Abstract
Expanded trinucleotide repeats cause Huntington’s disease (HD) and many other neurodegenerative disorders. There are no cures for these devastating illnesses and treatments are urgently needed. Each trinucleotide repeat disorder is the result of the mutation of just one gene, and agents that block expression of the mutant gene offer a promising option for treatment. Therapies that block expression of both mutant and wild-type alleles can have adverse effects, challenging researchers to develop strategies to lower levels of mutant protein while leaving adequate wild-type protein levels. Here, we review approaches that use synthetic nucleic acids to inhibit expression of trinucleotide repeat genes.
Many genes contain multiple consecutive trinucleotides (CGG, CAG, AGG, among others) [1]. Some of these repeats are genetically unstable and can increase in number during DNA replication from one generation to the next. For some genes, these expansions can interfere with normal function and cause disease [2]. There are at least 17 trinucleotide repeat disorders and together they afflict hundreds of thousands of people worldwide. The disorders are typically late onset, with symptoms becoming progressively worse. There are currently no curative treatments for any disease in this class.
These repeats can occur within the coding region of mRNA or within the 3′ or 5′ untranslated regions. The mechanisms causing the diseases differ and loss or gain of function can be involved. The direct trigger can be altered levels of wild-type protein, expression of mutant protein, or synthesis of toxic mutant RNA. In most cases, symptoms are caused by defects in central nervous system (CNS) function, but some diseases also affect muscle or other tissues.
Huntington’s disease (HD) is one of the most intensely studied trinucleotide repeat diseases and a major focus for therapeutic development [3]. It is an inherited neurodegenerative disorder with an incidence of five to ten per 100,000 individuals in most regions [4,5]. The disease is characterized by adult onset with symptoms including chorea, dystonia, and cognitive and psychiatric disturbances. These symptoms worsen progressively until death, 10–20 years after the initial onset of disease. Genetic testing can predict the disease in asymptomatic individuals, and strategies for palliative care exist, but there are no curative therapies to offer patients. Because of the devastating nature of the disease and lack of effective therapy, developing strategies to delay the onset of HD remains a major unmet medical need.
HD is caused by mutations within the gene encoding the huntingtin (HTT) protein [6]. HTT is a large protein with many potential functions that can act as a scaffolding protein. The mutations occur within a region containing the trinucleotide repeat CAG to create expanded trinucleotide repeats. Unaffected individuals will have fewer than 34 repeats in both alleles, with an average repeat length of less than 20. Individuals with more than 37 repeats in one allele are likely to develop HD, with 45 repeats being the average length in patients with HD. CAG encodes glutamine and expanded CAG repeats with HTT mRNA lead to expanded polyglutamine tracts within the HTT protein. Cellular defects caused by mutant HTT include abnormalities or dysfunctions of synapses, neurotrophic factors, apoptosis, altered post-translational modification and defects in protein folding or clearance [7,8].
Gene silencing as a therapeutic approach
Trinucleotide repeat disorders are not the only neurodegenerative diseases that have resisted development of effective treatments. Progress towards curative therapies for Alzheimer’s, Parkinson’s and other diseases has also been slow. The development of therapies for trinucleotide repeat diseases, however, has one advantage: each disease is caused by a defect in a single known gene [2,3]. In theory, agents that reduce the level of the mutant gene should alleviate the disease. Such reduction might be achieved using antisense oligonucleotides (ASOs) or duplex RNAs that target mRNA and inhibit expression of the disease gene. These gene-silencing agents function through mechanisms that are different from those used by small molecule drugs and could offer a strategy for controlling the expression of previously ‘non-drugable’ targets [9,10].
Nucleic acid-based gene silencing has been successfully used in animals. Treatment of mice with virally expressed small hairpin (sh)RNAs reduces mutant human HTT mRNA or protein [11–13]. Although inhibition of HTT expression was not complete, it was sufficient to improve motor coordination and survival. Partial suppression of wild-type HTT expression did not cause observable toxicity, but the full consequences of reduced wild-type HTT levels require further study [12,13]. Administration of synthetic small interfering (si)RNA yields similar results [14]. Several companies have preclinical research programs aimed at developing non-allele selective nucleic acids for HD therapy, including ISIS Pharmaceuticals (http://www.isispharm.com/), Alnylam Pharmaceuticals (http://www.alnylam.com/) and Prosensa (http://www.prosensa.eu/). An advantage of non-allele selective approaches is that only one compound would be needed to treat all patients with a given disease.
Mechanisms for gene silencing by nucleic acids
ASOs and duplex RNAs silence gene expression through different mechanisms [15]. Most ASOs being tested in clinical trials are ‘gapmers’; that is, they contain a DNA ‘gap’ of eight to ten bases flanked by three to five base regions of chemically modified bases. The DNA region forms an RNA–DNA hybrid upon binding an mRNA target; this hybrid can be a substrate for RNase H and leads to degradation of target mRNA. The flanking regions increase resistance to degradation by nucleases and increase affinity for binding to mRNA. If chemically modified bases are spread throughout the ASO, RNase H cannot induce mRNA cleavage. Instead, these ASOs bind mRNA and block progress of the ribosome.
siRNAs are duplex RNAs that, upon introduction into cells, interact with the proteins of the RNA-induced silencing complex (RISC) to recognize mRNA. If the siRNA is fully complementary to the mRNA target, it will induce cleavage of the target. If the siRNA contains centrally located mismatches, it will not induce cleavage and might silence gene expression by a mechanism more similar to steric-blocking ASOs.
Both ASOs and siRNAs are being tested in clinical trials. ASOs require only one nucleic acid strand, which could lead to less expensive drugs and possibly better biodistribution. siRNAs tap into natural pathways for gene silencing and might prove to be more potent. At present, there is no definitive answer for which strategy will be superior in the clinic and it is possible that their different properties will translate into one approach or the other being preferred for treating specific diseases.
The case for allele-selective inhibition
The ability of antisense oligonucleotides and duplex RNAs to slow progression of disease in animal models is encouraging because the data demonstrate that nucleic acids can penetrate and enter affected areas of the brain and that inhibition of mutant HTT expression has the potential to be an effective therapeutic strategy.
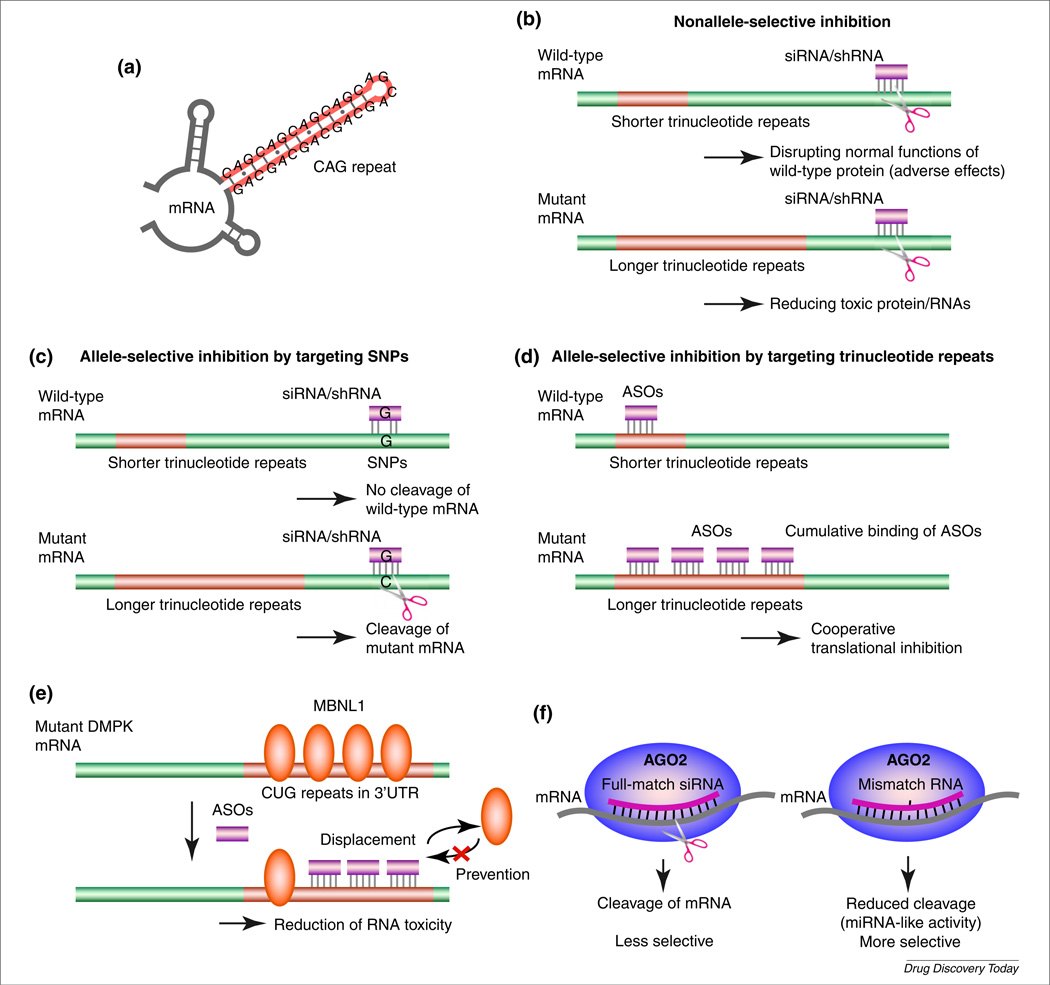
These ASOs and duplex RNAs, however, inhibit expression of mutant and wild-type HTT indiscriminately (Fig. 1b). HTT is known to play an essential role in embryogenesis, neurogenesis and normal adult function [16–22], and it is possible that inhibition of wild-type HTT might have an adverse effect on patients. This concern is amplified by the likelihood that treatment would need to be continued for a prolonged period. Therefore, although nonallele-selective strategies [11–13] might bring great benefits to patients, allele-selective approaches to inhibit expression of mutant variants might prove to be a superior alternative (Table 1).
FIGURE 1.
Mechanism of allele-selective inhibition. (a) Hairpin structure of CAG repeats. (b) Nonallele-selective inhibition of gene expression by small interfering (si) or short hairpin (sh) RNA. (c) Targeting single nucleotide polymorphisms (SNPs) with siRNA and/or shRNA. (d) Targeting expanded trinucleotide repeats with antisense oligonucleotides (ASOs). (e) Targeting expanded CUG repeats in dystrophia myotonica-protein kinase (DMPK) mRNA. (f) Targeting expanded trinucleotide repeats with mismatch-containing RNA. Abbreviations: AGO2, Argonaute 2; MBNL1, muscleblind-like protein 1; UTR, untranslated region.
TABLE 1.
Studies of allele-selective inhibition targeting triplet repeat genes
| Disease | Gene | Target | Oligonucleotides tested | Year | Refs |
|---|---|---|---|---|---|
| HD | HTT | SNPs | siRNA | 2006 | [26] |
| SNPs | siRNA | 2008 | [27] | ||
| SNPs | siRNA | 2009 | [29] | ||
| SNPs | siRNA | 2009 | [30] | ||
| Deletion polymorphism | siRNA | 2009 | [32] | ||
| CAG repeat | Modified LNA, among others | 2010 | [44] | ||
| CAG repeat | Mismatch dsRNA | 2010 | [46] | ||
| CAG repeat | ZNA (oligospermine-conjugated LNA) | 2011 | [60] | ||
| HD, SCA3 | HTT, ATX3 | CAG repeat | PNA, LNA, siRNA | 2009 | [42,49] |
| CAG repeat | Mismatch dsRNA | 2011 | [47] | ||
| SCA3 | ATXN3 | SNPs | siRNA | 2003 | [23] |
| SNPs | siRNA | 2004 | [25] | ||
| SNPs | shRNA | 2008 | [28] | ||
| CAG repeat | PNA, modified LNA, mismatch dsRNA | 2011 | [48] | ||
| SCA7 | ATXN7 | SNPs | shRNA, pri-miRNA mimic | 2009 | [31] |
Nucleic acids that target SNPs and/or deletion polymorphism
One approach to allele-selective silencing of gene expression takes advantage of single nucleotide polymorphisms (SNPs) and other polymorphisms to distinguish mutant and wild-type alleles. By targeting polymorphisms, double-stranded (ds)RNAs can be fully complementary to the mutant allele but have one or more mismatched bases relative to the wild-type allele [23] (Fig. 1c). Depending on the position of the mismatch, activity towards the wild-type allele can be reduced or abolished. Mismatches within base positions 2–8, the seed sequence necessary for efficient RNAi, appear to enhance allele selectivity [24].
Once an allele-specific SNP is identified, several different duplexes are tested with complementarity to different positions surrounding the SNP site to maximize discrimination and potency. Identifying potent and selective compounds requires systematic testing. For example, for some sequences, the mismatch might be in a position that induces little discrimination between wild type and mutant. Small changes in target sequence affect the efficiency of RNAi, so for other target sites, potency might be insufficient. The challenge will be to find a sequence that combines adequate potency and allele selectivity. Several laboratories have demonstrated that duplex RNAs that target SNPs can achieve allele-selective inhibition of trinucleotide repeat genes [23,25–31]. Another research group has reported allele-selective inhibition by targeting a deletion polymorphism associated with the mutant HTT gene [32].
One potential problem for attacking polymorphisms is that there is a limited number of target sites available for discrimination between wild type and mutant. Whereas hundreds of non-allele-selective RNAs can be targeted to different mRNA sequences to identify candidates with optimal potency, the need to be complementary to the SNP site restricts the number of candidates for testing [33]. Only a few duplexes will have the potential for adequate mismatch discrimination [34], further limiting the pool of compounds available for optimizing for tolerability and biodistribution.
Another disadvantage specific for SNP and deletion-targeting approaches is that all patients do not share common SNPs and/or deletions. For example, in a study of 327 patients with HD, it was found that 86% had at least one SNP, but these were found at 26 different sites [30]. Another study estimated that 75% of patients shared five SNPs [29]. Therefore, several drugs would be required to treat most of the population with HD.
Creating several drugs for a single relatively rare disease will be a challenge, but one that might not prove to be as difficult as some might assume. Similar to any drug candidate, duplex RNAs will need to be optimized for potency, biodistribution and tolerable off-target effects. However, all duplex RNAs are chemically similar so the development and clinical testing would not be as arduous as developing chemically unrelated compounds.
Nucleic acids that target expanded trinucleotide repeats
Another strategy exploits a universal difference between mutant and wild-type alleles: the mutant allele has more trinucleotide repeats [35] (Fig. 1d). The existence of additional repeats does not create the type of sequence differences that, as demonstrated for SNPs or deletion polymorphisms, might be exploited for allele-selective recognition. An oligonucleotide that is complementary to the mutant allele will also be complementary to the wild-type allele.
The expanded mutant repeat will, however, offer more binding sites for complementary oligonucleotides. It is also known that CAG repeat sequences can form hairpin structures [36,37]. The longer mutant repeat will form a longer secondary structure, and it is possible that these longer structures will be more conducive for binding to complementary oligonucleotides. It is impossible to predict whether these characteristics would yield selective recognition of a trinucleotide repeat gene inside cells because the intracellular structure of RNA cannot be determined; neither is it known with certainty which proteins associate with repeat RNAs. The hypothesis must be tested empirically by introducing oligomers into cells and assaying selectivity.
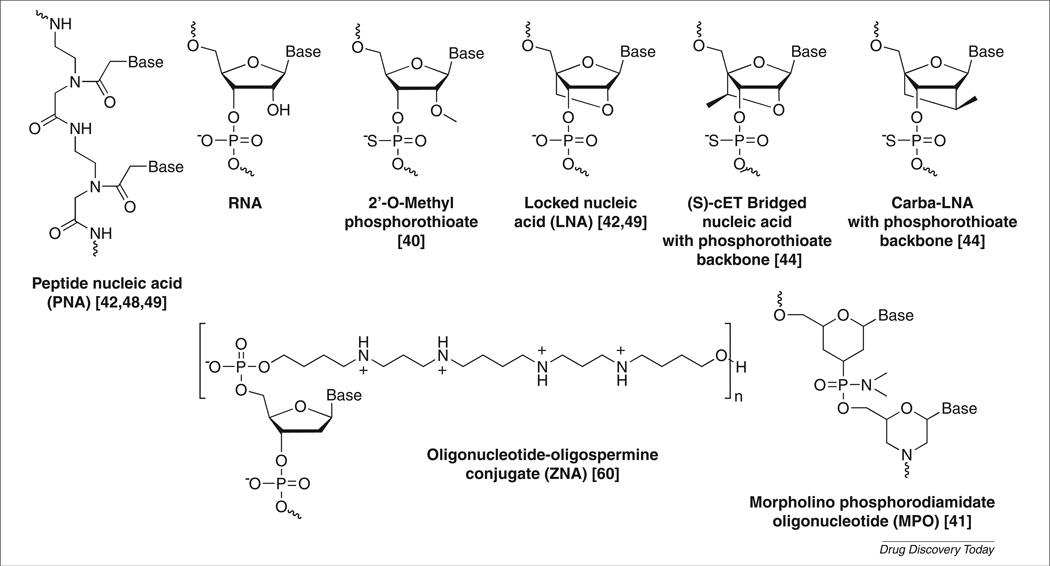
Myotonic dystrophy (dystrophia myotonica) type 1 (DM1) is caused by expansion of a CUG tract within the 3′-untranslated region (UTR) of dystrophia myotonica-protein kinase (DMPK) mRNA. In patients, expansions range from 50 to >2000 repeats [38]. The expanded repeat binds to muscleblind-like protein 1 (MBNL1) and reduces its effective concentration. This blocks the normal function of MBNL1 in modulating splicing. The expanded repeat also causes aberrant activation of protein kinase C (PKC), leading to upregulation of CUG binding protein (CUGBP1), which regulates alternative splicing [39]. Two laboratories found that either phosphorothioate 2′-O-methyl oligonucleotides [40] or morpholino oligomers [41] (Fig. 2) complementary to the CUG repeat reduced formation of toxic RNA–protein aggregates (Fig. 1e). MBNL1 function was restored and splicing defects were corrected in vivo.
FIGURE 2.
Chemical structures of unmodified or modified nucleic acids used for antisense oligonucleotides (ASOs) and/or double-stranded (ds)RNAs targeting trinucleotide repeat genes.
Our laboratory has been testing the repeat-targeting strategy for inhibition of HTT expression. For HTT, the mutation is an expansion of a CAG repeat that exists near the 5′ end of the protein-coding region of HTT mRNA. We first assayed peptide nucleic acid (PNA) (Fig. 2) oligomers complementary to CAG repeats [42]. PNAs are amide-linked oligonucleotide mimics known for their ability to invade nucleic acid structure.
To improve entry into cells, the PNAs were conjugated to the peptide d-Lys8. Because the amino acids are in the unnatural D configuration, it is probable that they will survive inside the cells and participate in target recognition through electrostatic interactions with the phosphodiester backbone of RNA. We observed that PNAs with d-Lys8 at the C-terminus showed 3.5-fold selectivity for inhibition of mutant HTT expression in a HD patient-derived cell line (69/17 repeats). Attaching the peptide to the N rather than C terminus of the PNA improved selectivity to >eightfold, demonstrating that selectivity can be increased by simple chemical modifications.
PNAs are not currently being developed for clinical use, so to facilitate development, we tested analogous oligonucleotides containing locked nucleic acids (LNAs) (Fig. 2) [42]. Introduction of LNA bases into oligonucleotides enhances affinity and allows binding strength to be tailored for a particular application [43]. LNAs are being used in phases I and II clinical trials [15]. Methods for their large-scale synthesis have been optimized and initial trial data suggest that they are well tolerated by patients. We observed potent and allele-selective (>sixfold) inhibition of HTT expression by LNAs and subsequently observed similar promising results with oligonucleotides containing (S)-cEt bridged nucleic acid and carba-LNA (Fig. 2) [44].
dsRNAs are another widely used approach to silencing gene expression. We observed that duplex RNAs that were fully complementary to the CAG repeat showed little or no allele selectivity [42]. We reasoned that this failure might have been because the RNAi was too powerful, overwhelming the differences in repeat length and secondary structure between the mutant and wild-type mRNA targets. To test this hypothesis, we weakened recognition by introducing mismatched bases within the central region of the RNA duplex. These mismatches would be predicted to weaken binding, disrupt cleavage of mRNA by argonaute 2 (AGO2) and shift the mechanism towards that used by miRNAs (Fig. 1f) [45].
The mismatch-containing RNAs were potent and selective (up to >40-fold) inhibitors of gene expression [46]. As many as four mismatches were tolerated without sacrificing potency and several different mismatch designs functioned equally well. The existence of several excellent inhibitors with different sequences provides molecular options for future development. Having diverse options might be important if a sequence being tested in vivo is found to have unacceptable sequence-specific adverse effects. Although most experiments were performed in patient-derived fibroblast cells with 69 repeats in the mutant allele (much higher that the 45 repeat mean), good selectivity was also observed in cell lines with 41 or 44 repeats.
The mechanistic reasons for this high selectivity have yet to be fully investigated, but it is possible that binding of multiple RNAs to the longer mutant repeat without cleavage leads to cooperative effects for translational inhibition. Similar allele selectivity was observed by Krzyzosiak and colleagues using RNA duplexes with mismatches at positions 13 and 16 [47]. Mismatched RNA duplexes targeting the expanded repeat of ataxin-3 were tested in cells derived from patients with Machado Joseph Disease [MJD; spinocerebellar ataxia type 3 (SCA3)] patients and showed >16-fold selectivity, suggesting that the approach can be applied to multiple expanded repeat genes [48].
Targeting expanded trinucleotide repeats has the potential to benefit entire patient populations for multiple diseases. However, there are also several disadvantages. Many genes contain CAG or CUG repeat regions [1] and some of these genes are known to be crucial for normal function. These other genes have relatively small numbers of CAG or CUG repeats and examination of the expression of repeat-containing genes, such as TATA box binding protein (TBP) and forkhead box P2 (FOXP2), has not revealed inhibition [42,46]. Nevertheless, specificity must be observed carefully during development.
As noted above for targeting SNPs, the number of compounds that are candidates for development is more limited than for oligomers that can target any sequence within the mRNA of a disease gene. Another issue is that the degree of allele selectivity of repeat-targeting oligomers depends on differences in repeat length between wild-type and mutant alleles [42,49].
It has also been noted that repeat-targeted RNAs have been reported to have lower potency than duplex RNAs that do not target repeats when comparisons are made between results from different laboratories [33]. This observation raises an important point. Strategies should be compared against one another and, ideally, this should be done directly. However, when values are taken from the literature and compared, the difference in protocols and assay systems should be carefully evaluated. Different transfection methods are used for different types of oligomer, such as PNA, LNA and siRNA, giving different potential for gene silencing. The use of different cell types might also affect potencies. Therefore, relative values should be evaluated with caution unless obtained through side-by-side assays.
In vivo delivery
The delivery of single-stranded antisense oligomers and duplex RNAs to tissues in vivo is widely recognized to be the biggest roadblock to clinical development. Unlike traditional small molecule drugs, which are <700 Da in molecular weight, antisense oligomers and siRNAs are 4000–15,000 Da. All duplex RNAs and most types of antisense oligomer also have charged backbones, which makes them even more different from small molecule drugs.
Despite the obstacles inherent in using large nucleic acids, progress is being made. For delivery to muscle, clinical trials of a systemically administered oligomer designed to alter splicing of dystrophin mRNA in Duchenne Muscular Dystrophy (DMD) has produced promising results in phases I and II clinical trials [50,51] and it is likely that similar progress can be made with oligomers designed to treat myotonic dystrophy.
For compounds designed to act in the CNS, the blood–brain barrier (BBB) presents an additional obstacle. For the foreseeable future, it is likely that compounds will need to be injected directly into the CNS [52–54].1 Given the severity of most triplet expansion diseases, this might be an acceptable mode for administration.
For antisense oligonucleotides, promising results for delivery of anti-HTT oligonucleotides to primate brain and downregulation of HTT expression have been reported at meetings [55] but have not yet appeared in the literature. However, similar molecules have been shown to repress expression of superoxide dismutase 1 (SOD1) in the brain and spinal cord of primates and a phase I clinical trial is ongoing. Results from eight patients after a 12-h intrathecal injection showed that the antisense oligonucleotide was well tolerated with no serious adverse events reported.1 Gene silencing by duplex RNAs has been reported in primate oligodendrocytes, suggesting the potential to treat CNS disorders with duplex RNA [53].
Clinically, it might be necessary to reduce HTT expression over a long period of time and the need for repeated injections would be a significant challenge. One strategy for avoiding the need to administer synthetic agents repeatedly would be virally expressed duplex RNAs, and these have been shown to reduce HTT expression in animals [11–13]. Another strategy would be to increase the ability of agents to penetrate the BBB [56].
Approaches that use duplex RNAs in vivo will also need to confront the fact that RNAi is an endogenous cellular pathway. Synthetic or virally expressed RNAs might disrupt normal function and cause unexpected off-target effects, and silencing approaches should be designed to minimize toxicities [57]. We note that preliminary clinical results are encouraging, with phase I trials of transthyretin (TTR) silencing for treatment of TTR-mediated amyloidosis showing repression of TTR expression with little accompanying toxicity.2
Concluding remarks
Effective treatments for trinucleotide-repeat diseases would have a major impact on the lives of thousands of patients. Developing small molecule drugs has proven difficult, and agents that directly affect expression of the disease gene might be the only option for bringing improved care to patients within the next decade. This realization has led to a growing focus on the properties of antisense oligomers and duplex RNAs (Box 1). A wealth of chemical alternatives exist that can help transform nucleic acid lead compounds into viable therapeutic candidates.
BOX 1.
Nucleic acids as a real therapeutic option
The concept of using nucleic acids as drugs to inhibit expression of disease genes has been around for well over two decades. Those who follow the field have observed repeated disappointments as one clinical trial after another has either not been pursued past phase I or phase II or has had a costly failure in phase III. The most recent example was the final failure of Genesense from Genta (http://www.genta.com/) after multiple phase III trials [58]. Over the past year, there has also been a series of downsizings, as one company after another closes or reduces their operations aimed at producing RNAi therapies. The casual observer would be justified in believing that the field was at a dead end.
There are good reasons for this slow progress. Oligonucleotides and duplex RNAs are unlike traditional small molecule drugs and an entire field of pharmaceutical science needed to be developed before a minimal understanding of rational drug development was achieved. Now, however, two decades of effort has begun to show results. Mipomersen is an antisense oligonucleotide that targets apolipoprotein B expression [59]. Multiple phase III trials have shown that it can be administered systemically and lower low-density lipoprotein (LDL) cholesterol in patients who respond inadequately to existing medications. This progress has important implications for antisense therapeutics in general because the clinical results demonstrate that oligonucleotides: (i) can be synthesized in the large quantities needed for systemic administration to several hundred patients; (ii) although not free of adverse effects, are well tolerated; and (iii) can reach target tissues and inhibit expression of their target genes. In this study, there was no need for a carrier or formulation as the oligonucleotides were administered in sterile water.
As with any drug, the path to the clinic for oligonucleotides will be challenging. Experience with mipomersen suggests that this path can be taken with apparent success.
It is clear that nucleic acids have the capacity to block the expression of disease genes. The primary unanswered question is whether silencing agents can be delivered to affected tissues in humans at concentrations sufficient to lower gene expression and alleviate the disease. Antisense oligonucleotides and siRNAs are potent silencing agents in mouse brains, but can they diffuse sufficiently in the much larger human brain to have an effect? If delivery is possible in humans, will the methods be tolerated by patients? Progress will be judged by the answers to these questions.
Assuming that nucleic acids can be delivered to a target tissue, they will need to reduce expression of the disease gene to a low enough level to produce a therapeutic effect without also producing unacceptable adverse effects. In some or all cases, it might be sufficient to block expression of both the mutant and wild-type alleles. Synthetic agents will always be only partially active, and the remaining wild-type protein might be sufficient to maintain normal function. Alternatively, normal function might not be necessary in the adult, especially if treatment times are measured in weeks or months. If allele-selective inhibition is desired, approaches that target polymorphisms or expanded trinucleotide repeats have both demonstrated that high selectivity and potency can be achieved.
Several excellent lead compounds for intractable diseases, such as HD, myotonic dystrophy and MJD, have been developed. There is no doubt that these strategies have weaknesses, and it is easy to envision reasons why they might fail during development. It is likely that patients and their families would more easily accept such failures than they would rationalizations for why strategies were not pursued to a clear conclusion.
Oligonucleotides and siRNAs are not a traditional approach to drug development. Similar to any other drug candidate, however, chemical modification, redesign and iterative testing and retesting in animals is likely to produce compounds with greater potency, tolerability and biodistribution. The field has been moving forward at an increasingly fast pace. If the coherent testing programs necessary for all serious drug development efforts are implemented and vigorously pursued, there is every reason to expect continued progress.
Acknowledgments
This work was supported by the US National Institutes of Health (NIGMS 73042), the Robert A Welch Foundation (I-1244), the CHDI Foundation, and a McKnight Foundation Neuroscience of Brain Disorders Award.
Footnotes
Miller, T. et al. (2011) Cohort 1 of a Phase 1, double-blind, placebo controlled, dose-escalation study of the safety, tolerability, and pharmacokinetics of ISIS 333611 administered intrathecally to patients with familial ALS due to SOD1 mutations. 2011 Annual Meeting of the American Academy of Neurology, April 9–16, 2011, Honolulu, USA [IN12–1.001].
Sah, D. (2011) Phase I safety, pharmacokinetic, and pharmacodynamic results for ALN-TTR01. VIIIth International Symposium on Familial Amyloidotic Polyneuropathy, November 20–22, 2011, Kumamoto, Japan.
References
- 1.Kozlowski P, et al. Trinucleotide repeats in the human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Morse RJ, et al. Pharmaceutical development for Huntington’s disease. In: Lo DC, Hughes RE, editors. Neurobiology of Huntington’s Disease: Applications to Drug Discovery. 2nd edn. CRC Press; 2011. pp. 197–224. [PubMed] [Google Scholar]
- 4.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 5.Finkbeiner S. Huntington’s disease. Cold Spring Harb. Perspect. Biol. 2011;3:a007476. doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez I, et al. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 8.Ehrnhoefer DE, et al. Convergent pathogenic pathways in Alzheimer’s and Huntington’s diseases: shared targets for drug development. Nat. Rev. Drug Discov. 2011;10:853–867. doi: 10.1038/nrd3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Souza EB, et al. Novel therapeutic modalities to address nondrugable protein interaction targets. Neuropsychopharmacology. 2009;34:142–158. doi: 10.1038/npp.2008.115. [DOI] [PubMed] [Google Scholar]
- 10.Scholefield J, Wood MJA. Therapeutic gene silencing strategies for polyglutamine disorders. Trends Genet. 2009;26:29–38. doi: 10.1016/j.tig.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Harper SQ, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudreau RL, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drouet V, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 14.DiFiglia M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasir J, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 17.Zeitlin S, et al. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 18.White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat. Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 19.Metzler M, et al. Huntingtin is required for normal hematopoiesis. Hum. Mol. Genet. 2000;9:387–394. doi: 10.1093/hmg/9.3.387. [DOI] [PubMed] [Google Scholar]
- 20.Woda JM, et al. Inactivation of the Huntington’s disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo. BMC Dev. Biol. 2005;5:17–28. doi: 10.1186/1471-213X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godin JD, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 22.Huang K, et al. Wild-type HTT modulates the enzymatic activity of the neuronal palmitoyl transferase HIP14. Hum. Mol. Genet. 2011;20:3356–3365. doi: 10.1093/hmg/ddr242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller VM, et al. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi Y, et al. Enhancement of allele-discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS ONE. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. Sequence-dependent and independent inhibition specific for mutant ataxin-3 by small interfering RNA. Ann. Neurol. 2004;56:124–129. doi: 10.1002/ana.20141. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz DS, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Bilsen PHJ, et al. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington’s disease patient-derived fibroblasts. Hum. Gene Ther. 2008;19:710–718. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 28.Alves S, et al. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado Joseph Disease. PLoS ONE. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfister EL, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombardi MS, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Scholefield J, et al. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS ONE. 2009;4:e7232. doi: 10.1371/journal.pone.0007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. Allele-specific silencing of mutant Huntington’s disease gene. J. Neurochem. 2009;108:82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sah DWY, Aronin N. Oligonucleotide therapeutic approaches for Huntingtin Disease. J. Clin. Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, et al. Profiling of mismatch-discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009;37:7560–7569. doi: 10.1093/nar/gkp835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzyzosiak WJ, et al. Triplet repeat RNA structure and is role as a pathogenic agent and therapeutic target. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 37.de Mezer M, et al. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muntoni F, Wood MJ. Targeting RNA to treat neuromuscular disease. Nat. Rev. Drug Discov. 2011;10:621–637. doi: 10.1038/nrd3459. [DOI] [PubMed] [Google Scholar]
- 39.Kuyumcu-Martinez NM, et al. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulders SAM, et al. Triplet-repeat oligonucletide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 44.Gagnon KT, et al. Allele-selective silencing of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, et al. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiszer A, et al. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, et al. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol. Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu J, et al. Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann. N.Y. Acad. Sci. 2009;1175:24–31. doi: 10.1111/j.1749-6632.2009.04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goemans NM, et al. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 51.Cirak S, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, does escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith RA, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Querbes W, et al. Direct CNS delivery of siRNA mediates robust silencing in oligodendrocytes. Oligonucleotides. 2008;19:23–29. doi: 10.1089/oli.2008.0165. [DOI] [PubMed] [Google Scholar]
- 54.Passini MA, et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3001777. 72ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gagnon KT. HD therapeutics – CHDI fifth annual conference. IDrugs. 2010;13:219–223. [PubMed] [Google Scholar]
- 56.Lichota J, et al. Macromolecular drug transport into the brain using targeted therapy. J. Neurochem. 2010;113:1–13. doi: 10.1111/j.1471-4159.2009.06544.x. [DOI] [PubMed] [Google Scholar]
- 57.McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein CA, Goel S. Therapeutic oligonucleotides – the road not taken. Clin. Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-11-2013. [DOI] [PubMed] [Google Scholar]
- 59.Adkim F, et al. Efficacy of apolipoprotein B synthesis inhibition in subjects with mild-to-moderate hyperlipidemia. Eur. Heart J. 2011 doi: 10.1093/eurheartj/ehr148. [DOI] [PubMed] [Google Scholar]
- 60.Gagnon KT, et al. Antisense and antigene inhibition of gene expression by cell-permeable oligonucleotide–oligospermine conjugates. J. Am. Chem. Soc. 2011;133:8404–8407. doi: 10.1021/ja200312y. [DOI] [PMC free article] [PubMed] [Google Scholar]