Abstract
To evaluate the mechanism of the development of therapeutic resistance after temozolomide treatment, we focused on changes in O6-methylguanine DNA methyltransferase (MGMT) and mismatch repair (MMR) between initial and recurrent glioblastomas. Tissue samples obtained from 24 paired histologically confirmed initial and recurrent adult glioblastoma patients who were initially treated with temozolomide were used for MGMT and MMR gene promoter methylation status and protein expression analysis using methylation-specific multiplex ligation probe amplification (MS-MLPA), methylation-specific polymerase chain reaction (MSP), and immunohistochemical staining. There was a significant decrease in the methylation ratio of the MGMT promoter determined by MS-MLPA, which was not detectable with MSP, and MGMT protein expression changes were not remarkable. However, there was no epigenetic variability in MMR genes, and a relatively homogeneous expression of MMR proteins was observed in initial and recurrent tumors. We conclude that the development of reduced methylation in the MGMT promoter is one of the mechanisms for acquiring therapeutic resistance after temozolomide treatment in glioblastomas.
Introduction
Glioblastoma still remains a devastating brain cancer with an unsatisfactory survival rate of 12 to 15 months despite recent advances in the best standard therapies, which include surgical resection, radiation, and temozolomide [1]. The majority of glioblastomas eventually progress or relapse, regardless of whether the O6-methylguanine DNA methyltransferase (MGMT) promoter in the tumor tissue is methylated [1,2]. The role of MGMT promoter methylation in glioblastoma as a prognostic biomarker is well established, and the status of MGMT is considered as a major determinant for temozolomide sensitivity [3]. MGMT removes methyl and chloroethyl groups from the O6 position of guanine produced by the alkylating agent, and overexpression of MGMT can rescue cancer cells from anticancer drugs [4]. However, MGMT activity, indirectly analyzed by MGMT promoter methylation using methylation-specific polymerase chain reaction (MSP) in initial tumor tissue, could not be used to predict the response to the recurrent tumor treated with temozolomide [5].
Additional important determinants related to temozolomide sensitivity are the DNA mismatch repair (MMR) genes. MMR functions in resistance to the alkylating agent; when O6-methylguanine is not repaired by MGMT before DNA replication, there is a resulting DNA gap, which if not corrected permanently leads to DNA double-strand breaks and apoptotic cell death [6]. Therefore, intact MMR is required for the death of cancer cells treated with an alkylating agent, such as temozolomide, in an MGMT-deficient condition. Previous reports indicated that there are low occurrences of MMR defects and consistent expression of MMR proteins in newly diagnosed adult glioblastoma samples [7–10]. However, studies evaluating MMR status in recurrent glioblastomas in relation to therapeutic resistance are sparse or reveal controversial results [11,12].
Therefore, the development of therapeutic resistance to temozolomide may harbor genetic changes or clone selections for resistant cells, including alterations in MGMT and/or MMR genes. To address this issue, we investigated alterations in the promoter methylation status and protein expression of MGMT and MMR genes in paired initial and recurrent glioblastoma tissues. All of the patients were initially treated with temozolomide. In the present study, we used the semiquantitative method of methylation-specific multiplex ligation probe amplification (MS-MLPA) for a closer evaluation of changes in the promoter methylation status of MGMT and MMR genes, and the results were compared with those of the standard method of MSP and protein expression levels evaluated by immunohistochemical (IHC) staining.
Materials and Methods
Study Population and Preparation of Tissue Samples
Clinical data and tissue samples were obtained from 24 paired histologically confirmed initial and recurrent adult glioblastoma patients who had undergone initial treatment of surgical resection, followed by a temozolomide-based regimen and reoperation at recurrence. The initial treatment protocol includes the following: standard concomitant radiation therapy and temozolomide followed by adjuvant temozolomide in 14 patients, neoadjuvant chemotherapy with nimustine and cisplatin followed by radiation therapy and adjuvant temozolomide in 6 patients, radiation therapy followed by adjuvant temozolomide in 3 patients, and temozolomide only in 1 patient. The mean age was 60.1 years (range, 30–73), and 15 patients (62.5%) were male. A median total dose of administered temozolomide before recurrence was 7800 mg (range, 1200–20,150), and the median period of relapse after initial surgery was 8 months (range, 2–55). Institutional review board approval was obtained for the data collection and study analysis.
Snap-frozen tumor tissues obtained during surgery and kept in liquid nitrogen at -80°C were used for MS-MLPA. DNA was isolated from the tissue samples using the QIAGEN DNA Mini Kit (QIAGEN Inc., Valencia, CA) according to the manufacturer's protocol. CpGenome Universal Methylated DNA and Unmethylated DNA (Chemicon, Millipore, Billerica, MA) were included as controls in each set of MS-MLPA and MSP experiments. Paraffin-embedded tissues were subjected to MSP and IHC staining.
Methylation-specific Multiplex Ligation Probe Amplification
The methylation status of the MGMT promoter and MMR genes were analyzed using the MS-MLPA probe mix prepared by MRC-Holland (Salsa MS-MLPA Kit ME011MMR version B1, Amsterdam, The Netherlands), which includes six probes for MLH1, four probes for MSH2, three probes for MSH6, three probes for PMS2, and six probes specific for the MGMT promoter region. The detailed procedure is described in previous publications [13,14]. A methylation-sensitive restriction enzyme, HhaI (R6441; Promega, Madison, WI), which cuts unmethylated GCGC sites, was applied to each set of samples. The resultant polymerase chain reaction fragments were separated by capillary gel electrophoresis (ABI Prism 7000/7700; Applied Biosystems, Foster City, CA). The methylation status was quantified using GeneMarker software (version 1.5; Soft Genetics, State College, PA). To evaluate the methylation status, the “methylation ratio” was calculated by dividing each normalized peak value of the HhaI enzyme-digested sample by that of the corresponding undigested sample. This value corresponds to the percentage of methylated sequences.
Methylation-specific Polymerase Chain Reaction
The methylation status of the MGMT promoter was also analyzed using MSP, and the results were compared with those of MS-MLPA. Prepared DNA was modified by sodium bisulfite treatment using an EZ DNA Methylation-Gold Kit (Catalogue No. D5005; Zymo Research, Orange, CA). The primer sequences used for the MGMT were given as follows: methylated forward, 5′ TTT CGA CGT TCG TAG GTT TTC GC 3′; methylated reverse, 5′ GCA CTC TTC CGA AAA CGA AAC G 3′; unmethylated forward, 5′ TTT GTG TTT TGA TGT TTG TAG GTT TTT GT 3′; unmethylated reverse, 5′ AAC TCC ACA CTC TTC CAA AAA CAA AAC A 3′. The annealing temperature was 64°C. The obtained polymerase chain reaction products were electrophoresed in 2% agarose gels and visualized under UV illumination after staining with ethidium bromide. The evaluation of the assay results was performed as described previously [13].
IHC Staining
Tissue expression of MMR proteins (PMS2, MLH1, MSH2, and MSH6) and MGMT protein were confirmed by IHC staining using conventional methods according to the manufacturer's protocols. The antibodies used were given as follows: MGMT (MS-470-P, 1:100; Neomarkers, Fremont, CA), PMS2 (1:50; Cell Marque, Rocklin, CA), MLH1 (1:50; DAKO, Glostrup, Denmark), MSH2 (1:200; Invitrogen, Camarillo, CA), and MSH6 (1:50, Cell Marque). IHC staining results were semiquantitatively graded as follows: no staining detected (0; no positive tumor cells), faint staining (1+; <10% positive tumor cells), moderate staining (2+; 10–50% positive tumor cells), and strong staining (3+; >50% positive tumor cells). Positive staining was defined as 2+ or 3+ nuclear staining based on the percentage of tumor cells showing immunoreactivity.
Statistical Analysis
The Wilcoxon signed rank test and McNemar test were used for nonparametric comparisons between the results of initial and recurrent samples. Statistical significance was accepted at probability values of less than 0.05. These statistical analyses were performed with the aid of IBM SPSS Statistics software (version 19.0; SPSS Inc., Chicago, IL).
Results
Changes in MGMT
A total of 24 paired cases of initial and recurrent glioblastoma samples were analyzed with MSP for MGMT promoter methylation status and IHC staining for MGMT protein expression, and the results are summarized in Table 1. The MGMT promoter was methylated in 8 (33%) patients and was unmethylated in 16 (67%) patients with initial glioblastoma. At recurrence, 22 of 24 patients (92%) showed identical results of MGMT promoter methylation levels measured by MSP, and their changes were statistically nonsignificant (P = 1.00). One patient exhibited a reversal of MGMT promoter status from methylated to unmethylated, and the opposite occurred in another patient. The MGMT protein was not expressed in 15 (63%) patients and was expressed in 9 (37%) patients with initial glioblastoma. Among all of the patients, 19 of 24 patients (79%) showed no changes in MGMT protein expression at tumor recurrence. Using the cutoff value of 10% positivity in tumor cells, four (17%) patients showed a new expression of MGMT protein, and the expression was lost in one (4%) patient at tumor recurrence. However, their changes were also statistically nonsignificant (P = .38).
Table 1.
Summary of MGMT Promoter Methylation Status (by MSP) and Protein Expression (by IHC Staining) in a Total of 24 Paired Cases of Initial and Recurrent Glioblastomas.
| Recurrent Glioblastoma | Identical with Initial Tumor | P Value (McNemar Test) | |||
| Initial glioblastoma, n (%) | MSP | Methylated | Unmethylated | ||
| Methylated | 7 (29%) | 1 (4%) | 22 (92%) | 1.00 | |
| Unmethylated | 1 (4%) | 15 (63%) | |||
| IHC staining | No expression | Expression | |||
| No expression | 11 (46%) | 4 (17%) | 19 (79%) | .38 | |
| Expression | 1 (4%) | 8 (33%) |
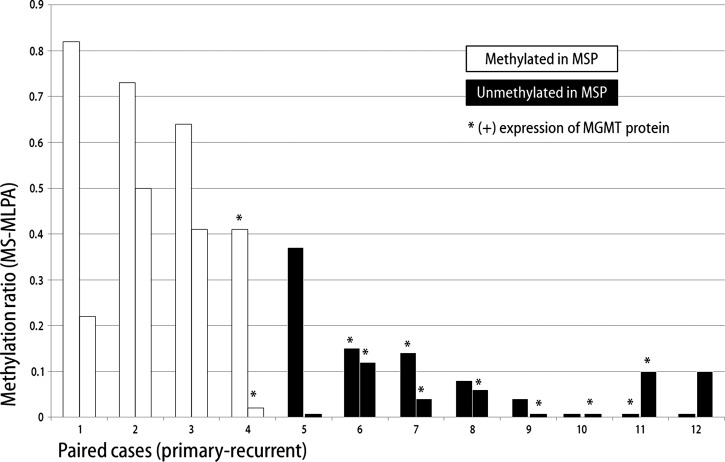
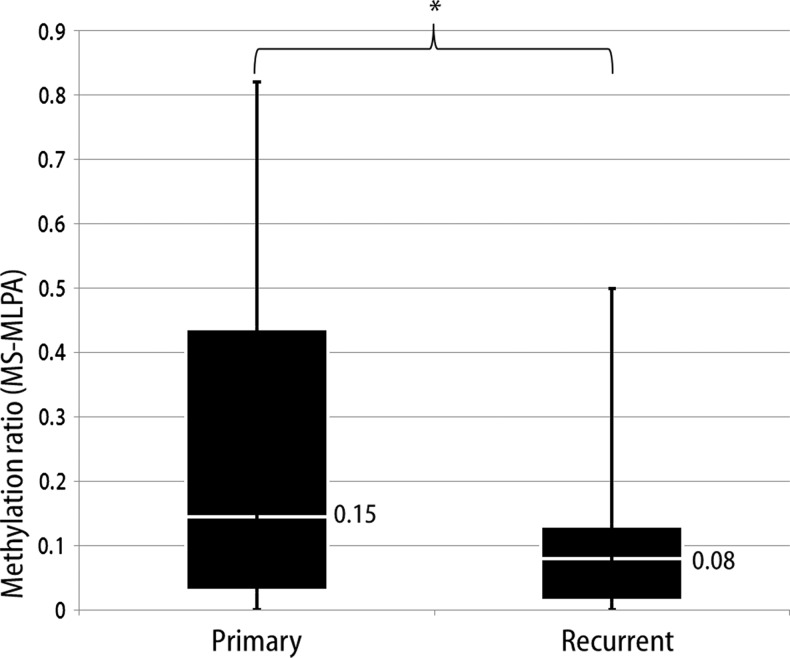
Among the study population, fresh frozen samples were available for a total of 12 paired cases, and MS-MLPA was performed with their 24 samples. Figure 1 shows the MGMT promoter methylation status expressed as the methylation ratio after MS-MLPA analysis of 12 paired initial and recurrent glioblastomas. There was a decrease in the methylation ratio of the MGMT promoter in recurrent tumor tissue observed in 9 of 12 cases (75%). Of the other three cases, all of which had methylation ratios of 0 at initial tumor, two cases showed an increase in the methylation ratio to 0.1, and one case showed no change. There was a significant decrease of the methylation ratio (P = .04, Figure 2) of the MGMT promoter between initial (median methylation ratio of 0.15) and recurrent glioblastomas (median methylation ratio of 0.08). These decreases in MGMT promoter methylation were undetectable using the MSP method in all 12 paired cases. Additionally, nonsignificant relationships were observed between MGMT protein expression and MGMT promoter methylation changes (P = .07), and the decreased methylation ratio was not significantly related to the temozolomide administered (P = 1.00).
Figure 1.
Changes in the methylation ratio of the MGMT promoter between initial and recurrent glioblastomas in 12 paired cases measured by MS-MLPA. The MSP results of the methylation status of the MGMT promoter are indicated by the bar colors, and MGMT protein expression is indicated by an asterisk.
Figure 2.
We observed a significant difference (*P = .04, Wilcoxon signed rank test) in the distribution of methylation ratios between initial and recurrent tumors (median methylation ratio: 0.15 vs 0.08) determined by MS-MLPA.
Changes in MMR
The results of IHC staining for MMR proteins (PMS2, MLH1, MSH2, and MSH6) in all 24 paired cases showed expression ofMMR proteins at initial tumor, and their expression was preserved at recurrent tumor in most of the cases. Although the changes of immunoreactivity at recurrence were relatively obvious in MSH2 (42%) and MSH6 (29%) compared with those in PMS2 (16%) and MLH1 (16%), none of the proteins reached significant levels (Figure W1 and Table W1). The MS-MLPA for MMR gene (PMS2, MLH1, MSH2, and MSH6) promoters demonstrated that all of 12 paired cases analyzed showed a methylation ratio of 0 in initial and recurrent tumors without exception.
Discussion
To maintain the effective anticancer effect of temozolomide, inactivation of the MGMT gene and intact expression of MMR genes in tumor cells are the key conditions in glioblastomas. There have been studies that attempt to explain the mechanism of therapeutic resistance in glioblastoma by evaluating MGMT and MMR gene status changes in initial and recurrent tumors [12,15–18]. However, inconsistent results have rendered answers for this issue that remain controversial.
After varied treatment failures in serial samples in glioblastoma, several studies have advocated for alterations in MGMT promoter methylation. Parkinson et al. observed changes in MGMT promoter methylation in 2 of 9 cases (all from unmethylated to methylated) analyzed by MSP and in 7 of 10 cases (increased methylation in 5 cases and decreased methylation in 2 cases) assessed by sequencing of 25 CpG dinucleotides in the MGMT promoter [15]. They also observed minimal changes in MGMT IHC staining [15]. Jung et al. reported changes in MGMT promoter methylation in 4 of 16 cases (from methylated to unmethylated in 3 cases and vice versa in 1 case) in the second operation samples of recurrent glioblastoma assessed by MSP [16]. They also showed that 15 of 18 specimens exhibited higher levels of MGMT protein expression compared to previous samples by IHC staining [16]. Brandes et al. observed that significant changes in the MGMT promoter methylation status during the course of glioblastoma occur more frequently in methylated (to unmethylated at recurrence in 8 of 13 cases, 61.5%) than unmethylated cases (to methylated at recurrence in 6 of 25 cases, 24%) assessed by MSP [17]. The concordance of MGMT promoter methylation assessed by MSP between initial and recurrent glioblastomas was 63% in their study [17].
Although the above-mentioned reports shared the result of alterations in MGMT promoter methylation at recurrence detected by the qualitative method of MSP, the frequency of alteration detection is relatively uncommon. In the present study, we also found only two cases (8%) of altered MGMT promoter methylation determined by MSP. However, when we employed the semiquantitative method of MS-MLPA, the majority of case pairs revealed alterations in methylation with the tendency of a decreased methylation ratio in spite of no changes detected by MSP. Therefore, progressive unmethylation of the MGMT promoter, presumably in association with increased expression of MGMT is a plausible mechanism for acquiring therapeutic resistance in glioblastoma after temozolomide treatment. The result of IHC staining for MGMT protein was not helpful for verifying this hypothesis. However, the value of semiquantitative MSMLPA analysis of MGMT promoter methylation was documented for the management of glioblastoma in previous studies [13,14,19]. Christmann et al. reported evidence supporting the results of the present study [18]. They showed significant up-regulation of MGMT enzyme activity in recurrent glioblastoma compared with initial tumor, and the difference was more evident in the temozolomide treatment group compared with the radiation therapy-only group [18]. Additionally, the correlation of MGMT enzyme activity and MGMT promoter methylation determined by MSP was reasonable only if the samples with a hemimethylated MGMT promoter were excluded [18]. Moreover, only a moderate correlation between MGMT enzyme activity and MGMT immunoreactivity was observed, indicating that IHC staining is only an approximate method for detecting the expression level of MGMT in glioblastomas [18].
In the present study, we could not find any evidence of MMR genes being involved in a possible mechanism for therapeutic resistance. Major MMR genes, such as PMS2, MLH1, MSH2, and MSH6, were invariably expressed in initial glioblastoma, and most of the genes were expressed in the recurrent tumor. However, there is a study showing a contradictory result to ours [12]. Among the other sporadic reports regarding MMR genes and therapeutic response or resistance in glioma, several studies mentioned the development of an inactivating mutation in MSH6 during the temozolomide treatment, which mediates therapeutic resistance in glioblastoma [20–22]. However, Maxwell et al. demonstrated contradictory results showing no role for MMR deficiency, includingMSH6 expression, for mediating temozolomide resistance in malignant glioma [11]. As the loss of MMR usually results in increase in insertion/deletion mutations particularly in repetitive sequence microsatellite DNA, microsatellite instability is a recognized surrogate biomarker for the loss ofMMR function [23]. Low frequency of microsatellite instabilities found in relapsed glioma samples in previously reported study also implies the limited role of MMR system in mediating treatment resistance [7]. However, still there is a possibility that MMR genes, especially MSH2 and MSH6, have some role for temozolomide resistance in a small subset of patients, allowing for our data showing decreased expression of MSH2 by 26% and that of MSH6 by 21% of the study population (Figure W1 and Table W1). Therefore, further evidence is still required to answer the controversial issue of the role of MMR genes in therapeutic resistance in glioblastoma after temozolomide treatment.
In summary, semiquantitatively measured reduced methylation of the MGMT promoter in recurrent glioblastoma after temozolomide treatment is one of the mechanisms for the acquisition of therapeutic resistance. MMR deficiency is not the major mechanism of temozolomide resistance in glioblastoma treatment, as MMR genes showed no variability in their unmethylated promoter statuses and they exhibited homogeneous expression in both initial and recurrent glioblastomas. Whether this genetic change is the result of clone selections for resistant cells or de novo modification of existing tumor cells is to be evaluated in further studies.
Supplementary Material
Footnotes
This study was supported by grants from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (Study No. A110749) and the Seoul National University Hospital Research Fund (Study No. 04-2011-0190). No other financial support or relationships that may pose conflict of interest exists.
This article refers to supplementary materials, which are designated by Table W1 and Figure W1 and are available online at www.transonc.com.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Felsberg J, Rapp M, Loeser S, Fimmers R, Stummer W, Goeppert M, Steiger HJ, Friedensdorf B, Reifenberger G, Sabel MC. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin Cancer Res. 2009;15:6683–6693. doi: 10.1158/1078-0432.CCR-08-2801. [DOI] [PubMed] [Google Scholar]
- 3.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 4.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Perry JR, Belanger K, Mason WP, Fulton D, Kavan P, Easaw J, Shields C, Kirby S, Macdonald DR, Eisenstat DD, et al. Phase II trial of continuous doseintense temozolomide in recurrent malignant glioma: RESCUE study. J Clin Oncol. 2010;28:2051–2057. doi: 10.1200/JCO.2009.26.5520. [DOI] [PubMed] [Google Scholar]
- 6.Kaina B, Christmann M. DNA repair in resistance to alkylating anticancer drugs. Int J Clin Pharmacol Ther. 2002;40:354–367. doi: 10.5414/cpp40354. [DOI] [PubMed] [Google Scholar]
- 7.Martinez R, Schackert HK, Plaschke J, Baretton G, Appelt H, Schackert G. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology. 2004;66:395–403. doi: 10.1159/000079488. [DOI] [PubMed] [Google Scholar]
- 8.Martinez R, Schackert HK, Appelt H, Plaschke J, Baretton G, Schackert G. Low-level microsatellite instability phenotype in sporadic glioblastoma multiforme. J Cancer Res Clin Oncol. 2005;131:87–93. doi: 10.1007/s00432-004-0592-5. [DOI] [PubMed] [Google Scholar]
- 9.Alonso M, Hamelin R, Kim M, Porwancher K, Sung T, Parhar P, Miller DC, Newcomb EW. Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res. 2001;61:2124–2128. [PubMed] [Google Scholar]
- 10.Eckert A, Kloor M, Giersch A, Ahmadi R, Herold-Mende C, Hampl JA, Heppner FL, Zoubaa S, Holinski-Feder E, Pietsch T, et al. Microsatellite instability in pediatric and adult high-grade gliomas. Brain Pathol. 2007;17:146–150. doi: 10.1111/j.1750-3639.2007.00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell JA, Johnson SP, McLendon RE, Lister DW, Horne KS, Rasheed A, Quinn JA, Ali-Osman F, Friedman AH, Modrich PL, et al. Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clin Cancer Res. 2008;14:4859–4868. doi: 10.1158/1078-0432.CCR-07-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, Schackert G, Kreth FW, Pietsch T, Loffler M, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129:659–670. doi: 10.1002/ijc.26083. [DOI] [PubMed] [Google Scholar]
- 13.Park CK, Kim J, Yim SY, Lee AR, Han JH, Kim CY, Park SH, Kim TM, Lee SH, Choi SH, et al. Usefulness of MS-MLPA for detection of MGMT promoter methylation in the evaluation of pseudoprogression in glioblastoma patients. Neuro Oncol. 2011;13:195–202. doi: 10.1093/neuonc/noq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeuken JW, Cornelissen SJ, Vriezen M, Dekkers MM, Errami A, Sijben A, Roots-Sprenger SH, Wesseling P. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87:1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 15.Parkinson JF, Wheeler HR, Clarkson A, McKenzie CA, Biggs MT, Little NS, Cook RJ, Messina M, Robinson BG, McDonald KL. Variation of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J Neurooncol. 2008;87:71–78. doi: 10.1007/s11060-007-9486-0. [DOI] [PubMed] [Google Scholar]
- 16.Jung TY, Jung S, Moon KS, Kim IY, Kang SS, Kim YH, Park CS, Lee KH. Changes of the O6-methylguanine-DNA methyltransferase promoter methylation and MGMT protein expression after adjuvant treatment in glioblastoma. Oncol Rep. 2010;23:1269–1276. doi: 10.3892/or_00000760. [DOI] [PubMed] [Google Scholar]
- 17.Brandes AA, Franceschi E, Tosoni A, Bartolini S, Bacci A, Agati R, Ghimenton C, Turazzi S, Talacchi A, Skrap M, et al. O6-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 2010;12:283–288. doi: 10.1093/neuonc/nop050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christmann M, Nagel G, Horn S, Krahn U, Wiewrodt D, Sommer C, Kaina B. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int J Cancer. 2010;127:2106–2118. doi: 10.1002/ijc.25229. [DOI] [PubMed] [Google Scholar]
- 19.Shah N, Lin B, Sibenaller Z, Ryken T, Lee H, Yoon JG, Rostad S, Foltz G. Comprehensive analysis of MGMT promoter methylation: correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6:e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.