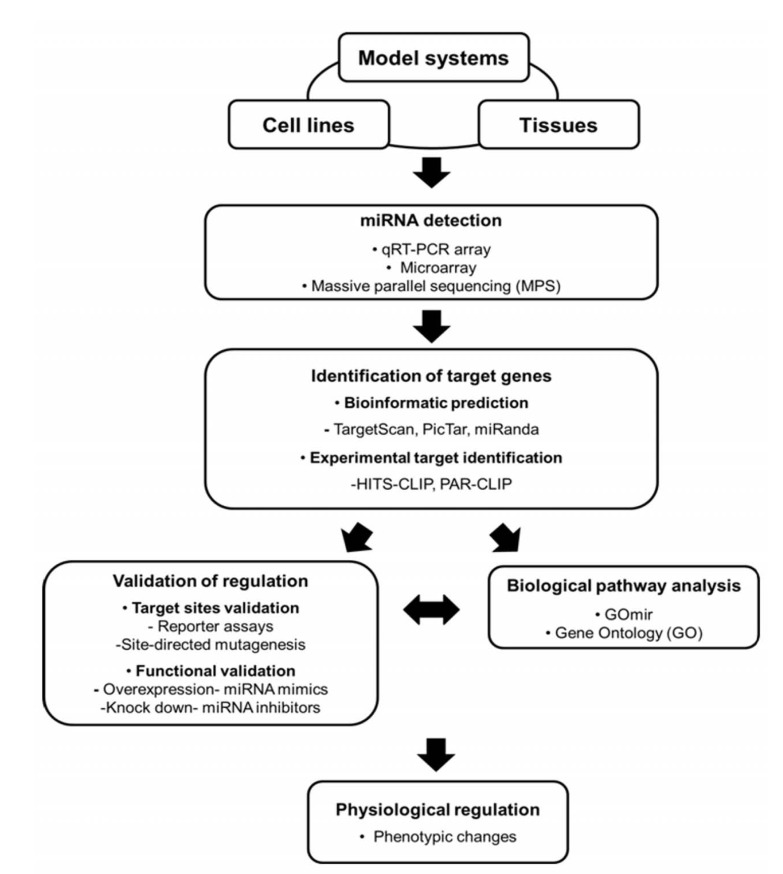
The discovery of miRNAs points to an entirely new regulatory module to control biological processes. Recent studies are linking altered miRNA function to a range of aging-related diseases and processes of aging. The increased availability and affordability of massively parallel sequencing offers a dramatically improved method to gain a high-resolution view of miRNA expression. In addition, high-throughput technologies allow the identification and the validation of miRNA target genes, providing new approaches to identify miRNA regulatory networks in aging Fig. (4). Identification of miRNAs that modulate aging will provide important mechanistic insights into the molecular basis of aging.
Fig. (4).
Summary of methodological workflow for studying miRNA function. The workflow summarizes the emerging high-throughput experimental approaches for the study of miRNA gene regulatory networks in aging. For miRNA discovery, high-throughput methods such as massively parallel sequencing or microarray can be utilized to identify differentially expressed miRNAs in a variety of aging models. Target sites of aging-related miRNAs are identified either by in silico analysis or experimental approaches such as HITS-CLIP and PAR-CLIP. Target sites are then validated by 3’ URT reporter assays or by assessing the anti-correlation between a miRNA and its target gene/protein levels in transfection experiments with miRNA minics, antagomir, or target protectors. Biological pathway analysis of target genes will provide insights into aging gene regulatory networks.
ACKNOWLEDGEMENTS
This work has been supported by grants to Dr. Y. Suh from the Glenn Award for Research in Biological Mechanisms of Aging, and from the US National Institute of Health (RO1 AG024391, PO1 AG027734, and PO1 AG17242).
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5 ):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2 ):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10(5 ):490–7. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat Struct Mol Biol. 2008;15(4 ):354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35(17 ):5944–53. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2 ):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132(21 ):4645–52. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 8.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589 ):2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 9.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670 ):594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 10.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20(14 ):1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44(1 ):120–33. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18(11 ):1218–26. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;19(3 ):364. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 14.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770 ):75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103(11 ):4034–9. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102(47 ):16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13(12 ):1102–7. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 18.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13(12 ):1108–14. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 19.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2 ):671–80. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 20.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21(4 ):533–42. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 21.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740 ):1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 22.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243(2 ):215–25. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308 ):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2 ):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 25.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36(2 ):181–8. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 26.Saugstad JA. MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab. 2010;30(9 ):1564–76. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1 ):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 28.McCormick KP, Willmann MR, Meyers BC. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence. 2011;2(1 ):2. doi: 10.1186/1758-907X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittmann J, Jack HM. New surprises from the deep--the family of small regulatory RNAs increases. ScientificWorldJournal. 2010;10:1239–43. doi: 10.1100/tsw.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18(7 ):1051–63. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2(2 ):e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, Homer N, Huentelman MJ. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5(10 ):887–93. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5(3 ):235–7. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5(3 ):e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316(5830 ):1481–4. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 36.Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35(15 ):e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith AM, Heisler LE, St Onge RP, Farias-Hesson E, Wallace IM, Bodeau J, Harris AN, Perry KM, Giaever G, Pourmand N, Nislow C. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010;38(13 ):e142. doi: 10.1093/nar/gkq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stiller M, Knapp M, Stenzel U, Hofreiter M, Meyer M. Direct multiplex sequencing (DMPS)--a novel method for targeted high-throughput sequencing of ancient and highly degraded DNA. Genome Res. 2009;19(10 ):1843–8. doi: 10.1101/gr.095760.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protoc. 2008;3(2 ):267–78. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- 40.Gunaratne PH, Coarfa C, Soibam B, Tandon A. miRNA data analysis: next-gen sequencing. Methods Mol Biol. 2012;822:273–88. doi: 10.1007/978-1-61779-427-8_19. [DOI] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33(Database issue ):D121–4. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue ):D105–10. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roubelakis MG, Zotos P, Papachristoudis G, Michalopoulos I, Pappa KI, Anagnou NP, Kossida S. Human microRNA target analysis and gene ontology clustering by GOmir, a novel stand-alone application. BMC Bioinformatics. 2009;10(Suppl 6):S20. doi: 10.1186/1471-2105-10-S6-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12(2 ):192–7. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36(Database issue ):D159–64. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43 ):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaya KD, Karakulah G, Yakicier CM, Acar AC, Konu O. mESAdb: microRNA expression and sequence analysis database. Nucleic Acids Res. 2011;39(Database issue ):D170–80. doi: 10.1093/nar/gkq1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long JM, Lahiri DK. Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders. Exp Neurol. 2012. [DOI] [PMC free article] [PubMed]
- 49.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2010;39(Database issue ):D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17(2 ):173–9. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18(2 ):237–44. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29(7 ):607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010;(41) doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4 ):271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 55.Lanceta J, Prough RA, Liang R, Wang E. MicroRNA group disorganization in aging. Exp Gerontol. 2010;45(4 ):269–78. doi: 10.1016/j.exger.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144(6 ):986–98. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310(5756 ):1954–7. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 58.de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs Both Promote and Antagonize Longevity in C-elegans. Current Biology. 2010;20(24 ):2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pincus Z, Smith-Vikos T, Slack FJ. MicroRNA Predictors of Longevity in Caenorhabditis elegans. PLoS Genet. 2011;7(9 ):e1002306. doi: 10.1371/journal.pgen.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibanez-Ventoso C, Driscoll M. MicroRNAs in C. elegans Aging: Molecular Insurance for Robustness? Curr Genomics. 2009;10(3 ):144–53. doi: 10.2174/138920209788185243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20(24 ):2159–68. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5(3 ):235–46. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 63.Kato M, Chen X, Inukai S, Zhao H, Slack FJ. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA. 2011;17(10 ):1804–20. doi: 10.1261/rna.2714411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One. 2008;3(7 ):e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang DE. MicroRNA Regulation and its Biological Significance in Personalized Medicine and Aging. Curr Genomics. 2009;10(3 ):143. doi: 10.2174/138920209788185216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bates DJ, Li N, Liang R, Sarojini H, An J, Masternak MM, Bartke A, Wang E. MicroRNA regulation in Ames dwarf mouse liver may contribute to delayed aging. Aging Cell. 2010;9(1 ):1–18. doi: 10.1111/j.1474-9726.2009.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue ):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith-Vikos T, Slack FJ. MicroRNAs and their roles in aging. J Cell Sci. 2012;125(Pt 1 ):7–17. doi: 10.1242/jcs.099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams AE, Perry MM, Moschos SA, Lindsay MA. microRNA expression in the aging mouse lung. BMC Genomics. 2007;8:172. doi: 10.1186/1471-2164-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li N, Bates DJ, An J, Terry DA, Wang E. Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol Aging. 2009;32(5 ):944–55. doi: 10.1016/j.neurobiolaging.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J, Liu Q, Zhang W, Li J, Li Z, Tang Z, Li Y, Han C, Hall SH, Zhang Y. Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta Biochim Biophys Sin (Shanghai) 2010;42(2 ):145–53. doi: 10.1093/abbs/gmp116. [DOI] [PubMed] [Google Scholar]
- 72.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5(5 ):e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454(7208 ):1065–71. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328(5976 ):321–6. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277(5328 ):942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 76.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514 ):107–10. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 77.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299(5606 ):572–4. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 78.van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4(2 ):79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 79.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Experimental gerontology. 2006;41(10 ):910–21. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 80.Greer EL, Brunet A. FOXO transcription factors in ageing and cancer. Acta physiologica. 2008;192(1 ):19–28. doi: 10.1111/j.1748-1716.2007.01780.x. [DOI] [PubMed] [Google Scholar]
- 81.Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL, Zhu JN, Liu XY, Zhang YY, Li Y, Lin SG, Yu XY. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem Biophys Res Commun. 2009;381(4 ):597–601. doi: 10.1016/j.bbrc.2009.02.097. [DOI] [PubMed] [Google Scholar]
- 82.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11(7 ):881–9. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sayed D, Abdellatif M. AKT-ing via microRNA. Cell Cycle. 2010;9(16 ):3213–7. doi: 10.4161/cc.9.16.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288 ):504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 85.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253 ):392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C elegans larval development, metabolism and life span. Development. 2004;131(16 ):3897–906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 87.Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751 ):1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 88.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14(10 ):885–90. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426(6967 ):620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 90.Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno NT, Hawkins SM, Anderson ML, Matzuk MM. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;24(2 ):447–63. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5(6 ):816–23. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kennedy BK, Smith ED, Kaeberlein M. The enigmatic role of Sir2 in aging. Cell. 2005;123(4 ):548–50. doi: 10.1016/j.cell.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 93.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404(1 ):1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A, Novelli G, Melino G, Lauro R, Federici M. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120(15 ):1524–32. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 95.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010;285(17 ):12604–11. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23(11 ):1876–84. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104(7 ):879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andersen RV, Wittrup HH, Tybjaerg-Hansen A, Steffensen R, Schnohr P, Nordestgaard BG. Hepatic lipase mutations,elevated high-density lipoprotein cholesterol, and increased risk of ischemic heart disease: the Copenhagen City Heart Study. J Am Coll Cardiol. 2003;41(11 ):1972–82. doi: 10.1016/s0735-1097(03)00407-8. [DOI] [PubMed] [Google Scholar]
- 99.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170(6 ):1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47(1 ):5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, Dong X, Qin S, Zhang C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119(2 ):87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31(22 ):2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han M, Toli J, Abdellatif M. MicroRNAs in the cardiovascular system. Curr Opin Cardiol. 2011;26(3 ):181–9. doi: 10.1097/HCO.0b013e328345983d. [DOI] [PubMed] [Google Scholar]
- 104.Hulsmans M, De Keyzer D, Holvoet P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. Faseb J. 2011. [DOI] [PubMed]
- 105.Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem. 2010;285(26 ):20281–90. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100(3 ):416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 107.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42(6 ):1137–41. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48 ):18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hamrick MW, Herberg S, Arounleut P, He HZ, Shiver A, Qi RQ, Zhou L, Isales CM, Mi QS. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem Biophys Res Commun. 2010;400(3 ):379–83. doi: 10.1016/j.bbrc.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guller I, Russell AP. MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J Physiol. 2010;588(Pt 21 ):4075–87. doi: 10.1113/jphysiol.2010.194175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol. 2008;315(2 ):418–25. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer's and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14(10 ):2495–505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol. 2011;11:34. doi: 10.1186/1471-213X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen JF, Callis TE, Wang DZ. microRNAs and muscle disorders. J Cell Sci. 2009;122(Pt 1 ):13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1 ):306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 117.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4 ):239–50. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vinciguerra M, Sgroi A, Veyrat-Durebex C, Rubbia-Brandt L, Buhler LH, Foti M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology. 2009;49(4 ):1176–84. doi: 10.1002/hep.22737. [DOI] [PubMed] [Google Scholar]
- 119.Bork S, Horn P, Castoldi M, Hellwig I, Ho AD, Wagner W. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol. 2010;226(9 ):2226–34. doi: 10.1002/jcp.22557. [DOI] [PubMed] [Google Scholar]
- 120.Kinoshita M, Ono K, Horie T, Nagao K, Nishi H, Kuwabara Y, Takanabe-Mori R, Hasegawa K, Kita T, Kimura T. Regulation of adipocyte differentiation by activation of serotonin (5-HT) receptors 5-HT2AR and 5-HT2CR and involvement of microRNA-448-mediated repression of KLF5. Mol Endocrinol. 2010;24(10 ):1978–87. doi: 10.1210/me.2010-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, Selimyan R, Egan JM, Smith SR, Fried SK, Gorospe M. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2010;31(4 ):626–38. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. Febs J. 2009;276(8 ):2348–58. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23(6 ):925–31. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang Z, Bian C, Zhou H, Huang S, Wang S, Liao L, Zhao RC. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2010;20(2 ):259–67. doi: 10.1089/scd.2010.0072. [DOI] [PubMed] [Google Scholar]
- 125.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Consortium D, Investigators M, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1 ):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 2012;133(2 ):142–50. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grillari J, Grillari-Voglauer R. Novel modulators of senescence, aging, and longevity: Small non-coding RNAs enter the stage. Exp Gerontol. 2010;45(4 ):302–11. doi: 10.1016/j.exger.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 128.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209 ):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3(11 ):e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]