Abstract
This report describes the synthesis and properties of a series of polyvalent side chain peptide – synthetic polymer conjugates designed to block the CD4 binding site on gp120 and inhibit HIV-1 entry into a host cell. The peptide sequences in the conjugates are based on the CDR H3 region of the neutralizing anti-HIV-1 antibody IgG1 b12. Using a consecutive ester-amide/thiol-ene post-polymerization modification strategy, a library of polymer conjugates was prepared. Evaluation of the HIV-1 inhibitory properties revealed that mid-sized polymer conjugates displayed the highest antiviral activity, while shorter and longer conjugates proved to be less efficacious inhibitors. The lower molecular weight conjugates may not have sufficient length to span the distance between two neighboring gp120 containing spikes, while the higher molecular weight conjugates may be compromised due to a higher entropic penalty that would accompany their binding to the viral envelope. Although the IC50 values for these polymer conjugates are higher than that of the parent IgG1 b12 antibody, the strategy presented here may represent an interesting antiviral approach due to the attractive properties of such polymer therapeutics (relatively inexpensive production and purification costs, high thermal and chemical stability in storage conditions, long half life in biological tissues, low immunogenicity, protection from proteolytic degradation).
Keywords: polyvalency, peptide – polymer conjugate, HIV, inhibition, post-polymerization modification
INTRODUCTION
Infection by the human immunodeficiency virus type 1 (HIV-1) is a global health problem with more than 33 million people affected worldwide. Despite ongoing efforts, no known cure has been developed to date to combat this infection, which causes acquired immune deficiency syndrome (AIDS).1 However, a number of therapeutics have been developed that significantly delay the onset of AIDS and improve the quality of life and life expectancy of these patients. The four main treatment strategies are distinguished by the stage of the HIV life cycle that is targeted: (i) membrane fusion and viral entry, (ii) reverse transcription, (iii) integration and (iv) maturation/proteolysis.2
HIV-1 entry inhibitors are attractive therapeutics as they block the initial stages of viral infection (cellular attachment and membrane fusion), as opposed to the other classes of antivirals that disrupt lifecycle events occurring after the virus has successfully penetrated the cell membrane. HIV-1 entry inhibitors block the function of the viral glycoprotein Env, which is composed of gp120 and gp41 subunits that are arranged as a trimer of heterodimers on the virion surface (gp1203/gp413).3,4 The gp120 subunits interact with cellular CD4 and a chemokine receptor (primarily CCR5 or CXCR4) to coordinate a series of structural changes in the gp41 trimer that culminates in the fusion of the viral and cellular membranes. An HIV-1 virion is thought to contain ~14 copies of the Env trimer on its surface, although a significant number of these adopt nonnative or misfolded forms incapable of promoting viral entry.5,6
One approach in the development of HIV-1 entry inhibitors involves the use of polypeptides derived from the HR1 and HR2 regions of gp41.7–10 These peptides act as competitive inhibitors that disrupt the interaction of the HR1 and HR2 domains required for gp41-mediated membrane fusion. A prominent example of a fusion inhibitor is T-20 (Fuzeon®).11 This peptide drug is FDA approved, but, partly due to its high cost of production and requirement for parenteral administration, it is primarily used as salvage therapy for HIV-1 infections refractory to standard antiviral therapy.12 Another approach involves the use of small molecules that bind either CXCR4 or CCR5 receptors and block their interaction with gp120.13 The FDA-approved entry inhibitor maraviroc binds CCR5 and specifically prevents infection of CCR5-tropic HIV-1. A major drawback to maraviroc therapy is its ineffectiveness in individuals infected with HIV-1 that utilize CXCR4.12
Given the trimeric nature of Env and its multiple copies on the virion surface, an inhibitor that presents multiple ligands attached to a polymeric scaffold might be an effective antiviral agent. The interaction between one entity containing multiple ligands and a different entity containing multiple receptors is referred to as polyvalency and can result in an extremely high binding strength (avidity) compared to the corresponding monovalent interaction (affinity).14,15 Polyvalency has been successfully employed in the development of inhibitors against influenza,16,17 anthrax toxin18–20 and cholera toxin.21 The concept of polyvalency has also been exploited to combat the HIV-1 virus. One approach is based on the use of natural sulfated polysaccarides22,23 or synthetic sulfated polymers24–26 that bind to positively charged residues on the gp120 envelope protein and block its normal function in HIV-1 entry.27 More targeted approaches that involve specific ligand – receptor interactions have also been explored for the design of polyvalent HIV-1 entry inhibitors, albeit to a limited extent.28–30 In one example, Li et al. have described the use of bi-28 and trivalent29 CD4 miniproteins. In the bivalent CD4 miniproteins, two polypeptides that mimic the CD4 receptor were linked by a poly(ethylene glycol) (PEG) spacer. Depending on the PEG spacer length, the efficacy of the bivalent miniproteins increased up to 21-fold as compared to the peptide alone.28 In the trivalent miniproteins, the peptides were attached to Kemp's acid or triazacyclododecane via a short alkyl (-(CH2)2- or -(CH2)10-) or oligo(ethylene glycol) spacer (-(CH2CH2O)2- or -(CH2CH2O)12-).29 Irrespective of the spacer length, the trivalent miniproteins were found to show anti-HIV activities that were comparable to the bivalent analogues. More recently, multivalent benzoboroxole functionalized methacrylamide copolymers that bind 4,6-diols of mannopyranose residues were synthesized to inhibit Env function by complexation with a high mannose region of gp120.30 As compared to a small molecule analogue, the multivalent copolymers showed a ten times higher binding strength for gp120. This latter study is the only current example of a linear polyvalent polymer that selectively targets specific regions on the HIV-1 gp120.
This contribution reports the design, synthesis, chemical properties and antiviral activities of a series of polyvalent peptide – synthetic polymer conjugates that present multiple copies of the CDR H3 loop from the HIV-1 neutralizing antibody b12. This region of IgG1 b12 interacts with the CD4 binding site on gp120 and possesses weak antiviral activity in the absence of the remainder of the antibody. Using a consecutive ester-amide/thiol-ene post-polymerization modification strategy, a library of 11 side chain peptide – polymer conjugates with degrees of polymerization ranging from 199 to 539 and peptide-ligand densities between 19 and 49 per polymer chain was prepared. In HIV-1 infection experiments, the antiviral activities were found to vary with polymer length, but not significantly with peptide-ligand loading density. The polymer conjugates showed significantly improved inhibitory activity as compared to the peptide alone. The strategy undertaken here represents a novel approach to develop new HIV-1 entry inhibitors that takes advantage of the multiple copies of Env on the surface of the virion.
EXPERIMENTAL SECTION
Materials
All Fmoc-amino acids, hydroxybenzotriazole (HOBT.H2O) and O-benzotriazole-N,N,N’,N’-tetramethyl-uronium-hexafluorophosphate (HBTU) were purchased from Iris Biotech (Marktredwitz, Germany). Azobisisobutyronitrile, AIBN (99%), triethylamine (99 %), piperidine (99 %), diisopropylethylamine (99 %), triisopropylsilane (99 %), ethan-1,2-dithiol (99 %) and dioxane (99 %) were obtained from Sigma-Aldrich (Buchs, Switzerland). Hexane (99 %), diethyl ether (99 %) and trifluoroacetic acid (99 %) were obtained from Merck (Darmstadt, Germany). HPLC grade trifluoroacetic acid (99.5 %) was obtained from Fisher Scientific (Wohlen, Switzerland). Gradient grade methanol (99.5 %), tetrahydrofuran (99%) and dimethylformamide (99 %) were obtained from VWR (Nyon, Switzerland) and were dried by passing through columns of molecular sieves using a PureSolv 400 apparatus and finally stored under N2. MilliQ water (18.2 MΩ cm at 25 °C) was obtained from a Millipore MilliQ machine equipped with a 0.22 µm filter. AIBN (recrystallized from hot methanol) as well as 4-cyanopentanoic acid dithiobenzoate (chain transfer agent, CTA)31 were stored at −20 °C prior to use. Pentafluorophenyl methacrylate (PFMA) was prepared according to Eberhardt et al.32 and stored at 4 °C. Dulbecco’s Modified Eagle’s Medium (DMEM), penicillin/streptomycin and L-glutamine were purchased from Mediatech (USA); puromycin from Fisher Scientific (USA); and fetal bovine serum from Atlanta Biologicals (USA). Transfection reagent Lipofectamine and Plus reagent were purchased from Invitrogen (USA). The luciferase assay system was purchased from Promega (USA). The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: HIV-1 gp120 monoclonal antibody IgG1 b12 from Drs. Dennis Burton and Carlos Barbas and HOS CCR5 target cells from Dr. Nathaniel Landau. The 293T cells were obtained from the American Type Culture Collection (ATCC).
Methods
1H-NMR spectra were acquired with a Bruker Avance-400 spectrometer operating at 400 MHz using a relaxation time (t1) of 10 s using either CDCl3 or MeOD as solvents. Chemical shifts are reported relative to the residual proton resonance of the solvent. Size exclusion chromatography (SEC) was performed using N,N’-dimethylformamide with 0.5 g.L−1 LiCl as the eluent on a Waters Alliance GPCV 2000 system equipped with refractive index, differential viscometer and light scattering detectors. Samples were eluted at a flow rate of 0.6 mL.min−1 over TSK-Gel Alpha 2500 + 3000 + 4000 columns. Molecular weights of the samples were determined relative to poly(methyl methacrylate) (PMMA) standards ranging from 600 g.mol−1 to 685000 g.mol−1. A universal calibration analysis was performed thereby accounting for the intrinsic viscosity of the samples. FT-IR spectra were recorded on a Nicolet 6700 Fourier transform infrared spectrometer (Thermo Scientific) equipped with an attenuated total reflectance set up. The FT-IR spectrometer was purged with nitrogen gas and each spectrum represents an average of 16 spectra corrected for a background.
Procedures
Synthesis of poly(pentafluorophenyl methacrylate)s 1–3
The synthesis of poly(pentafluorophenyl methacrylate)s 1–3 was performed as described by Gibson et al.33 In a typical experiment, 1 g (4 mmol) pentafluorophenyl methacrylate in 2.2 mL dioxane was added to a 10 mL Schlenk tube. For polymer 3, 0.1 mL (8 µmol) of a freshly prepared 80 µmol/mL dioxane stock solution of 4-cyanopentanoic acid dithiobenzoate and 0.1 mL (4 µmol) of a freshly prepared 40 µmol/mL dioxane solution of AIBN were added to the monomer solution to yield a molar ratio of monomer : CTA : initiator of 500 : 1 : 0.5. The monomer : CTA and CTA : initiator ratios for polymers 1 and 2 are listed in Table 1. A stirrer bar was added to the Schlenk tube and the mixture was degassed by three freeze-pump-thaw cycles followed by the addition of nitrogen gas. Subsequently, the Schlenk tube was immersed in an oil bath at 90 °C for 2 hours. Following this, the reaction mixture was cooled down using an ice bath. A 25 µL aliquot was taken from the crude polymerization mixture and diluted with 0.5 mL CDCl3 in an NMR tube for the determination of conversion. The remainder of the polymerization mixture was precipitated in 40 mL hexane and reprecipitated from tetrahydrofuran (THF) into hexane two times. The precipitate was vacuum dried overnight to yield a dry pink polymer product. Yields: 65 – 75 %. 1H-NMR (400 MHz, CDCl3) δppm of PPFMA polymers with RAFT dithiobenzoate end group 1, 2, and 3: 7.87 (m-Ar, end-group), 7.56 (p-Ar, end group), 7.43 (o-Ar, end group), 2.53 (backbone CH2), 1.53 (backbone CH3). FT-IR (νcm−1): 1515 (sh, ring C=C stretch), 1776 (sh, C=O stretch), 2880–3020 (w, C-H stretch).
Table 1.
Physicochemical properties of the poly(pentafluorophenyl methacrylate) (PPFMA) precursor polymers.
| Polymer | [M]:[CTA] (−) |
[CTA]:[I] (−) |
Conversiona (%) |
Mn, thb (g.mol−1) |
DPn, thb (−) |
Mn, SECc (g.mol−1) |
DPn, SECc (−) |
Mw/Mnc (−) |
Mn, NMRd (g.mol−1) |
DPn, NMRd (−) |
Mn, PHPMAe (g.mol−1) |
DPn, PHPMAe (−) |
Mw/Mne (−) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 250 | 2 | 73 | 46000 | 182 | 23000 | 91 | 1.69 | 50100 | 199 | 24900 | 175 | 1.45 |
| 2 | 350 | 2 | 70 | 61700 | 245 | 28700 | 147 | 1.70 | 67800 | 269 | 34800 | 253 | 1.47 |
| 3 | 500 | 2 | 71 | 89500 | 355 | 51600 | 204 | 1.72 | 93700 | 372 | 47800 | 336 | 1.57 |
| 4 | 800f | n.a. | 76 | 153000 | 608 | 62000 | 246 | 1.38 | n.a. | n.a. | 76000 | 539 | 1.41 |
Determined by 1H-NMR.
Theoretical number-average molecular weight/degree of polymerization determined from conversion (1H-NMR) and [M]:[CTA].
Number-average molecular weight/degree of polymerization/polydispersity from DMF SEC relative to PMMA standards.
Number-average molecular weight/degree of polymerization determined from 1H-NMR spectroscopy from comparison of the RAFT agent end group to the -CH2 backbone peaks.
Number-average molecular weight/degree of polymerization/polydispersity determined from DMF SEC after post-polymerization modification with 2-hydroxypropylamine to yield poly(2-hydroxypropyl methacrylamide).
Monomer to initiator ratio used was 800 : 1.
Synthesis of poly(pentafluorophenyl methacrylate) 4
PPFMA 4 was obtained via free radical polymerization using pentafluorophenyl methacrylate (1 g, 4 mmol) and AIBN (0.8 mg, 5 µmol) in 3 mL of dioxane. The mixture was degassed by three freeze-pump-thaw cycles followed by subsequent addition of nitrogen gas. The Schlenk tube was immersed for 16 hours in an oil bath at 90 °C. After polymerization, the mixture was cooled down using an ice-bath and a small aliquot (25 µL) was withdrawn from the crude polymerization mixture and diluted with 0.5 mL CDCl3 in an NMR tube to determine the conversion. The remainder of the polymerization mixture was precipitated into 40 mL hexane and reprecipitated from tetrahydrofuran (THF) into hexane two times followed by evacuation overnight to yield a dry pink polymer product. Yield: 74 %. 1H-NMR (400 MHz, CDCl3) δppm of PPFMA 4: 2.53 (backbone CH2), 1.53 (backbone CH3). FT-IR (νcm−1): 1515 (sh, ring C=C stretch), 1776 (sh, C=O stretch), 2880–3020 (w, C-H stretch).
RAFT chain transfer agent end group exchange
Prior to post-polymerization modification, the CTA end-group of the PPFMA polymers was exchanged using a similar procedure as described by Perrier et al.34 For polymer 3 (89500 g/mol determined by conversion in 1H-NMR, see Table 1), the dithiobenzoate CTA end group was exchanged through the addition of 30 mol eq. (27 mg, 167 µmol) of AIBN to 500 mg (5.6 µmol) of PPFMA in 2.5 mL dioxane. After purging the solution with nitrogen, the temperature was increased to 80 °C for 3 hours. After the reaction, the polymer was precipitated into hexane to yield a white polymer product, which was dried in vacuo. 1H-NMR spectroscopy revealed complete removal of the dithiobenzoate end-group (see Supporting Information Figure S1). Yield: 90 – 100 %.
Post-polymerization modification of poly(pentafluorophenyl methacrylate)
For the synthesis of poly(2-hydroxylpropyl methacrylamide) (PHPMA), PPFMA (100 mg, 0.4 mmol of PFMA groups) was dissolved in 4.5 mL of DMF followed by the addition via a syringe of 0.4 mL (1 mol. eq., 0.4 mmol) from a 1 mmol/mL DMF stock solution of 2-hydroxypropylamine and 0.1 mL (4 mol. eq., 1.6 mmol) from a 16 mmol/mL DMF stock solution of triethylamine (Et3N). For PPFMA precursor polymer 4, stirring in DMF at 40 °C accelerated complete dissolution.
For the post-polymerization modification to form copolymers of allyl methacrylamide and 2-hydroxypropyl methacrylamide P(AllMA-co-HPMA), PPFMA (100 mg, 0.4 mmol PFMA groups) was dissolved in 4.4 mL DMF. To obtain random copolymers 6, 9 and 12, a 0.25 mL DMF solution containing 0.32 mmol (0.8 mol eq., 24 µL) 2-hydroxypropylamine was prepared followed by the addition of a 0.25 mL DMF solution containing 0.08 mmol (0.2 mol eq., 3 µL) allylamine and added to the DMF solution containing PPFMA. To obtain random copolymers 5, 8, 11 and 14, a 0.25 mL DMF solution containing 0.2 mmol (0.5 mol eq., 15 µL) 2-hydroxypropylamine was prepared followed by the addition of a 0.25 mL DMF solution containing 0.2 mmol (0.5 mol eq., 9 µL) allylamine and added to the DMF solution containing PPFMA. After the addition of the 2-hydroxypropylamine and allylamine containing solutions, 16 mmol Et3N (4 mol eq., 221 µL) was added to the reaction mixture.
For each post-polymerization modification experiment, the Schlenk tube was immersed in an oil-bath at 70 °C for 16 hours after which the mixture was cooled using an ice-bath followed by precipitation in diethyl ether. The post-modified polymer product was reprecipitated five to seven times from MeOH to yield an off-white powder. The polymers were stored at 4 °C until future use. Yield: 50 – 80 %. 1H-NMR (400 MHz, MeOD) δppm of P(AllMA-co-HPMA) copolymers 5, 6, 8, 9, 11, 12 and 14: 5.85 (CH2CH=CH2), 5.20 (CH=CH2), 3.80–3.90 (CHOH from HPMA + CH2NH from AllMA), 3.12 (CH2NH from HPMA), 2.53 (backbone CH2), 0.90–1.45 (backbone CH3, CH3CH(OH)). FT-IR (νcm−1): 1657 (sh, amide C=O stretch), 2930 (m, asymmetrical CH2 stretch), 2971 (m, symmetrical CH2 stretch), 3078 (w, C=C stretch) 3354 (br, O-H + N-H stretch). 1H-NMR (400 MHz, MeOD) δppm of PHPMA polymers 7, 10, 13 and 15: 3.90 (CHOH), 3.12 (CH2NH), 2.53 (backbone CH2), 0.90 – 1.45 (backbone CH3, CH3CH(OH)). FT-IR (ν cm−1): 1657 (sh, amide C=O stretch), 2930 (m, asymmetrical CH2 stretch), 2971 (m, symmetrical CH2 stretch), 3360 (br, O-H + N-H stretch).
Peptide synthesis
The CDR H3 peptide (Ac-CPGK-ARVGPYSWDDSPQDNYYMDVW-Am) as well as a scrambled control sequence (Ac-CPGK-DRMDNWYPVDAYPSGDQSVWY-Am) (2991 g.mol−1) were synthesized via Fmoc solid phase peptide synthesis using a CEM Liberty® automated microwave synthesizer. Peptide synthesis was performed at the 0.25 mmol scale on a Rink Amide-AM resin with a loading of 0.71 mmol/g. A typical synthetic cycle consisted of Fmoc deprotection, amino acid coupling and washing steps before and after the amino acid coupling step. Fmoc deprotection was performed in two consecutive steps by addition of 5 mL 20 % (v/v) piperidine solution containing 0.1 M HOBT.H2O in DMF with 40 W for 0.5 minutes and 54 W microwave power for 6 minutes, respectively, reaching a maximum operating temperature of 75 °C. After 5-fold washing of the resin with 10 mL DMF, 5 mL (0.2 M) amino acid, 2 mL (0.5 M) activator (HOBT.H2O and HBTU) and 1 mL (2 M) base (DIEA) was added to the resin in the reaction vessel to yield a respective molar ratio of 4 : 4 : 8 relative to the resin. Couplings were carried out for 7 minutes using a microwave power of 24 W and a maximum operating temperature of 75 °C. After the last amino acid was coupled and Fmoc deprotected, acetylation at the N-terminus was performed using 5 mL of 20 % (v/v) acetic anhydride in DMF for 5 minutes at a maximum operating temperature of 60 °C. The peptides were cleaved from the resin for 3 – 5 hours using 5 mL of TFA : triisopropylsilane : EDT : H2O in the approximate volume ratio 92.5 : 2.5 : 2.5 : 2.5. The crude peptides were precipitated 5 times in cold diethyl ether and freeze dried prior to purification.
Peptide Purification
The peptides were purified using a Waters semi-preparative HPLC equipped with an Atlantis® C-18 reverse phase column and a Waters 400 fraction collector. The crude peptide samples were purified using a 40 % methanol/60 % water (with 0.1 % TFA) to 60 % methanol/40 % water (with 0.1 % TFA) gradient over 20 minutes at 20 mL min−1. Both the original and the scrambled peptides eluted at 13.5 minutes. The mass of the purified peptides was determined using electron spray ionization mass spectrometry (ESI-MS). The purified peptide solutions were reduced in volume by rotary evaporation, freeze dried to yield a white powder, which was stored at −20 °C before use.
Thiol-ene post-polymerization modification of the poly(allyl methacrylamide-co-2-hydroxypropyl methacrylamide) copolymers
Thiol-ene additions were performed in DMF in a 10 mL Schlenk tube under nitrogen using AIBN as a radical source. A molar ratio of carbon-carbon double bonds from the allyl methacrylamide moiety, C=C : AIBN of 1 : 0.3 was used. To vary the grafting density of the peptide to the copolymer, a molar ratio of C=C : peptide of 1 : 1.6 was used to obtain sample 20, a molar ratio of 1 : 2 was used to obtain samples 16–19, 21, 23, 25 and a molar ratio of 1 : 3 was used to obtain samples 22, 24 and 26 (Table 3). As a typical example, the synthesis of copolymer 21 was achieved by adding 1 mL of DMF to a 10 mL Schlenk tube containing 3.0 mg copolymer 11 (11.5 µmol C=C bonds for 48 % allyl methacrylamide modified copolymer) and 68 mg CDR H3 peptide (2991 g/mol, 2 mol eq., 23 µmol). In a separate vial, a freshly made 1 mL stock solution of AIBN (5.7 mg, 34.5 µmol) in DMF was prepared of which 0.1 mL (0.3 mol eq., 3.5 µmol) was added to the copolymer 11-peptide solution in DMF described above. The solution was degassed by three freeze-pump-thaw cycles followed by purging with nitrogen gas. The reaction was heated at 70 °C for 16 hours after which a second 0.1 mL portion of a freshly prepared, degassed 5.7 mg/mL DMF solution of AIBN was added under a stream of nitrogen gas. The mixture was then heated at 70 °C for a further 18 hours after which it was cooled down to 4 °C and precipitated in cold diethyl ether. The precipitate was dissolved in 10 mL MilliQ water with 0.1 % NH3(aq) and dialyzed against MilliQ using a SpectraPor dialysis membrane (MWCO 6000–8000) for 3 days. In a final step, the solution was removed from the dialysis bag and was freeze dried in a 50 mL falcon tube under vacuum to yield a fluffy white powder. Yields: 12–35 %. Post-polymerization modification conversions were determined by comparing the integrals of the 22 protons derived from tryptophan and tyrosine side chains to the CH2CH=CH2 proton at 5.85 ppm, which expresses the amount of peptides per allyl group conversion. These values were then recalculated to express the conversion and number of peptides per entire copolymer. 1H-NMR spectra of the peptide modified copolymers are provided in the Supporting Information in Figures S2 and S3.
Table 3.
Properties of the peptide modified copolymers.
| Peptide- copolymer |
Derived from copolymer |
Copolymer DPa (−) |
Eq. of peptide for thiolene reactionb |
Allyl-MA in copolymer (%)c |
Allyl conversion (%)d |
Peptide per copolymer (%)e |
Number of peptides per copolymerf (−) |
|---|---|---|---|---|---|---|---|
| 16 | 5 | 199 | 2.0 | 38 | 23.5 | 10.9 | 18 |
| 17 | 6 | 199 | 2.0 | 16 | 37.5 | 7.2 | 12 |
| 18 | 8 | 269 | 2.0 | 47 | 15.1 | 7.5 | 19 |
| 19 | 9 | 269 | 2.0 | 20 | 33.4 | 7.6 | 18 |
| 20 | 11 | 372 | 1.6 | 48 | 8.4 | 4.2 | 15 |
| 21 | 11 | 372 | 2.0 | 48 | 18.0 | 9.1 | 32 |
| 22 | 11 | 372 | 3.0 | 48 | 21.9 | 11.1 | 39 |
| 23 | 12 | 372 | 2.0 | 20 | 25.7 | 5.6 | 19 |
| 24 | 12 | 372 | 3.0 | 20 | 54.1 | 11.7 | 40 |
| 25 | 14 | 539* | 2.0 | 47 | 11.9 | 6.0 | 30 |
| 26g | 11 | 372 | 3.0 | 48 | 27.5 | 13.9 | 49 |
Number average degree of polymerization of the PPFMA precursor as determined from 1H-NMR spectroscopy from comparison of the protons on the RAFT agent end group to the -CH2 backbone peaks.
Note: the number average degree of polymerization of copolymer 14 is determined by DMF SEC.
Number of peptide equivalents with respect to allyl groups used during the thiol-ene reaction.
Determined with 1H-NMR spectroscopy.
Calculated from 1H-NMR spectra as: number of peptides/original number of allyl units.
Calculated from 1H-NMR spectra as: number of peptides per polymer /(total number of original allyl units prior to thiol-ene addition + number of HPMA units).
The number of peptide chains per copolymer based on the total polymer DP derived from 1H-NMR presented in Table 1.
Copolymer 26 functions as a negative control with the scrambled peptide grafted on to the copolymer via thiol-ene reaction.
HIV-1 production
HIV-1 pseudotyped with EnvHXB2 or EnvJRFL were produced in 293T cells cotransfected with an Env-deficient HIV-1NL4-3 genome (pNL4-3E−V−Luc+) and an Env-expressing plasmid (pEBB_EnvHXB2 or pEBB_EnvJRFL) using the Lipofectamine Plus system (see Champagne et al. and references therein).35 Cells were incubated at 37 °C with 5 % CO2 in complete DMEM media (cDMEM containing DMEM + Pen/Strep + L-glutamine + 10% fetal bovine serum) for 24 hours before media was replaced with fresh cDMEM. After an additional 24 hour incubation, virus-containing media was collected by aspiration and centrifuged (5 minutes, ~500 × g) to remove cell debris. Viruses were either utilized immediately in infection assays or stored in 1.5 mL aliquots at −80 °C for later use.
Infectivity assay
HOS CCR5 cells in cDMEM + puromycin were plated on a 48 well plate (50000 cells / well in 500 µL) and incubated for 24 hours at 37 °C with 5 % CO2. Phosphate-buffered saline stock solutions of polymers were prepared and sample concentrations were determined by absorption at 280 nm using an extinction coefficient of 15595 M−1.cm−1 corresponding to a single peptide (or scrambled peptide) based on reference values for tryptophan and tyrosine.36 From these stock solutions, 10-fold dilution series were generated in cDMEM at 2x final concentration for immediate use in inhibition experiments. Media from the 48 well plates was aspirated, and the 2x concentrated solutions were applied to HOS CCR5 cells (100 µl/well) in duplicate, followed by an equal amount of HIV-1-containing media. Viral cultures were incubated 24 hours at 37 °C with 5 % CO2, after which an extra 500 µL of cDMEM was added to each well. After an additional 24 hour incubation, media in each well was removed by aspiration and and 100 µL of lysis buffer (25 mM TRIS pH 8.0, 1 mM EGTA, 1 mM DTT, 10 % glycerol and 1 % triton-X) was added. Cell lysate (50 µL) was analyzed for expression of a luciferase reporter construct by addition of luciferin (50 µL, Luciferase Assay System, Promega) and luminescence detection by a FlUOStar Optima (BMG Labtech, Offenburg, Germany). In all assays, a positive control for infection inhibition (1000 nM C37 peptide derived from HIV-1 gp41)35 and three negative controls (scrambled peptide, scrambled peptide attached to a polymer (sample 26) and no added peptide) were also evaluated. At the concentration used, the C37 peptide inhibitor completely inhibited HIV-1 infection. The relative infectivity was calculated as quotient of the luciferase signals determined in the presence and absence of inhibitor.
RESULTS AND DISCUSSION
Design and synthesis of the polyvalent side chain peptide - synthetic polymer conjugates
The design of the side chain peptide – synthetic polymer conjugates discussed in this contribution is based on the work by Saphire et al. who identified an amino acid sequence within the so-called CDR H3 region of the antibody IgG1 b12, which was proposed to be involved in blocking the CD4 binding site of the viral envelope glycoprotein gp120.37 Blocking binding of the CD4 receptor to gp120 neutralizes the virus and reduces the probability of virus - target cell membrane fusion. This proposal was validated by coupling the CDR H3 peptide to bovine serum albumin (BSA). The resulting protein-peptide conjugates were also found to be capable of viral neutralization.37
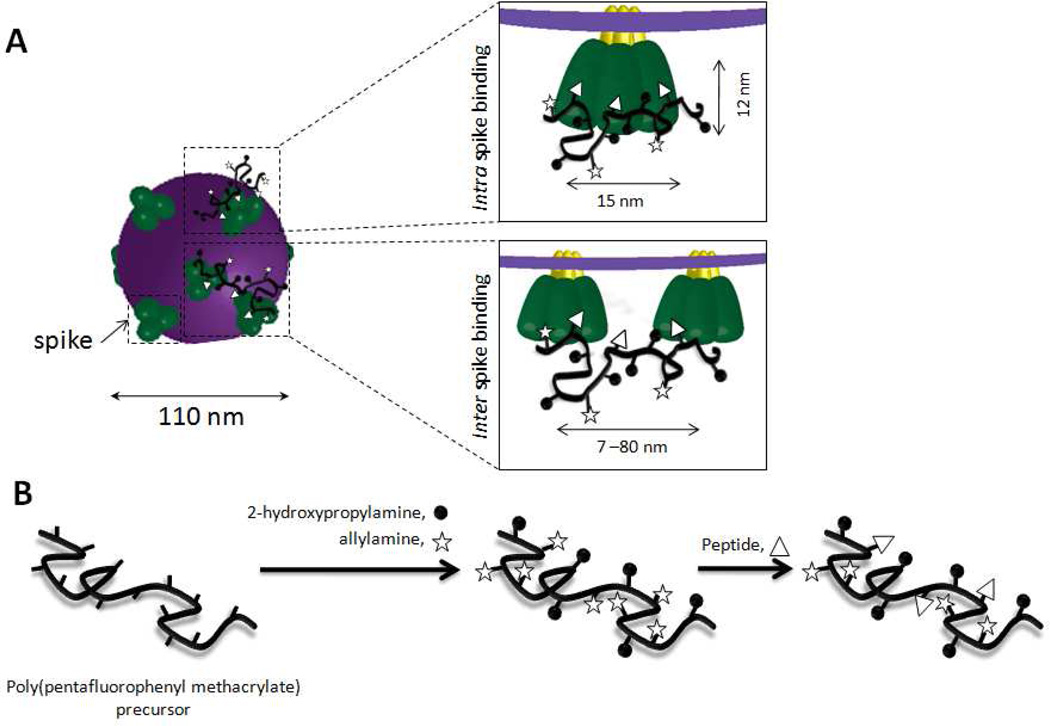
The CD4-binding region of gp120 is an attractive target for polyvalent peptide - synthetic polymer inhibitors. On its surface, HIV-1 presents envelope glycoprotein spikes, which expose three copies of gp120. For the HIV-1 virion an average diameter of 110 nm and an average spike density of ~14 spikes per virion have been determined by electron tomography (Figure 1A).6 Each spike has a height of 12 nm and a maximal width of 15 nm.38 The use of CDR H3 peptide-synthetic polymer conjugates allows us to explore possible polyvalency effects at two length scales; (i) two or three CDR H3 peptides that are attached to a common synthetic polymer may bind simultaneously to a single glycoprotein spike, (ii) if sufficiently high molecular weight peptide-synthetic polymer conjugates are used, a single polyvalent inhibitor might be able to span two spikes (Figure 1A). In addition to blocking the CD4 binding site by the side chain CDR H3 peptide sequences, the polymer backbone of the proposed polyvalent conjugates may also contribute to preventing virus-host cell entry by sterically preventing access of the viral envelope proteins to the host cell surface.
Figure 1.
(A) Schematic representation of binding of the proposed polyvalent inhibitors to gp120 in the envelope glycoprotein spikes that are exposed on the surface of the HIV-1 virion. The dimensions of the spike proteins are described in Ref 38 and the dimensions of HIV-1 as well as the spacing of spikes is derived from Ref 6. (B) Schematic outline of the synthesis of the proposed side chain CDR H3 peptide-synthetic polymer inhibitors.
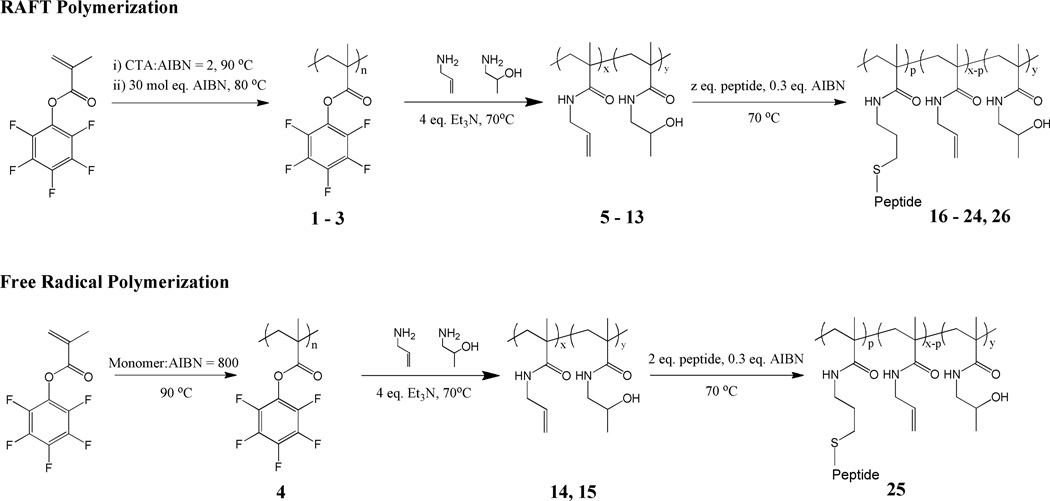
The synthetic strategy for the preparation of the side chain peptide – synthetic polymer conjugates is schematically outlined in Figure 1B. Further details are given in Scheme 1. The envisioned side chain peptide – synthetic polymer conjugates were prepared via consecutive ester-amide/thiol-ene post-polymerization modification of poly(pentafluorophenyl methacrylate) (PPFMA). This three-step process starts with polymerization of pentafluorophenyl methacrylate followed by post-polymerization modification of the resulting PPFMA with allylamine and 2-hydroxypropylamine. The final step involves thiol-ene coupling of the CDR H3 peptide via the thiol group on the N-terminal cysteine residue to the allyl side chains of the precursor copolymer.39 Application of this post-polymerization modification strategy enables the generation of a library of copolymers that are uniform in the degree of polymerization and polydispersity, while differing in the relative composition of allyl methacrylamide (i.e. peptide content) and 2-hydroxypropyl methacrylamide.40
Scheme 1.
The first step towards the envisioned peptide – synthetic polymer conjugates involves the synthesis of activated ester41 containing PPFMA precursor polymers 1–4. The molecular weight characteristics of these polymers are summarized in Table 1. While PPFMA polymers 1–3 were prepared using RAFT polymerization,42 sample 4 was obtained via conventional free radical polymerization. Sample 4 was prepared via conventional free radical polymerization in an attempt to prepare precursor side chain reactive polymers with degrees of polymerization that are difficult to access using RAFT conditions. All RAFT polymerizations were terminated at around 70 % conversion. At longer reaction times, the RAFT polymerization did no longer show first-order kinetics, which is indicative for a loss of the controlled/”living” character of the RAFT polymerization (Supporting information, Figure S4). By varying the monomer to chain transfer agent ratio ([M] : [CTA]), three PPFMA precursor polymers with different molecular weights were prepared. All PPFMA precursor polymers could be dissolved in DMF at least up to 25 mg/ mL. Dissolution could be accelerated by stirring at 40 °C. After RAFT polymerization, samples 1–3 were treated with an excess AIBN in order to exchange the dithiobenzoate end group into an isobutyronitrile group. This step was necessary in order to prevent reaction of possible thiol polymer end groups with the allyl side chains during the final thiol-ene addition of the peptide (vide infra). As shown in Table 1, the degrees of polymerization of samples 1–3 as determined via NMR are in fairly good agreement with the expected degrees of polymerization. However, the degrees of polymerization calculated from DMF SEC analysis of the PPFMA polymers deviated from and were significantly lower than the expected degrees of polymerization. This has been observed before and has been described by Gibson et al.33 More accurate DMF SEC data of the PPFMA precursor polymers could be obtained after post-polymerization modification of 1–4 into the corresponding poly(2-hydroxypropyl methacrylamide) (PHPMA) polymers (see Table 1).
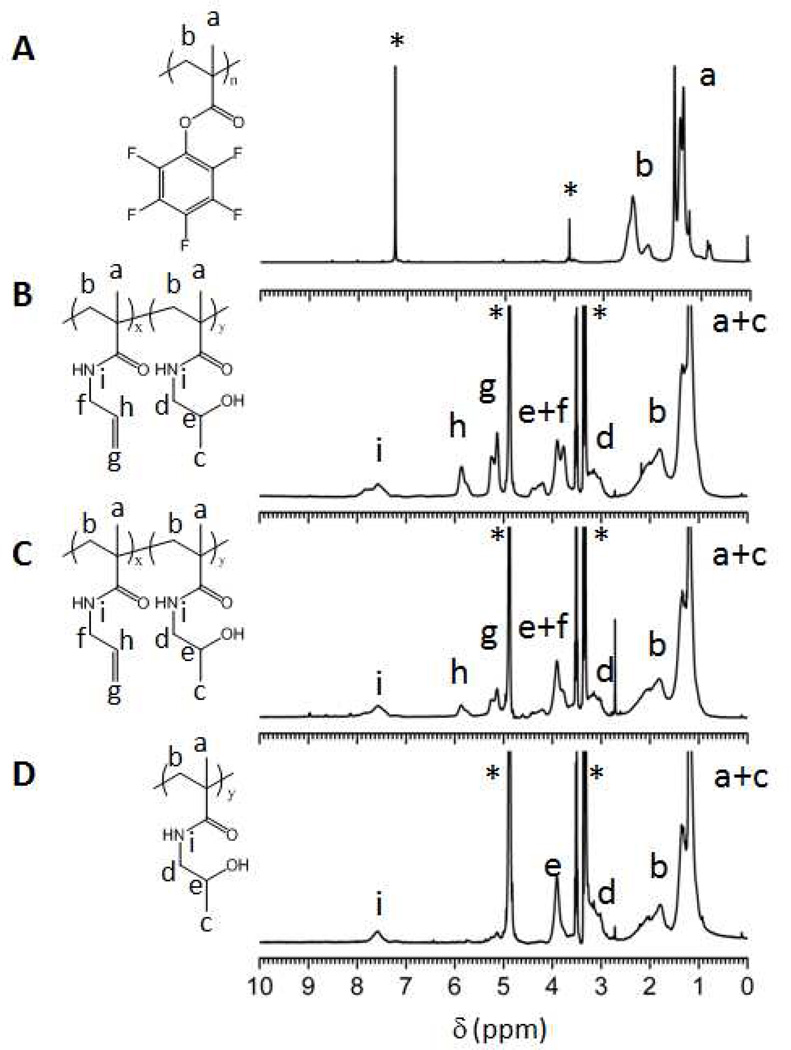
The PPFMA precursor polymers 1–4 were subsequently subjected to a first post-polymerization modification and treated with 1 molar equivalent (amine groups with respect to PFMA residues) of a 50/50 or 80/20 (mole/mole) mixture of 2-hydroxypropylamine and allylamine. SEC chromatograms and 1H-NMR spectra of the post-modified polymers are included in the Supporting Information (Figure S5 – S9). The degree of modification of the postmodified polymers, i.e. the relative number of the different repeat units was determined by 1H-NMR spectroscopy by comparing the integrals of the =CH- group at 5.85 ppm (peak ‘h’ in Figure 2) of the allyl methacrylamide units and one of the two protons of the –CH2NH- group at 3.12 ppm of the hydroxypropyl methacrylamide units (peak ‘d’ in Figure 2) with that of the - CH2- group at 1.8 ppm peak ‘b’ in Figure 2) of the copolymer backbone. As indicated in Table 2, the post-polymerization modification reaction proceeded very well with 82 – 95 % of the pentafluorophenyl ester groups being converted into allyl or 2-hydroxypropyl methacrylamide units. These conversions are in good agreement with earlier reported results on the post-polymerization modification of PPFMA.33 The remaining 5 – 18 % of the repeat units are probably either methacrylic acid repeats (due to hydrolysis of pentafluorophenyl ester groups during post-polymerization modification or during work up) but could also include cyclized glutarimide units that may result from nucleophilic attack of amides to neighboring pentafluorophenyl ester units in a way similar to that reported for poly(methacryloxysuccinimide) by Wong and Putnam.43 DMF SEC analysis of the post-modified polymers afforded number-average molecular weights that corresponded well with the expected values and also did not reveal any significant changes in polydispersity upon post-polymerization modification (Table 2).
Figure 2.
1H-NMR spectra of (A) PPFMA precursor polymer 3 in CDCl3, (B) post-modified copolymer P(AllMAx-co-HPMAy) 11 in MeOD where x = 178 and y = 171, (C) post-modified copolymer P(AllMAx-co-HPMAy) 12 in MeOD where x = 74 and y = 268, and (D) post-modified polymer 13 P(HPMAy) in MeOD where y = 345. * indicates the residual solvent peaks.
Table 2.
Physicochemical properties of the poly(allyl methacrylamide-co-hydroxypropyl methacrylamide) copolymers.
| Copolymer | Precursor PPFMA polymer |
Feed Ratioa All-NH2 / HP- NH2 (% / %) |
AllMAb (%) |
AllMAb (n) |
HPMAb (%) |
HPMAb (n) |
PFMA conversion (%) |
DPc (−) |
Mn, thd (g.mol−1) |
Mn, SECe (g.mol−1) |
Mw/Mne (−) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 1 | 50 / 50 | 38 | 76 | 44 | 88 | 82 | 199 | 21900 | 24500 | 1.35 |
| 6 | 1 | 20 / 80 | 16 | 32 | 68 | 135 | 84 | 199 | 23000 | 24500 | 1.32 |
| 7 | 1 | 0 / 100 | 0 | 0 | 88 | 175 | 88 | 199 | 24700 | 24900 | 1.45 |
| 8 | 2 | 50 / 50 | 47 | 126 | 48 | 129 | 95 | 269 | 33900 | 28900 | 1.41 |
| 9 | 2 | 20 / 80 | 20 | 54 | 69 | 186 | 89 | 269 | 33000 | 33500 | 1.42 |
| 10 | 2 | 0 / 100 | 0 | 0 | 92 | 247 | 92 | 269 | 34800 | 36000 | 1.47 |
| 11 | 3 | 50 / 50 | 48 | 178 | 46 | 171 | 94 | 372 | 46400 | 43000 | 1.52 |
| 12 | 3 | 20 / 80 | 20 | 74 | 72 | 268 | 92 | 372 | 47000 | 44400 | 1.50 |
| 13 | 3 | 0 / 100 | 0 | 0 | 92 | 345 | 92 | 372 | 48600 | 47800 | 1.57 |
| 14 | 4 | 50 / 50 | 47 | 253 | 45 | 243 | 92 | 539f | 65788 | 69500 | 1.48 |
| 15 | 4 | 0 / 100 | 0 | 0 | 91 | 490 | 91 | 539f | 69200 | 76000 | 1.41 |
Feed ratio of allyl amine (All-NH2) and 2-hydroxypropylamine (HP-NH2) in the post-polymerization modification reaction.
Percentage/number of allyl methacrylamide and 2-hydroxypropyl methacrylamide units determined by 1H-NMR spectroscopy after post-polymerization modification of PPFMA with either 2-hydroxypropylamine or a feed mixture of 50 or 80 mol. % 2-hydroxypropylamine and 50 or 20 mol. % allylamine, respectively.
Number average degree of polymerization of the PPFMA precursor as determined from 1H-NMR spectroscopy from comparison of the protons on the RAFT agent end group to the -CH2 backbone peaks.
Theoretical number-average molecular weight of the post-modified copolymers using the number-average degree of polymerization of the PPFMA precursor and the number of allyl methacrylamide and 2-hydroxypropyl methacrylamide repeat units, respectively as determined from 1H-NMR spectroscopy.
Number-average molecular weight/polydispersity determined from DMF SEC.
Number average degree of polymerization of the PPFMA determined from DMF SEC after post-modification with 2-hydroxypropylamine to yield poly(2-hydroxypropyl methacrylamide).
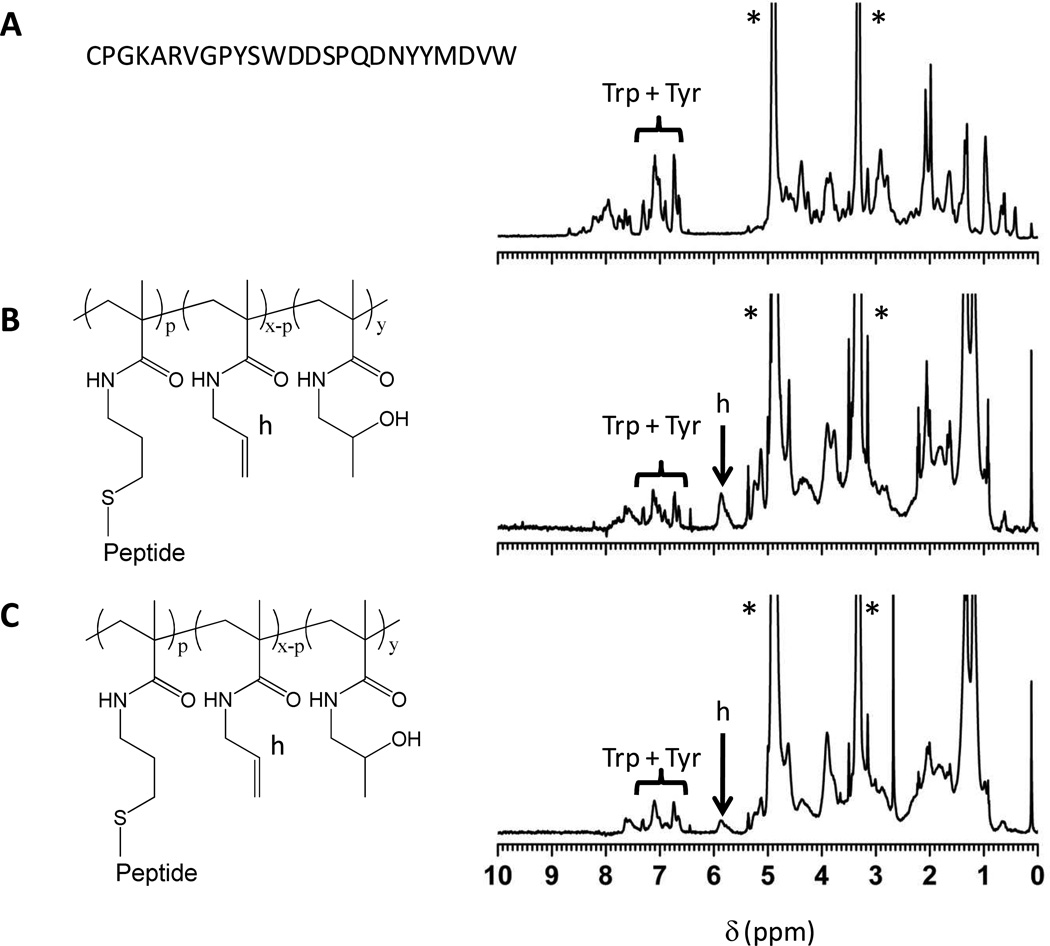
In a final step, the poly(allyl methacrylamide-co-2-hydroxypropyl methacrylamide) copolymers were modified with the CDR H3 peptide as well as with a scrambled control sequence via a thiol-ene coupling reaction44 using the thiol group of the N-terminal cysteine residue of the peptide. The thiol-ene post-polymerization modification reaction was carried out using 1.6 – 3.0 mole equivalents of peptide to allyl methacrylamide repeats. All samples required gentle agitation for dissolution. The resulting peptide-polymer conjugates were soluble up at least up to 200 µM (based on a peptide concentration for conjugate 25) in 10 mM phosphate buffered saline, pH 7.4 (higher concentrations were not evaluated). The degree of modification of the allyl methacrylamide repeat units was determined by 1H-NMR spectroscopy by comparison of the integral of the =CH- group of the allyl methacrylamide repeat units at 5.85 ppm (signal ‘h’ in Figure 3) with that of the 22 aromatic protons of the peptide’s tyrosine and tryptophan residues between 6.60 and 7.40 ppm (see Figure 3 and Table 3). 1H-NMR spectra of all synthesized peptide – polymer conjugates are included in Figures S2 and S3 of the Supporting Information. From the allyl group conversion, the percentage of repeat units bearing a peptide sequence as well as the total number of side chain peptides per polymer were calculated, which are included in Table 3. Comparison of the results on copolymers 20–22 and 23–24 indicates that increasing the excess of peptide with respect to allyl groups increases the degree of modification. The degree of polymerization of the copolymer or the relative number of allyl methacrylamide repeats, however, had little or no influence on the conversion of the allyl methacrylamide residues with peptides.
Figure 3.
1H-NMR spectra in MeOD of (A) CDR H3 peptide, (B) peptide modified copolymer 20 P(AllMA-co-HPMA) where p = 15, x = 178 and y = 171, (C) peptide modified copolymer 23 P(AllMAco-HPMA) where p = 19, x = 74 and y = 268,* indicates residual solvent peaks.
HIV-1 entry inhibition by polyvalent side chain peptide - polymer conjugates
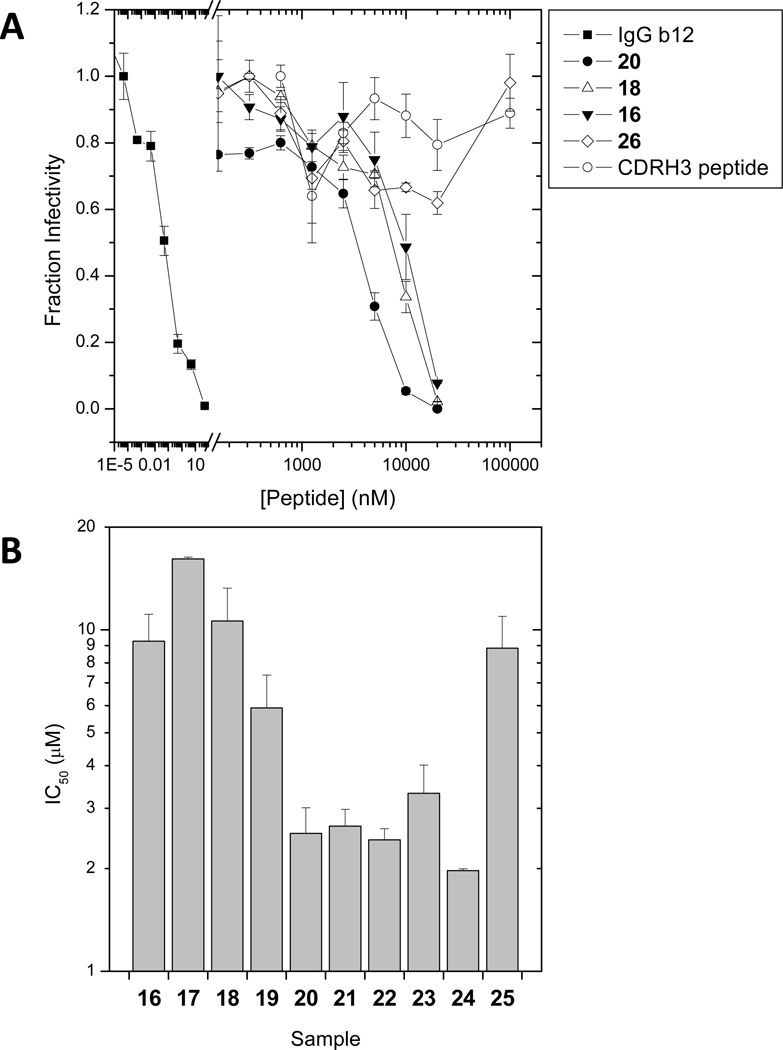
The biological activity of the peptide – polymer conjugates was tested in a single-cycle HIV-1 infectivity experiment using HOS CCR5 cells that express the CD4 receptor as well as both the CCR5 and CXCR4 coreceptors.45 The presence of both major coreceptors allowed us to evaluate the antiviral efficacies against both X4- and R5-tropic strains of HIV-1 (HXB2 and JRFL, respectively). The inhibitory activities of the copolymers were evaluated 48 hours post-infection by determining the amount of a luciferase reporter construct expressed as a result of productive infection. As controls, inhibition by the free CDR H3 peptide, the IgG1 b12 antibody as well as a copolymer containing the scrambled CDR H3 peptide was evaluated. For all samples, fraction infectivity was calculated as the luciferase activity measured in presence of inhibitor relative to that measured in its absence. Figure 4 summarizes the antiviral activities observed for the full series of synthesized copolymers. The CDR H3 peptide – polymer conjugates showed inhibition efficacies in the low micromolar range, whereas neither the CDR H3 peptide alone nor the copolymer containing the scrambled peptide showed antiviral activity up to 100 µM (Figure 4A). The inhibitory activity of the side chain peptide-polymer conjugates reflects the polyvalent nature of these samples. It should be noted that the reported concentrations reflect the total peptide content of solution in order to directly compare copolymers of different lengths and peptide densities. The actual molarity of each copolymer species is 12- to 49-fold lower depending on the average number of peptides conjugated per polymer.
Figure 4.
(A) HIV-1HXB2 viral infectivity inhibition by the IgG1 b12 antibody and side chain peptidesynthetic polymer conjugates 16, 18, and 20. As controls, a copolymer containing a scrambled peptide (26) and the CDRH3 peptide alone are included. The error bars are the standard error observed within one experiment. (B) Efficacy of the polyvalent peptide copolymers against HIV-1 HXB2 wild type on HOS CCR5 cells. The error bars depict the standard error over three or more independent experiments.
From the titration experiments illustrated in Figure 4A, IC50 values for the different peptide – synthetic polymer conjugates were determined and compared in Figure 4B. The data show a clear influence of the number-average degree of polymerization on antiviral activity. The lowest IC50 values (~ 2 µM) were observed for copolymers with a number-average degree of polymerization (DP) of 372, whereas for copolymers with smaller (199 or 269) or larger (539) DP, IC50 values of ~ 10 µM were found. From the perspective of total peptide concentration, the peptide conjugation density of the copolymers did not have a significant effect on IC50 values. However, when considered from the perspective of the actual copolymer concentration, increasing peptide conjugation density did result in a small decrease in IC50 values. For instance, comparing copolymers 20, 21 and 22 (polymer length 372, peptide loading density 15, 32 and 39, respectively), the IC50 values based on copolymer concentration were 170 nM, 78 nM, and 64 nM. The same trends in antiviral activity were observed when the experiments were repeated with HIV-1JRFL virions (see Supporting Information Figure S10).
The chain length dependence of the IC50 values as suggested by the data in Figure 4 could reflect the opposing contributions of the enthalpy and entropy of binding46 to the overall free energy. While the degree of polymerization of polymer conjugates 16–19 may not be sufficient to bridge the average distance between two neighboring Env spikes, the degree of polymerization of longer chain analogues 20–24 may be sufficient to span this distance. While the longer copolymer (conjugate 25) would be sufficient to bind multiple spikes, the increased degree of polymerization would necessarily increase the entropic penalty of polyvalent binding, compromising the IC50 value. From the perspective of copolymer concentration, the peptide-polymer conjugates are about 2–3 orders of magnitude less active as compared to the IgG1 b12 antibody, for which an IC50 of 0.08 nM was determined. Thus, polyvalency and the CDR H3 peptide are not sufficient to recapitulate the potency of the antibody, suggesting that context (i.e., tertiary structure) plays an important role in antibody binding. Whereas the bivalent IgG1 b12 antibody has a defined tertiary structure and, as a consequence, presents the CDR H3 domains in a relatively defined conformation, the peptide-polymer conjugates have an unordered solution conformation, which likely presents a higher entropic barrier towards binding. Furthermore, additional regions on the IgG1 b12 antibody outside the CDR H3 domain contribute to binding gp120,37,47 which could further add to the observed differences in efficacy between the IgG1 b12 antibody and the polymer conjugates.
Although the polyvalent peptide-synthetic polymer conjugates presented herein are less potent as compared to IgG1 b12, the measured efficacies are on the same order of magnitude as a variety of other polyvalent HIV-1 entry inhibitors that have been reported, Li et al. have shown that producing a bivalent CD4M9 conjugate with various PEG spacers increases the efficacy as compared to the monovalent peptide 4.6 to 21 fold depending on spacer length with the medium length spacer having a maximum efficacy of 0.12 µM.28 Li et al. have also shown that trivalent samples possess similar efficacies in the high nanomolar up to low micromolar range (0.4 – 3.0 µM).29 In spite of the lower efficacy as compared to the IgG1 b12 antibody, the peptide – synthetic polymer conjugates presented herein represent an interesting alternative therapeutic, especially for microbicidal applications.48 The synthesis and purification strategies presented in this contribution are cost effective compared to the high costs and purification challenges associated with large scale industrial production of therapeutic antibodies. The chemical nature of the copolymers is extremely similar to gels utilized for microbicidal applications and is likely to achieve high solubility. The copolymers bind directly to the outside HIV-1 virions and are not required to penetrate into host tissues or interact with cellular receptors as many HIV-1 targeted microbicides in development must. The large size of the copolymers should decrease the rate of their egress out of the gel and away from the site of microbicidal application. In addition, the conjugation of peptides/proteins to a water-soluble, biocompatible synthetic polymer (such as poly(ethylene glycol) (PEG)49–51 or PHPMA52,53) is a well-established strategy to decrease proteolytic sensitivity, reduce immunogenicity and increase overall half-life in biological tissues, which may also help to compensate for the lower efficacy as compared to e. g. the IgG1 b12 antibody. Polymer conjugation, however, is not expected to help to overcome the susceptibility of the virus to mutate, which would render the conjugates (but also the IgG1 b12 antibody) ineffective. These potential advantages and limitations for the specific conjugates discussed herein will have to be corroborated in future experiments.
CONCLUSIONS
This contribution has presented the design, synthesis and HIV-1 inhibitory properties of a library of polyvalent side chain peptide – synthetic polymer conjugates. The peptide sequences in these conjugates represent the CDR H3 region of the IgG1 b12 antibody previously identified as a key domain involved in binding of the IgG1 b12 antibody to the HIV envelope protein gp120, which forms the basis for the anti-HIV properties of this antibody. Using a consecutive ester-amide/thiol-ene post-polymerization modification strategy, a library of peptide – synthetic polymer conjugates covering a broad range of molecular weights and peptide contents was prepared. The HIV-1 inhibitory activity of these conjugates was assessed on 2 HIV-1 strains, viz. HXB2 and JRFL. The HIV-1HXB2 infection assays revealed IC50 values of 2–3 µM for the mid-sized polymer conjugates and of 10 µM (concentrations reflect total peptide content) for the lower and higher molecular weight analogues. These results probably reflect the opposing contributions of the enthalpically favorable CDR H3 – gp120 interaction and the entropic penalty that accompanies binding of the polymer conjugates to the gp120 envelope protein to the overall free energy of the HIV-1 inhibition process. While the degree of polymerization of the lowest molecular weight polymer conjugates may not be sufficient to span the distance between two neighboring gp120 spikes, the higher entropic penalty that goes with binding of the highest molecular weight conjugate may result in an increase in IC50 value as compared to the mid-sized conjugates. Although the polymer conjugates presented in this contribution are not as effective as the IgG1 b12 antibody they may still represent an interesting therapeutic alternative, especially for possible microbicidal applications.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the European Commission, FP6 project ‘NanoBioPharmaceutics’ (NMP4-CT-2006-026723 to H.-A. K.) and by the National Institutes of Health, U.S.A. (R01 GM66682 to M. J. R.). The authors would like to thank Dr. Harald Wutzel for the synthesis of the chain transfer agent, Dr. Tuan Q. Nguyen and Cindy Kanel for performing the DMF SEC analysis and Angela Stauffer for performing an HIV infectivity assay.
Footnotes
Supporting Information Available: Additional SEC chromatograms and 1H-NMR spectra as well as viral infectivity titrations of the polymer conjugates. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Cohen J. Science. 2007;318:1360–1361. doi: 10.1126/science.318.5855.1360. [DOI] [PubMed] [Google Scholar]
- 2.Mehellou Y, De Clercq E. J. Med. Chem. 2010;53:521–538. doi: 10.1021/jm900492g. [DOI] [PubMed] [Google Scholar]
- 3.Eckert DM, Kim PS. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 4.Root MJ, Steger HK. Curr. Pharm. Des. 2004;10:1805–1825. doi: 10.2174/1381612043384448. [DOI] [PubMed] [Google Scholar]
- 5.Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. J. Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi E, Finotto M, Ingallinella P, Hrin R, Carella AV, Hou XS, Schleif WA, Miller MD, Geleziunas R, Pessi A. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12903–12908. doi: 10.1073/pnas.0502449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, Tvermoes NA, Matthews TJ, Greenberg ML, Delmedico MK. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12772–12777. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melikyan GB. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilton JC, Doms RW. Antiviral Res. 2010;85:91–100. doi: 10.1016/j.antiviral.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volberding PA, Deeks SG. Lancet. 2010;376:49–62. doi: 10.1016/S0140-6736(10)60676-9. [DOI] [PubMed] [Google Scholar]
- 13.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Antimicrob. Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mammen M, Choi SK, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Vance D, Martin J, Patke S, Kane RS. Adv. Drug Delivery Rev. 2009;61:931–939. doi: 10.1016/j.addr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Mammen M, Dahmann G, Whitesides GM. J. Med. Chem. 1995;38:4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 17.Honda T, Yoshida S, Arai M, Masuda T, Yamashita M. Bioorg. Med. Chem. Lett. 2002;12:1929–1932. doi: 10.1016/s0960-894x(02)00330-x. [DOI] [PubMed] [Google Scholar]
- 18.Basha S, Rai P, Poon V, Saraph A, Gujraty K, Go MY, Sadacharan S, Frost M, Mogridge J, Kane RS. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13509–13513. doi: 10.1073/pnas.0509870103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Nature Biotechnol. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 20.Karginov VA, Mestorovich EM, Moayeri M, Leppla SH, Bezrukov SM. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15075–15080. doi: 10.1073/pnas.0507488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polizzotti BD, Maheshwari R, Vinkenborg J, Kiick KL. Macromolecules. 2007;40:7103–7110. doi: 10.1021/ma070725o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito M, Baba M, Sato A, Pauwels R, De Clerq E, Shigeta S. Antiviral Res. 1987;7:361–367. doi: 10.1016/0166-3542(87)90018-0. [DOI] [PubMed] [Google Scholar]
- 23.Shaunak S, Gooderham NJ, Edwards RJ, Payvandi N, Javan CM, Baggett N, MacDermot J, Weber JN, Davies DS. Br. J. Pharmacol. 1994;113:151–158. doi: 10.1111/j.1476-5381.1994.tb16187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusconi S, Moonis M, Merrill DP, Pallai PV, Neidhardt EA, Singh SK, Willis KJ, Osburne MS, Profy AT, Jenson JC, Hirsch MS. Antimicrob. Agents Chemother. 1996;40:234–236. doi: 10.1128/aac.40.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kensinger RD, Yowler BC, Benesi AJ, Schengrund CL. Bioconjugate Chem. 2004;15:349–358. doi: 10.1021/bc034156a. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy TD, Karellas P, Henderson SA, Giannis M, O'Keefe DF, Heery G, Paul JRA, Matthews BR, Holan G. Mol. Pharm. 2005;2:312–318. doi: 10.1021/mp050023q. [DOI] [PubMed] [Google Scholar]
- 27.Batinic D, Robey FA. J. Biol. Chem. 1992;267:6664–6671. [PubMed] [Google Scholar]
- 28.Li H, Song H, Heredia A, Le N, Redfield R, Lewis GK, Wang LX. Bioconjugate Chem. 2004;15:783–789. doi: 10.1021/bc049960r. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Guan Y, Szczepanska A, Moreno-Vargas AJ, Carmona AT, Robina I, Lewis GK, Wang LX. Bioorg. Med. Chem. 2007;15:4220–4228. doi: 10.1016/j.bmc.2007.03.064. [DOI] [PubMed] [Google Scholar]
- 30.Jay JI, Lai BE, Myszka DG, Mahalingam A, Langheinrich K, Katz DF, Kiser PF. Mol. Pharm. 2010;7:116–129. doi: 10.1021/mp900159n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitsukami Y, Donovan MS, Lowe AB, McCormick CL. Macromolecules. 2001;34:2248–2256. [Google Scholar]
- 32.Eberhardt M, Mruk R, Zentel R, Théato P. Eur. Polym. J. 2005;41:1569–1575. [Google Scholar]
- 33.Gibson MI, Fröhlich E, Klok HA. J. Polym. Sci., Part A Polym. Chem. 2009;47:4332–4345. [Google Scholar]
- 34.Perrier S, Takolpuckdee P, Mars CA. Macromolecules. 2005;38:2033–2036. [Google Scholar]
- 35.Champagne K, Shishido A, Root MJ. J. Biol. Chem. 2009;284:3619–3627. doi: 10.1074/jbc.M809269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saphire EO, Parren PWHI, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singha NK, Gibson MI, Koiry BP, Danial M, Klok H-A. Biomacromolecules. 2011;12:2908–2913. doi: 10.1021/bm200469a. [DOI] [PubMed] [Google Scholar]
- 40.Gauthier MA, Gibson MI, Klok H-A. Angew. Chem. Int. Ed. 2009;48:48–58. doi: 10.1002/anie.200801951. [DOI] [PubMed] [Google Scholar]
- 41.Theato P. J. Polym. Sci., Part A Polym. Chem. 2008;46:6677–6687. [Google Scholar]
- 42.Boyer C, Bulmus V, Davis TP, Ladmiral V, Perrier S. Chem. Rev. 2009;109:5402–5436. doi: 10.1021/cr9001403. [DOI] [PubMed] [Google Scholar]
- 43.Wong SY, Putnam D. Bioconjugate Chem. 2007;18:970–982. doi: 10.1021/bc0603790. [DOI] [PubMed] [Google Scholar]
- 44.Hoyle CA, Bowman CN. Angew. Chem. Int. Ed. 2010;49:1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 45.Kahle KM, Steger HK, Root MJ. PLoS Pathog. 2009;5:e1000674. doi: 10.1371/journal.ppat.1000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammen M, Shakhnovich EI, Whitesides GM. J. Org. Chem. 1998;63:3168–3175. [Google Scholar]
- 47.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lederman MM, Jump R, Pilch-Cooper HA, Root M, Sieg SF. Retrovirology. 2008;5:116. doi: 10.1186/1742-4690-5-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasut G, Veronese FM. Progr. Polym. Sci. 2007;32:933–961. [Google Scholar]
- 50.Klok H-A. Macromolecules. 2009;42:7990–8000. [Google Scholar]
- 51.Alconcel SNS, Baas AS, Maynard HD. Polym. Chem. 2011;2:1442–1448. [Google Scholar]
- 52.Ulbrich K, Subr V. Adv. Drug Delivery Rev. 2010;62:150–166. doi: 10.1016/j.addr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 53.Kopeček J, Kopečková P. Adv. Drug Delivery Rev. 2010;62:122–149. doi: 10.1016/j.addr.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.