Abstract
In many streptococci, quorum sensing utilizes secreted, linear peptides that engage cognate receptors to coordinate gene expression among members of a local population. Streptococcus mutans employs the secreted peptides CSP and XIP to stimulate production of antimicrobial bacteriocins and to induce development of competence for genetic transformation. Recent progress in the field reveals that pathways not only monitor the presence of signal emitters but also sense environmental factors. Both kinds of information are integrated by regulatory networks that then generate multiple outcomes, even among parallel cells growing in identical conditions. In this issue of Molecular Microbiology, Son and coworkers investigate how two medium types shape cellular responses to CSP and XIP pheromones in individuals across a population (Son et al., 2012). Their findings characterize restrictive properties of media differing in peptidic fragment content and reveal unusual signaling properties that contribute to bimodal responses of gene expression.
Keywords: Quorum Sensing, Pheromones, Bacteriocins, Mutacins, Protease, Bimodal distribution, CDM
In a classic Gary Larson cartoon, two amoeboid microbes are drawn to resemble a stereotypical housewife looming over a stereotypical lazy husband watching TV. The nagging partner exclaims to the other, “Stimulus, response. Stimulus, response; don’t you ever think?!” Microbes are constantly presented with stimuli. Some are cues from their environment, and others are self-generated commands. Responses, however, do not always follow simple if-then logic rules. In many cases, complex responses reflect probabilistic determination and interfering signals.
Self-generated commands include secreted signaling molecules, or pheromones, used by quorum sensing pathways to coordinate gene regulation across a population. In the streptococci, quorum sensing utilizes linear peptides that combine with cognate receptors to serve this role. Peptide signaling in S. mutans has attracted special attention due to its role in regulating plaque biofilms that contribute prominently to dental caries, but it also regulates bacteriocin production and competence development. Two signaling pathways transmitting information from the secreted peptides CSP and XIP to regulators that stimulate development of competence for genetic transformation have been recognized for 11 and 2 years, respectively (Li et al., 2001, Mashburn-Warren et al., 2010).
The two quorum sensing pathways differ both in basic architecture and in mechanism of signaling. Different classes of receptor are used to detect the two peptides; CSP is detected by a two-component signal transduction histidine kinase at the cell surface, while XIP is transported to the cytoplasm to interact with a cytoplasmic Rgg-type transcriptional regulator. Here we discuss recent progress that begins to reveal how these signaling pathways can sense environmental factors as well as monitor the presence of signal emitters, and how even a uniform environment can initiate multiple parallel developmental paths to create neighboring cells of very different types.
The CSP signal originates from ComC, a 46-amino-acid polypeptide containing a leader sequence with a characteristic double-glycine motif commonly used as a processing site in non-lantibiotic bacteriocins and related peptide pheromones (Havarstein et al., 1995). During secretion, thought to occur via ClsAB (Petersen & Scheie, 2000), ComC is processed at the GG motif, and the C-terminal 21-amino-acid peptide is exported. Binding of CSP by the ComD histidine kinase is thought to lead to phosphorylation, and thus activation, of the response regulator ComE. A ComE-binding sequence, conserved among several species of Streptococcus, is located upstream of comC, nlmAB (mutacin IV), cipB (mutacin V), and several other putative bacteriocin genes (Hung et al., 2011, Kreth et al., 2007, van der Ploeg, 2005, Ween et al., 1999). These direct interactions of ComE with mutacin promoter regions are consistent with a regulatory role for CSP as the primary controller of mutacin production. What remains unclear is the pathway that connects CSP to expression of SigX (also known as ComX), the single streptococcal alternative sigma factor, which drives transcription of a regulon containing over a dozen genes for the effector proteins of genetic transformation. Interestingly, deletion of cipB, one of the mutacins regulated by ComE, dramatically reduces CSP-dependent transformation and down-regulates the SigX regulon (Dufour et al., 2011). Though it is not clear how CipB contributes to sigX transcription, this finding adds to the list of possible regulators involved in the pathway.
In contrast, another known peptide-mediated quorum-sensing system in S. mutans, encoded by comR-comS, is linked quite directly to competence development. ComS, a ribosomally-derived 17-amino-acid polypeptide, is secreted and processed to a mature form, designated XIP. Steps of its secretion and processing remain unknown. Interestingly, ComRS orthologs are found in every sequenced genome of pyogenic, mutans, and bovis streptococci (more than 18 species to date), and in each case the ComS C-terminal moiety contains a conserved double-tryptophan motif, suggesting a conserved function. A synthetic XIP consisting of the C-terminal seven amino acids of ComS induces a robust increase in sigX transcription when added to cells grown in CDM (Mashburn-Warren et al., 2010). This response requires the Opp oligopeptide permease, suggesting that the peptide is imported to the cytoplasm. Once inside the cell, XIP likely interacts directly with ComR to induce transcription of its two known targets, comS (thus providing positive feedback to the regulatory loop) and sigX. Without comR or comS, competence development does not transpire, even when cells are stimulated with CSP (Mashburn-Warren et al., 2010). In contrast, comE mutations do not affect the response of sigX to added XIP. Therefore, the ComR-ComS pathway is necessary and sufficient for induction of SigX, which in turn, is essential for competence development.
Though described as distinct regulatory pathways, the CSP and XIP systems are in fact linked, though by means that remain unclear, and converge at sigX. While the simple models described above present a basis for understanding how the quorum-sensing systems contribute to competence and bacteriocin production independently, recent findings, including a study that appears in this issue of Molecular Microbiology, indicate there are many surprises hidden in these pathways, with important pieces of the puzzle still missing.
In recent reports, several pieces of the puzzle have been filled in. For instance, processing of S. mutans CSP had been assumed to follow the same path as seen for CSP in S. pneumoniae, where cleavage at the double-glycine motif directly releases a mature signal. Indeed, synthetic CSP with the predicted 21-amino-acid length is active when added to S. mutans cultures, consistent with this assumption. However, a processed form of CSP without the three C-terminal amino acids, termed 18-CSP, also accumulates in competent culture supernatants, and synthetic 18-CSP has a 10-fold higher specific activity than the 21-CSP counterpart (Petersen et al., 2006). Moreover, a recently discovered membrane-associated protease called SepM appears to be responsible for this final processing step, which is effected outside the cell (Hossain & Biswas, 2012). In sepM mutants, 21-CSP remains unprocessed and is inactive, indicating that it is an extracellular intermediate in CSP maturation. Secretion and processing steps of XIP biosynthesis remain entirely unknown, but the predicted mature form of ComS, proposed to be seven amino acids in length, was recently identified directly from culture supernatants by mass spectrometry, showing that cells are able to generate this unmodified form of the peptide naturally and substantiating the notion that native XIP is the 7-mer.
Degradation of streptococcal signaling peptides is also now being investigated more thoroughly. In studies with S. pneumoniae, it was found that competence was promoted by manipulations that increased the frequency of translational errors and thereby the endogenous production of misfolded proteins, which relieved negative effects that the extracellular chaperone and serine protease HtrA places on the competence pathway (Stevens et al., 2011). More recently, it was shown that CSP can be a substrate of HtrA, perhaps because CSP’s amphipathic structure resembles misfolded proteins, which are natural substrates for HtrA. Interestingly, CSP was proteolysed by HtrA unless other substrates were present in the culture fluids that competed with CSP for the protease (Cassone & Sebert, 2012), offering a possible solution to the longstanding puzzle of why sera or BSA are required for development of competence in S. pneumoniae in vitro. During times of stress, higher-priority protein substrates are preferentially targeted by HtrA, which then leaves CSP intact. In S. mutans, it is also known that HtrA has a negative effect on competence, but these effects can be suppressed by addition of CSP to cultures (Ahn et al., 2005). Therefore it seems possible that in this species too, HtrA may directly modulate the availability of CSP.
Accumulation of CSP in cultures of S. mutans promotes competence development, but at most 30–50% of cells develop this capacity, reflecting a bimodal distribution of competence gene expression in the population. Bimodality sometimes develops from bistable pathways containing stochastic noise in regulatory components that are coupled to hypersensitive feedback loops (Dubnau & Losick, 2006). At least three examples of bimodal distributions have been noticed within CSP-treated S. mutans cultures, competence induction, indicated by sigX-gfp expression, being the first one recognized (Perry et al., 2009). Secondly, autolysis events were found among a subset of cells in which sigX was induced (Lemme et al., 2011, Perry et al., 2009). In one report, cell death (a consequence of imbalanced mutacin V and immunity protein ratios) occurred only in cells where the level of SigX was low compared to that in surviving cells, implicating a role of SigX in the autolytic decision (Lemme et al., 2011). Thirdly, ComE protein levels also diverged into two distinguishable groups in response to CSP. This distinction does not arise from variable exposures to CSP, since bacteriocin production was expressed evenly throughout the population (Lemme et al., 2011). If bi-stability accounts for the bimodal distribution of ComE levels, then it may be that ComE participates in a positive auto-feedback loop. Since no recognizable ComE binding sites have been found upstream of comE, such a positive feedback likely occurs through an additional unknown component capable of regulating comE expression. Concurrent bimodality in comE and sigX expression lends weight to the possibility these phenomena are coupled, and will be interesting to learn how their patterns of expression are determined.
Growth medium components, it turns out, have a large impact on pheromone responses in S. mutans. A chemically defined medium (CDM), devoid of any exogenous peptides, facilitates a high frequency of natural transformation when cultures are grown to high densities, even without exogenously added CSP or XIP (Wenderska et al., 2012, Desai et al., 2012). In complex media, such as BHI or THY, transformation frequency is strongly dependent on addition of CSP or is increased in the presence of horse serum, perhaps consistent with HtrA effects noted above. However, in CDM, CSP signaling is ineffective, and though XIP was readily identified in CDM, CSP accumulation could not be verified, suggesting either it was not produced or it is highly unstable (Khan et al., 2012). A review of the literature on transformation in S. mutans shows that responses to CSP have always been studied in complex media, while responses to XIP have been studied exclusively in CDM. Son et al. (Son et al., 2012) appear to be the first to compare the two responses directly, using a single recipient strain and identical experimental and physical conditions. Their results confirm and extend inferences that peptide signaling is qualitatively different in the two types of environments. In this report, they use a gfp reporter to compare expression of sigX in response to CSP and XIP between the two medium types. In BHI, but not in CDM, S. mutans is responsive to exogenously supplied CSP. The opposite is true for XIP; it induces sigX expression in CDM but is much less effective in BHI. Nonetheless, CSP induction of sigX in BHI requires an intact ComRS pathway. Because BHI is non-conducive to signaling by XIP, this latter finding appears paradoxical. How can XIP serve as a signal in a medium in which it is ineffective? Inhibition of XIP signaling in BHI is likely due to competition by non-specific peptidic fragments for access to the Opp transporter. Moreover, inactivation of opp does not block CSP induction of sigX in BHI. How can these findings be explained? The authors suggest that non-secreted and unprocessed ComS might be able to bypass the quorum-sensing aspect of the circuit in BHI, and instead, directly interacts with ComR. If comRS is capable of operating under this kind of internal feedback, and if peptides present in the complex medium can modulate the strength of that feedback, then the different signaling behaviors seen in BHI versus CDM are readily understood at the molecular level. While formally consistent with available data, this scenario would call for a new pathway linking ComS and ComR within the cell and seems surprising considering the cells have developed a rather complex system to produce XIP; simply to disregard the quorum-sensing pathway is a bit like going through the trouble of organizing a parliament but voting only after a decision has been made.
Equally puzzling is the failure of CSP to stimulate sigX in CDM. Are cells even able to sense CSP in CDM conditions at all? This question has not yet been tested. Perhaps CSP is unstable in these conditions. Perhaps HtrA is able to inactivate CSP before it stimulates the ComDE pathway. Perhaps the presence of misfolded proteins in BHI (a medium produced from heat-extracted animal tissues), or the addition of heat-treated sera in other competence media, interferes with degradation of CSP by HtrA. Alternatively, perhaps a component of BHI enhances SepM’s ability to process 21-CSP to the mature pheromone, 18-CSP.
The current article (Son et al., 2012) also highlights the phenomenon of bimodal expression of SigX, but finds that bimodality is strictly dependent on medium composition. In concordance with prior reports of cells grown in batch cultures, bacteria stimulated with CSP and sustained in flow-cell conditions in BHI also fail to reach greater than 50% competence levels. Stimulation with XIP, on the other hand, confers 100% compliance in sigX activation when in CDM. Through experiments on a variety of mutants (comS, opp) as well as mathematical modeling, Son et al. demonstrate that an internal feedback loop in comRS is sufficient to explain how the composition of the medium could drive this transition between uniform and bimodal activation of sigX. In addition to ComE’s bimodal expression pattern, the positive feedback loop built into the ComR-ComS pathway sets up the potential for another branch point. The study adds compelling reasons to interrogate the linkage between ComE and ComRS levels. The ability to monitor individual cells over time and control medium and pheromone concentrations now provide the means to study how kinetic flow of information contained in upstream regulators, like ComE and CipB, influences downstream components like ComR and SigX.
Altogether, the recent new results on streptococcal peptide signals begin to provide a view of multiple ways an apparently simple peptide signal can be used to integrate various aspects of a cell’s circumstance beyond the mere presence of other secreting cells. Looking forward, several questions may motivate particularly informative studies. How do the many other sensory pathways, like CiaRH, VicRK, and HdrMR influence the quorum sensing pathways? How are bacteriocins able to affect competence development? Can the knowledge gained from CSP and XIP pathways in S. mutans be applied to the numerous other streptococci that contain one or both of these peptide signals? Finally, how well do growth conditions used in the laboratory resemble that found in clinically-relevant situations? Answers to these questions will resolve many question marks that currently populate existing models of signaling.
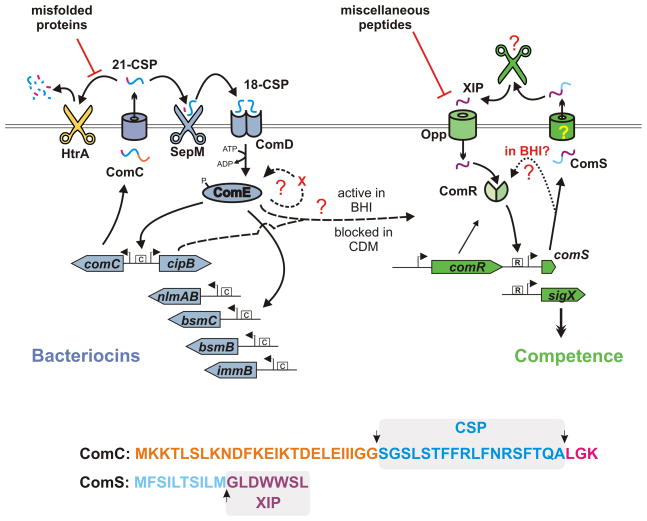
Figure 1.
Model of CSP and XIP quorum sensing in Streptococcus mutans. ComC (whose sequence is included at the bottom and color coded to illustrate processing sites) is secreted as a pre-peptide termed 21-CSP. CSP can be degraded by HtrA in a process affected by misfolded proteins. Productive processing by SepM generates the mature signal, 18-CSP, that binds to ComD leading to phosphorylation of ComE. ComE-P binds inverted repeat sites, indicated by a boxed “C”, enhancing transcription of bacteriocin genes. A positive feedback loop affecting ComE expression is proposed to include an additional regulatory component that establishes a bimodal expression pattern. ComS is secreted and processed by unknown components, but the mature form of the pheromone, XIP, comprises the last seven amino acids of ComS. XIP is imported via the oligopeptide permease (Opp), but transport is blocked by competing peptides present in complex media. Within the cell, an XIP-ComR complex binds to sites indicated by boxed “R” elements, activating transcription of comS and sigX. ComR and ComS are required to induce SigX, and SigX is required for transformation. In the report by Son et al. (Son et al., 2012), medium components are shown to modulate both CSP and XIP systems. In CDM, CSP does not induce competence, while XIP does so efficiently. In BHI, CSP stimulates competence, but, paradoxically, XIP signaling is blocked, yet comS remains required to stimulate sigX. For this reason it is proposed that ComS bypasses secretion and processing to interact with ComR directly.
References
- Ahn SJ, Lemos JA, Burne RA. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol. 2005;187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone M, Sebert ME. Degradation of the Pneumococcal Competence-Stimulating Peptide by the HtrA Protease. Abstract K-276. American Society for Microbiology General Meeting; San Francisco, CA. 2012. [Google Scholar]
- Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. Development of Competence for Genetic Transformation of Streptococcus mutans in a Chemically Defined Medium. J Bacteriol. 2012;194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Dufour D, Cordova M, Cvitkovitch DG, Levesque CM. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol. 2011;193:6552–6559. doi: 10.1128/JB.05968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- Hossain MS, Biswas I. An Extracelluar Protease, SepM, Generates Functional CSP in Streptococcus mutans UA159. J Bacteriol. 2012 doi: 10.1128/JB.01381-12. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung DC, Downey JS, Ayala EA, Kreth J, Mair R, Senadheera DB, Qi F, Cvitkovitch DG, Shi W, Goodman SD. Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J Bacteriol. 2011;193:3642–3652. doi: 10.1128/JB.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MO, Thiede B, Petersen FC. Extracellular Identification of a Processed Type II ComR/ComS Pheromone of Streptococcus mutans. J Bacteriol. 2012;194:3781–3788. doi: 10.1128/JB.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Hung DC, Merritt J, Perry J, Zhu L, Goodman SD, Cvitkovitch DG, Shi W, Qi F. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology. 2007;153:1799–1807. doi: 10.1099/mic.0.2007/005975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemme A, Grobe L, Reck M, Tomasch J, Wagner-Dobler I. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol. 2011;193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol. 2009;72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Fimland G, Scheie AA. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol Microbiol. 2006;61:1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x. [DOI] [PubMed] [Google Scholar]
- Petersen FC, Scheie AA. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol Immunol. 2000;15:329–334. doi: 10.1034/j.1399-302x.2000.150511.x. [DOI] [PubMed] [Google Scholar]
- Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. 2012 doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KE, Chang D, Zwack EE, Sebert ME. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. MBio. 2011;2 doi: 10.1128/mBio.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg JR. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol. 2005;187:3980–3989. doi: 10.1128/JB.187.12.3980-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ween O, Gaustad P, Havarstein LS. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol. 1999;33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- Wenderska I, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett. 2012 doi: 10.1111/j.1574-6968.2012.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]