Abstract
At the time of implantation, the early mouse embryo consists of three distinct cell lineages: the epiblast (EPI), primitive endoderm (PrE), and trophectoderm (TE). Here we will focus on the EPI and PrE cell lineages, which arise within the inner cell mass (ICM) of the blastocyst. Though still poorly understood, our current understanding of the mechanisms underlying this lineage allocation will be discussed. It was originally thought that lineage choice was strictly controlled by the position of a cell within the ICM. However, it is now believed that the EPI and PrE lineages are defined both by their position and by the expression of lineage-specific transcription factors. Interestingly, these lineagespecific transcription factors are initially co-expressed in early ICM cells, suggesting an initial multi-lineage priming state. Thereafter, lineage-specific transcription factors display a mutually exclusive salt-and-pepper distribution that reflects cell specification of the EPI or PrE fates. Later on, lineage segregation and likely commitment are completed with the sequestration of PrE cells to the surface of the ICM, which lies at the blastocyst cavity roof. We discuss recent advances that have focused on elucidating how the salt-and-pepper pattern is established and then resolved within the ICM, leading to the correct apposition of cell lineages in preparation for implantation.
10.1 Preimplantation Development Involves Two Cell Fate Decisions
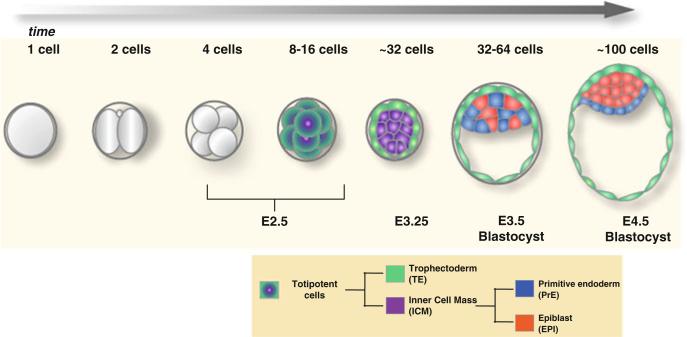
Before implanting into the maternal uterus, the mouse embryo consists of three molecularly distinct spatially segregated cell lineages: the epiblast (EPI) that lies within the interior of the inner cell mass (ICM) of the blastocyst. The EPI is encapsulated by two tissues: the primitive endoderm (PrE) an epithelium located on the surface of the ICM, which lies in contact with the blastocyst cavity; and the trophectoderm (TE) comprising the epithelial surface of the blastocyst, which lies in contact with the external environment (Fig. 10.1) (reviewed by Arnold and Robertson 2009; Nowotschin and Hadjantonakis 2010; Rossant and Tam 2009; Zernicka-Goetz et al. 2009). The EPI is the pluripotent lineage within mammalian embryos and so will give rise to most of the fetus, whereas the TE and PrE predominantly give rise to extraembryonic tissues, namely, the fetal portion of the placenta and the endodermal component of the visceral and parietal yolk sacs, respectively. Thus two cell fate decisions take place before blastocyst formation to ensure the proper specification and spatial segregation of the extraembryonic lineages from the pluripotent epiblast.
Fig. 10.1. Mouse preimplantation development leading to blastocyst formation.
During preimplantation development, the mouse embryo undergoes cleavages that culminate in the proper segregation of three lineages at the blastocyst stage. This process involves two cell fate decisions: the first decision occurs when the inner cell mass (ICM) is segregated from the extraembryonic trophectoderm (TE); the second decision occurs within the ICM and involves the segregation of the pluripotent epiblast (EPI) from the extraembryonic primitive endoderm (PrE)
After fertilization, the embryo undergoes three rounds of cell division, leading to the eight-cell stage. At this time, the blastomeres generally appear morphologically indistinguishable and have the ability to contribute to any of the three lineages of the blastocyst, as has been assessed in chimera experiments (Kelly 1977; Suwinska et al. 2008; Tarkowski and Wroblewska 1967). However, several studies have argued for an existing heterogeneity between the blastomeres at the four- and eight-cell stages, resulting from previous asymmetric cell divisions at the animal part of the oocyte (Gardner 1996). This heterogeneity is evident at the level of differential epigenetic modifications at the four-cell stage, as well as at the level of expression and kinetics of certain transcription factors (e.g., Oct4; discussed later) (Plachta et al. 2011; Torres-Padilla et al. 2007). Moreover, each of these eight blastomeres has acquired an apical-basal polarity as a result of compaction, a process in which cell–cell contacts increase (Johnson and Ziomek 1981). The first fate decision, involving the segregation of the TE lineage from the ICM, takes place after the third embryonic cell division. It relies on a cascade of cell divisions taking place at the 8- to 16- and 16- to 32-cell stage transitions. These divisions can be either symmetric or asymmetric, depending on the orientation of the mitotic spindle with respect to the apical–basal (inside–outside) polarity of the blastomeres. Symmetric divisions generate two daughter cells that remain on the outer surface of the embryo and contribute to TE, whereas asymmetric divisions produce one cell that stays on the outer surface, giving rise to TE and one cell that becomes internalized and contributes to the ICM.
This “inside–outside” model is based on the two rounds of asymmetric divisions; it was first introduced more than 40 years ago and could explain observations from experiments where spatial rearrangements have an effect on cell fate (Tarkowski and Wroblewska 1967). However, more recent studies have challenged the positional model suggesting that it may only provide a mechanism underlying the first fate choice and have thus argued for more determinants to be taken into account. First, acquisition of cell polarity affects cell fate, perhaps earlier than the emergence of inside and outside cells (Jedrusik et al. 2008; Johnson and Ziomek 1981; Plusa et al. 2005). Second, TE and ICM identities are linked to key molecular determinants such as differential Hippo signaling, which depends on the position of the cell and regulates the expression of certain lineage-specific transcription factors. Hippo signaling remains inactive in outer cells, leading to the translocation of the transcriptional cofactor Yap/Taz in the nucleus, which then binds the transcription factor TEAD4 and results in the expression of the TE-specific transcription factor Cdx2; importantly, an additional target gene directed by TEAD4 is Gata3, which encodes for another TE-specific transcription factor (Ralston et al. 2010). Conversely, the Hippo signaling pathway becomes activated in inside cells, promoting phosphorylation and exclusion of Yap/Taz from the nucleus, which results in repression of Cdx2 (Nishioka et al. 2009, 2008; Ralston and Rossant 2008; Strumpf et al. 2005; Yagi et al. 2007). Third, expression of the pluripotency-linked transcription factor Oct4, which has been shown to repress Cdx2 activity, ensures the TE versus ICM segregation (Chambers and Smith 2004; Niwa et al. 2005). To this end, a recent study suggested that seemingly “equivalent” blastomeres at the eight-cell stage display differential Oct4 transcription factor kinetics; those with slow kinetics mostly undergo asymmetric divisions, contributing mostly to ICM, whereas those with fast kinetics divide symmetrically, giving rise to outer TE cells (Plachta et al. 2011). It should however be noted that this study involved the widespread misexpression of a photoactivatable Oct4–GFP fusion protein, which might not fully recapitulate the endogenous behavior of the Oct4 promoter or protein. Therefore, the identity of the two lineages emerging from the first fate decision is linked to the levels of expression as well as the kinetics of lineage-specific transcription factors, which play an equally important role for the second fate decision, as will be discussed extensively in this chapter.
The second fate decision occurs within the ICM and results in the segregation of the extraembryonic PrE lineage from the pluripotent EPI. The segregated PrE lineage acquires an epithelial morphology and lies on the surface of the ICM, contacting the blastocyst cavity. On the other hand, the EPI lineage is located in the interior of the ICM likely having no contact with the blastocyst cavity or the outer environment. In this chapter, we focus on recent findings aimed at elucidating the mechanisms underlying this lineage allocation event within the ICM.
10.2 Challenging the Positional Model for Determining Cell Fate Choice in the ICM: The Role of Lineage-Specific Transcription Factors
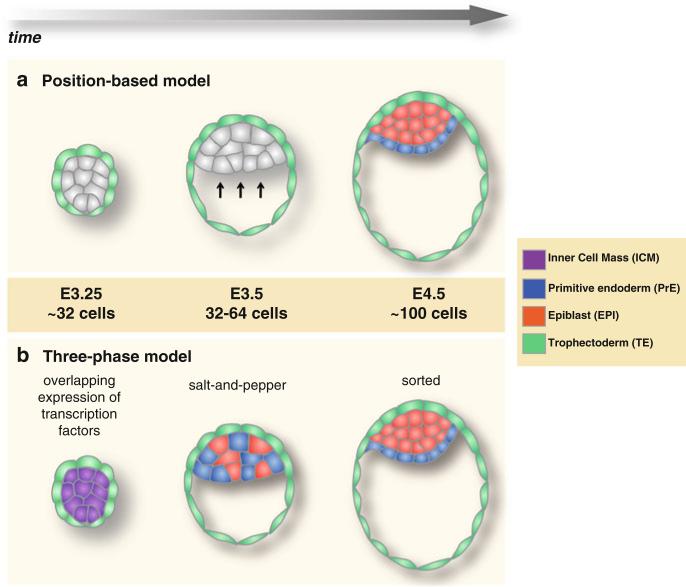
Drawing parallels with the “inside–outside” model underlying the first cell fate decision, it was originally thought that cells within the ICM are segregated in corresponding lineages based on their positions, such that inner cells give rise to EPI, whereas outer ICM cells, which face the blastocyst cavity, give rise to PrE. Moreover, PrE specification was thought to be linked with positional cues acting on the ICM cells at the blastocyst cavity roof (Fig. 10.2) (Enders et al. 1978). This positional model was supported by studies which have demonstrated that during the formation of embryonic bodies from cultured embryonic stem (ES) cells, a PrE layer is generated on the surface (Becker et al. 1992; Martin and Evans 1975; Murray and Edgar 2001). Recently however, this strictly positional-based model has been challenged from several studies over the last years (Chazaud et al. 2006; Plusa et al. 2008). These studies have shown that, cells within the ICM express certain EPI-specific or PrE-specific transcription factors. Interestingly, these transcription factors start to be expressed early, at around the 32-cell stage, irrespectively of the position of cells within the ICM, perhaps suggesting a multi-lineage priming state (Fig. 10.2). As the two nascent lineages emerge, PrE cells begin to exclusively express transcription factors such as Gata4, Gata6, Sox7, and Sox17, whereas EPI cells express pluripotency-associated factors such as Nanog, Sox2, and Oct4.
Fig. 10.2. Models for lineage allocation within the ICM.
(a) In the positional-based model, all ICM cells are morphologically and molecularly equal, being able to contribute to either EPI or PrE. Lineage allocation is strictly controlled by cell position: cells positioned in the interior of the ICM give rise to EPI, whereas cells that underlie the blastocyst cavity form PrE, possibly as a result of positional induction. (b) In the three-phase-based model, lineage allocation is controlled by the sequential activation of key transcription factors as well as dynamic cell movements within the ICM. Initially, EPI- and PrE-specific transcription factors are co-expressed in all cells of the ICM. That is followed by a mutually exclusively expression pattern, where cells expressing EPI-or PrE-specific transcription factors are distributed in a salt-and-pepper fashion irrespectively of their position within the ICM. Finally, randomly positioned PrE precursor cells will sort to the surface of the ICM ensuring the spatial segregation of the EPI and PrE lineages before implantation
Gata4 and Gata6 are two transcription factors of the Gata family, which are expressed in the PrE. Gata6 is one of the first lineage-specific transcription factors to be observed, being expressed at around the 16- to 32-cell stage, in an overlapping manner with the expression of the EPI-specific factor Nanog. Gata4 is expressed later, at around the 64-cell stage, when the “salt-and-pepper” distribution of transcription factors is evident and thus, when cells are more likely fated to form PrE. Mouse mutant embryos for either Gata4 or Gata6 usually form PrE and die at postimplantation stages and, in the case of Gata6 mutants, they exhibit defects in the visceral endoderm, a tissue derivative of PrE (Koutsourakis et al. 1999; Kuo et al. 1997; Molkentin et al. 1997; Morrisey et al. 1998). Therefore, PrE formation is not compromised in either single mutant embryos, perhaps hinting at a functional redundancy between these two transcription factors. One might predict that a double Gata4; Gata6 mutant might exhibit a phenotype in PrE formation. Gata6-expressing cells also co-express the transcription factor Sox17, which is required for the maintenance of the PrE lineage (Artus et al. 2011; Niakan et al. 2010). Another transcription factor, Sox7, is expressed at the final stages of preimplantation development, when PrE cells are spatially segregated on the blastocyst cavity roof (Artus et al. 2011). This observation reveals that PrE cells positioned on the cavity are molecularly distinct with those that have yet to sort to their final position. In addition to these PrE-specific transcription factors, other PrE markers have been identified. For example, Pdgfrα is a marker of PrE cells, and a Pdgfrα::H2B-GFP reporter has been used as a tool for single-cell resolution live imaging experiments (Plusa et al. 2008).
The EPI-specific factors Oct4 and Nanog start being expressed in all cells at the eight-cell stage, and at around the 32-cell stage, they are co-expressed in all cells along with the PrE-specific Gata6. Nanog persists within the ICM, and thereafter becomes exclusive in EPI cells until the late blastocyst stage when its expression declines and later becomes re-established in the germline (Chambers et al. 2003; Mitsui et al. 2003). Nanog has been shown to be required for the maintenance of pluripotency in mouse ES cells, as well as in the ICM (Chambers et al. 2007). Interestingly, Nanog expression is also required for the maintenance of the PrE lineage (Messerschmidt and Kemler 2010). Along with Nanog, Oct4 is another key transcriptional regulator, expressed by pluripotent cells (Chambers and Smith 2004). Oct4 is a POU-domain transcription factor that is co-expressed with Cdx2 in all blastomeres of the early embryo. Oct4 regulates the activity of Cdx2, thus promoting ICM over TE fate during the first fate decision (Niwa et al. 2005). Moreover, Oct4 mutant embryos do not form an ICM and inner blastomeres acquire a TE identity (Nichols et al. 1998). As mentioned previously, the kinetics of Oct4 were suggested to play a significant role during the first cell fate choice involving cells dividing symmetrically or asymmetrically and thus contributing to TE or ICM, respectively (Plachta et al. 2011). As with Nanog, Oct4 expression becomes restricted within the ICM and is enriched in EPI cells; however, unlike Nanog, low levels of expression Oct4 are observed in PrE cells, even at the late blastocyst stage. Oct4 does eventually become restricted and is expressed specifically in the germline (Palmieri et al. 1994). Another early ICM-specific, and thereafter EPI-specific, transcription factor is Sox2. A recent study showed that, along with Oct4, Sox2 is one of the first markers of the emerging ICM. However unlike Oct4 and Nanog, Sox2 is not expressed by all blastomeres early on, instead it was shown to be specifically upregulated in cells that internalize first during the divisions occurring between the 8- to 16-cell stages (Guo et al. 2010). Later on, Sox2 is downregulated in ICM cells that contribute to PrE but remains expressed in TE cells (as opposed to Oct4). Disruption of Sox2 results in preimplantation lethality, emphasizing its importance on the formation and maintenance of multipotent cell lineages (Avilion et al. 2003).
It is now widely accepted that the EPI versus PrE lineage decision is likely to be linked to the expression of lineage-specific transcription factors rather than strictly determined by cell position alone. Several studies, supported by experimental data obtained by live embryo imaging, have indicated that lineage allocation within the ICM involves three distinct, but successive phases involving: (1) initial co-expression of lineage-specific transcription factors (at around the 32-cell stage), (2) subsequent mutually-exclusive expression and salt-and-pepper distribution of EPI- and PrE-precursor cells (at around the 64-cell stage), and (3) finally dynamic cell movements leading to the final sorting and spatial segregation of the EPI and PrE cell lineages (at around the 100-cell stage) (Fig. 10.2) (Chazaud et al. 2006; Meilhac et al. 2009; Plusa et al. 2008). Therefore, cells within the ICM initiate expression of lineage-specific transcription factors prior to the formation of the blastocyst cavity, suggesting that initially they may acquire a state of multi-lineage priming, and thereafter develop a propensity to form the EPI or PrE lineage before the induction of any positional cues. An important step during this process occurs when transcription factors transition their expression from a homogeneous to a mutually-exclusive pattern, where cells expressing PrE-specific factors cease to express EPI-specific ones and vice versa, resulting in the emergence of a salt-and-pepper distribution of nascent PrE and EPI progenitors.
During the next sections, we will focus on how this mutually exclusive pattern, which marks the first point on EPI versus PrE cell fate decision, takes place and is acquired within the ICM cell population. Specifically, recent studies have aimed at answering the following questions: Is the establishment of a salt-and-pepper distribution influenced by signaling cues? Do ICM cells acquire this pattern of expression randomly, or is there a lineage bias that influences the subsequent cell fate decision?
10.3 Salt-and-Pepper Expression of Lineage-Specific Transcription Factors
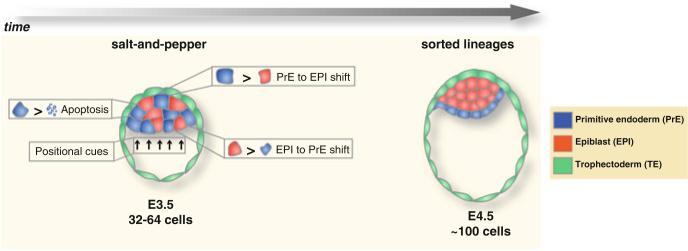
At around the morula stage (16–32 cells), the EPI-specific transcription factor Nanog starts being expressed homogeneously within inner blastomeres and is excluded from outer cells (Chazaud et al. 2006; Plusa et al. 2008). A similar pattern of expression has also been observed for the earliest observed PrE-specific transcription factor, Gata6. This overlapping expression is evident until the 64-cell stage. By this stage, the EPI and PrE markers are being expressed in a nonoverlap-ping mutually exclusive manner, which is also referred to as a salt-and-pepper distribution (Chazaud et al. 2006; Plusa et al. 2008). By using the PrE-specific marker Pdgfrα, live imaging experiments in embryos carrying a Pdgfrα::H2B-GFP knock-in reporter, comprising a human histone H2B fusion to GFP targeted to the Pdgfrα locus (Hadjantonakis and Papaioannou 2004), have confirmed that Gata6-positive PrE precursors do not express Nanog and are positioned randomly within the ICM in a salt-and-pepper distribution (Plusa et al. 2008). At the 64- to 100-cell stage transition, the majority of GFP-positive cells not already residing on the cavity roof move toward the surface of the ICM, subsequently becoming committed to the PrE. A minor population of internal GFP-positive cells will likely either downregulate the Pdgfrα::H2B-GFP reporter or apoptose, and so be eliminated from the embryo. In this way, the salt-and-pepper distribution precedes the formation of the PrE epithelium, suggesting that the specification of EPI and PrE precursor cells takes place before the subsequent cell sorting and epithelialization of the PrE lineage, which in turn depends on positional cues relative to the blastocyst cavity.
The salt-and-pepper distribution marks the first step of PrE versus EPI lineage allocation within the ICM. Cells expressing PrE but not EPI markers are fated to contribute to the PrE lineage rather than the EPI during normal development (Chazaud et al. 2006). It is however debatable whether these cells though committed to a specific lineage may exhibit plasticity dependent on context. This raises the question of whether cells possess a less restricted developmental potential, not necessarily exclusively reflecting the lineage for which they exhibit marker-specific expression. Recent experiments support such a hypothesis, indicating that, at the salt-and-pepper stage, individual ICM cells, and in particular those of the PrE, exhibit greater plasticity than generally appreciated when isolated from embryos and reintroduced into chimeras, and that this plasticity is lost once the cells have sorted to their respective tissue layers and the PrE begins to epithelialize (Grabarek et al. 2012).
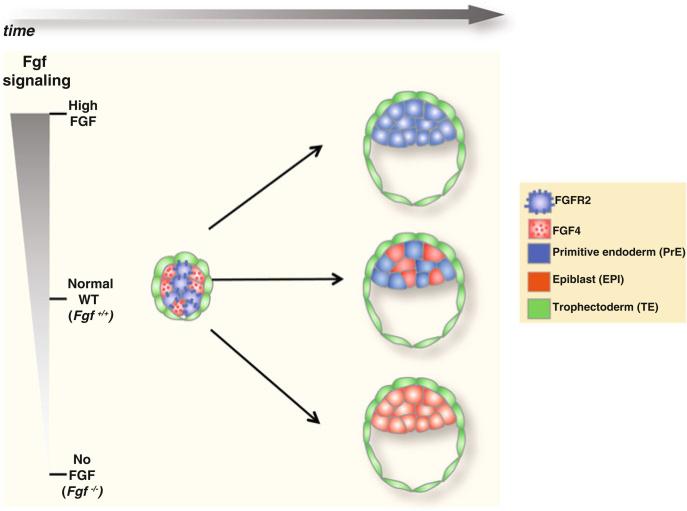
Lineage allocation and the developmental potential within the ICM have been shown to be influenced by FGF/MAPK signaling (Fig. 10.3) (reviewed in Lanner and Rossant 2010). Activation of FGF/MAPK signaling induces the expression of PrE-specific transcription factors, such as Gata6 (Li et al. 2004; Morrisey et al. 1998). The analysis of mouse mutants has revealed that the ligand Fgf4, the receptor Fgfr2 and the adaptor protein Grb2, which mediate FGF/MAPK signaling, are all required for PrE formation (Arman et al. 1998; Cheng et al. 1998; Feldman et al. 1995; Wilder et al. 1997). Perturbations in FGF/MAPK signaling greatly influence the balance between the EPI and PrE lineages in the ICM: excess of Fgf4 converts all ICM cells to adopt a PrE identity (Yamanaka et al. 2010), whereas in Fgf4 mutants, all ICM cells become Nanog positive and PrE is not formed (Piliszek A, MK and AKH unpublished observations). A comparable defect is observed in embryos lacking the adaptor protein Grb2 (Chazaud et al. 2006). In support of these observations on mutants with perturbations in Fgf signaling, chemical inhibition of FGF signaling results in all ICM cells adopting an EPI fate (Nichols et al. 2009). Therefore, it has been proposed that cells expressing Fgfr2 and thus receiving signal through binding of the Fgf4 ligand are fated to form PrE, whereas cells that do not express Fgfr2 but instead secrete Fgf4 will be EPI precursors.
Fig. 10.3. Lineage allocation in the ICM depends on FGF/MAPK signaling.
At around the 32-cell stage, when EPI- and PrE-specific transcription factors are expressed homogeneously in all ICM cells, a differential expression of the ligand Fgf4 and the receptor Fgfr2 starts to become evident. EPI precursor cells express and secrete Fgf4, whereas PrE precursor cells express Fgfr2. Binding of Fgf4 to Fgfr2 activates the FGF/MAPK signaling pathway, which is required for the induction of PrE-specific transcription factors, thus promoting PrE formation. When the signaling is not active, such as in Fgf4 mutants or by inhibiting the receptor Fgfr2, PrE is not formed and all ICM cells contribute to EPI. On the contrary, in the presence of excess Fgf4, all ICM cells contribute to PrE
One interesting question pertains the stage at which this differential FGF signaling takes place with respect to the expression of EPI- and PrE-specific transcription factors. A recent single-cell expression analysis study showed that, at around the 32-cell stage, there is a reciprocal Fgf4/Fgfr2 ratio of expression in the ICM cells with approximately half cells predominantly expressing the ligand and the other half expressing the receptor (Fig. 10.3) (Guo et al. 2010). Interestingly, this reciprocal ligand/receptor expression is evident prior to the emergence of the salt-and-pepper distribution of transcription factors, indicating that FGF/MAPK signaling might act upstream of the differential expression of lineage-specific transcription factors. Consistent with this hypothesis, in Fgf4 mutant embryos, even though the initial phase of transcription factor co-expression is observed, the salt-and-pepper distribution is not established and all ICM cells shift to the EPI lineage (Piliszek A, MK and AKH unpublished observations). This suggests that FGF signaling is not required for the initial expression of lineage-specific transcription factors, although it does regulate the salt-and-pepper patterning and thus cell fate choice.
It is tempting to speculate how the heterogeneity in FGF signaling might be established prior to the emergence of a salt-and-pepper distribution of lineage-fated precursors. First, might it be possible that the initial homogeneous expression of factors such as Nanog, Oct4 and Gata6 play a role? It is worth mentioning that Fgf4 is produced in the ICM under the control of the early ICM factors Oct4 and Sox2 (Yuan et al. 1995), suggesting that an initial expression of these factors is needed for the ligand to be expressed and secreted. Second, could this heterogeneity in Fgf4/Fgfr2 expression be stochastic? This hypothesis fits well with the heterogeneity observed in ES populations, which exhibit stochastic fluctuations in the levels of pluripotency-linked transcription factors, such as Nanog (Chambers et al. 2007; Kalmar et al. 2009; Singh et al. 2007). Third, might it be possible that FGF signaling is influenced by cues from other signaling pathways? As mentioned earlier, the Hippo signaling pathway plays a role in the first (TE versus ICM) cell fate decision, by converting cell-density signals into cell growth control and gene activity (Nishioka et al. 2009; Yagi et al. 2007). In low-density/outside cells, the transcriptional cofactor Yap/Taz mediates the Hippo signaling pathway by its translocation to the nucleus, which results in the establishment of Tead4 activity. Tead4 will then direct the expression of Cdx2, which is the first lineage-specific transcription factor marking the TE. By contrast, in high density/inside cells, Yap/Taz remains in the cytoplasm, and thus Cdx2 is not expressed. Therefore, Oct4 is not downregulated and these cells adopt an ICM identity. Interestingly, recent studies have connected the Hippo signaling pathway with the TGF-b/Smad activity (Varelas et al. 2010). Considering the significant degree of cross-talk between signaling pathways during late development (Guo and Wang 2009), it is tempting to speculate that these pathways might influence the initial ratio of expression of Fgf4/Fgfr2 in early ICM cells. Furthermore, if existing, such a link would connect the first and second cell fate decisions, a hypothesis that has been put forward over the last years (Bruce and Zernicka-Goetz 2010). Finally, does the heterogeneity in FGF signaling reflect a lineage bias? An interesting observation was that cells that are internalized early (between the 8- and 16-cell stages) were shown to express Sox2 (Guo et al. 2010). Since Sox2 directs the expression of the ligand Fgf4 in these cells (Chen et al. 2008; Yuan et al. 1995), this might then signal to Fgfr2-expressing cells that are internalized later setting up a lineage bias earlier than believed (i.e., in the transition between the 16- and 32-cell stages). Therefore, at around the 32-cell stage, the reciprocal Fgf4/Fgfr2 pattern of expression starts to become evident. Consistent with these observations, cells that are internalized first are more likely to contribute to EPI, whereas cells that are internalized later are fated to become PrE. This attractive “time inside–time outside” model was recently put forward by Zernicka-Goetz and colleagues and will be discussed in the next section in greater detail (Morris et al. 2010). It is worth mentioning that, although this model may explain the upregulation of Fgf4 in the early-internalized cells, the mechanisms underlying its downregulation as well as the upregulation of Fgfr2 in the latter internalized cells remain unknown.
10.4 Two Models for the Establishment of the “Salt-and-Pepper” Pattern of Expression in the ICM
EPI and PrE lineage-restricted precursors are first identified at the 64-cell stage, when EPI- and PrE-specific transcription factors, such as Nanog and Gata6, respectively, are observed in a mutually exclusive pattern. As mentioned previously, the establishment of this salt-and-pepper distribution of lineage progenitors greatly depends on signaling cues, in particular the FGF/MAPK pathway. Nevertheless, an interesting observation is that nascent EPI and PrE cells appear to be positioned in a salt-and-pepper distribution within the ICM at this stage. Recent studies have attempted to elucidate how this distribution emerges: does it occur in a random way, or is it biased by the developmental history of a cell? Two models have been put forward with studies providing support for both the random-based as well as the lineage bias-based models (Fig. 10.4).
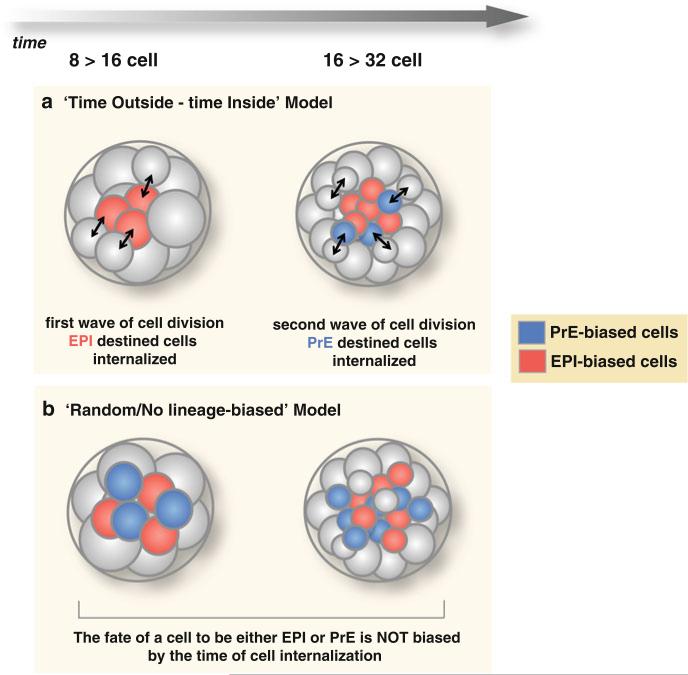
Fig. 10.4. Two models for the establishment of the salt-and-pepper distribution of cells in the ICM.
The two “waves” of asymmetric cell divisions, which take place at the 8- to 16-cell and 16- to 32-cell stages, generate the inner cells of the ICM. (a) In the “time-outside/time-inside” model, cells that are internalized with the first wave are biased to form EPI, whereas cells that are internalized with the second wave are biased to form PrE. (b) In the “random/no lineage-biased” model, cells could contribute to EPI or PrE irrespectively of whether they have internalized during the first or the second wave
Two “waves” of asymmetric cell divisions (8- to 16-cell and 16- to 32-cell stages) generate the inner cells that contribute to the ICM (Chazaud et al. 2006; Chisholm and Houliston 1987). It has been proposed that the history of a cell, namely whether it was internalized during the first or the second wave of cell divisions, could influence its propensity to contribute to EPI or PrE. Experimental support for this hypothesis (Morris et al. 2010) was reported through experiments which used noninvasive lineage tracing of cells from the eight-cell to late blastocyst stage. This study demonstrated that cells internalized during the first wave exhibited a greater bias toward the EPI, whereas cells internalized with the second were biased to form PrE. Moreover, the few cells that are internalized during the third wave of asymmetric divisions (32- to 64-cell stage) invariably contribute to PrE. Contributions to EPI or PrE were scored based on the morphology of cells by the late blastocyst stage. Based on these observations, Morris and colleagues put forward the “time-outside/time-inside model,” suggesting that the greater the time an ICM cell spends on the outside of an embryo the greater its propensity to differentiate, whereas with more time spent inside, their pluripotency is preserved and thus, they contribute to EPI.
The “time-outside/time-inside” model invoking a cellular memory was challenged by the findings of another study that showed that the waves of asymmetric divisions generate EPI- and PrE-precursors in an apparently random fashion, irrespectively of whether the cells were internalized early or late (Yamanaka et al. 2010). Yamanaka et al. injected blastomeres at the eight-cell stage with mRNA for nuclear and membrane markers, which allowed them to monitor whether inner cells were generated during the first or the second wave of asymmetric divisions. The fate of these cells in postimplantation stages (i.e., contribution to either established EPI or PrE-derived tissues) was then assayed after uterine transfer to pseudo-pregnant females. In contrast to Morris and colleagues, Yamanaka et al. reported that cells internalized from either the first or the second wave contributed to either EPI or PrE without being biased to form a certain lineage.
Recently, an informed debate has taken place over the conclusions drawn from different studies (Bruce and Zernicka-Goetz 2010). It has been suggested that the discrepancies between studies, may be due to technical differences, for example different methods have been used for lineage tracing. However, an absolute correlation between lineage bias and timing of internalization is not evident. Morris et al. reported that 25 % of cells internalized with the first wave (i.e., cells biased to form EPI) in fact contribute to PrE, whereas a 15 % of cells internalized with the second wave (i.e., cells biased to form PrE) can instead contribute to EPI (Morris et al. 2010). Therefore, even though a lineage bias could certainly be a factor, the cells of the early ICM seem to exhibit some developmental plasticity, and additional mechanisms are likely required for their final commitment to a specific lineage. Most notably, as mentioned previously, FGF/MAPK signaling influences cell fate choice within the ICM. The study of Yamanaka and colleagues lent further support for the critical role of FGF signaling by showing that inhibition of FGF/MAPK signaling results in all ICM cells shifting to a Nanog-positive EPI fate. Conversely, removal of the FGF/MAPK inhibitor resulted in the restoration of the PrE lineage, indicating that the ICM cells retain a highly dynamic and plastic state. Interestingly, the effects of FGF signaling inhibition could be observed after the salt-and-pepper pattern of expression, suggesting that the developmental plasticity in the ICM is evident even after that stage. Nevertheless, the plasticity was progressively lost after E4.0, indicating that by the late blastocyst stage, cells have committed to a certain lineage (Yamanaka et al. 2010). Therefore, commitment likely becomes fixed as, or soon after, cells have undergone their final sorting process (discussed in the next section). Consistent with these observations, recent experimental data have further shown that lineage plasticity is retained at the salt-and-pepper stage and is lost much later, just before implantation (Grabarek et al. 2012). Interestingly, this stage coincides with when Oct4 has begun to be excluded from cells of the PrE lineage after they have sorted (Grabarek et al. 2012).
10.5 The Final Cell Sorting and Spatial Segregation
After the salt-and-pepper distribution at around the 64-cell stage, dynamic cell movements and rearrangements occur within the ICM and lead to the spatial segregation (aka “sorting”) of the PrE precursor cells toward the surface of the ICM, facing the blastocyst cavity (Fig. 10.5) (Gerbe et al. 2008; Meilhac et al. 2009). By using a PrE-specific GFP reporter for expression of Pdgfrα, live imaging experiments have unraveled the cell behaviors that take place during the sorting process (Plusa et al. 2008): (1) GFP-positive cells (i.e., PrE precursors) that are positioned at an inner location in the ICM, tend to move toward the surface of the cavity. These cells later contribute to form the established PrE epithelial layer. However, some other GFP-positive cells, located at similar inner positions, do not move close to the cavity and instead undergo apoptosis. Interestingly, a substantial number of these cells have also been shown to evade the apoptotic pathway downregulating GFP expression and convert to an EPI fate. (2) GFP-positive cells that are located close to the surface of the cavity already from an early stage rarely move. These observations suggest that a positional signal might be important to retain these cells to their location. (3) Some GFP-negative cells (i.e., EPI precursors) that lie next to the cavity have been shown to upregulate GFP expression and eventually contribute to PrE. Therefore, at this case, cells have shifted their fate from EPI to PrE, possibly due to positional cues originating at the surface of the cavity.
Fig. 10.5. Cell sorting ensures the spatial segregation of PrE and EPI.
After the salt-and-pepper distribution of cells within the ICM, dynamic cell rearrangements take place, involving the movement of EPI and PrE precursor cells to inner locations of the ICM and surface of the blastocyst cavity, respectively. Some PrE precursor cells that are located deep into the ICM may undergo apoptosis or convert to an EPI fate. On the contrary, some EPI precursor cells that lie next to the blastocyst cavity might convert to a PrE identity. Moreover, positional signals from the blastocyst cavity (such as Wnt9α) might induce the migration of PrE precursor cells toward the cavity and the maintenance of PrE identity in these cells
Based on the studies described above, it is becoming widely accepted that dynamic cell rearrangements, involving actin-dependent cell movements and/or apoptosis, as well as possible PrE-to-EPI or EPI-to-PrE cell lineage conversions occur during the final sorting. After sorting, cells are committed to their prospective lineages, confirmed by their inability to contribute to chimeras. Indeed, the sorted PrE cells are morphologically and molecularly distinct from the unsorted PrE precursor cells. The sorting cells gradually polarize as they move next to the surface of the blastocyst cavity, showing a characteristic epithelial morphology (Gerbe et al. 2008); moreover, they start to express the transcription factor Sox7, which is an exclusive marker for these PrE-committed cells (Artus et al. 2011). Activation of Sox7 in these cells (as well as exclusion of Oct4) marks the point where developmental plasticity has been lost and commitment to PrE has occurred (Grabarek et al. 2012).
The mechanisms underlying the final cell sorting have not yet been fully elucidated. The fact that sorted cells are polarized is critical. Indeed, important markers were shown to localize in the apical surface (positioned adjacent to the blastocyst cavity) of the sorted cells (Gerbe et al. 2008). One of them, the low-density lipoprotein receptor-related protein Lrp2, is expressed by PrE-precursor cells at around the salt-and-pepper stage and then localizes specifically at the apical surface of the sorted PrE-committed cells (Gerbe et al. 2008). The cargo protein adaptor Dab2 also shows a distinct localization on the apical surface of the sorted cells (Yang et al. 2002). Dab2 binds to Lrp2 and it has been suggested that Dab2 recruits Lrp2 to the apical surface after sorting. Interestingly, disruption of Dab2 results in preimplantation embryos in which PrE precursor cells specified within the ICM are unable to sort to their final position adjacent to the cavity (Yang et al. 2002). However, the mechanisms through which Dab2 facilitates sorting remain unknown. It is possible that it could play a critical role in transporting protein(s) important for the movement of these cells to the surface of the ICM. To this end, mutation of the Dab2 interaction partner integrin β1 leads to PrE formation failure, suggesting that cell adhesion changes, mediated by Dab2 and its integrin partners, play a significant role in the sorting process (Fassler and Meyer 1995; Stephens et al. 1995). Overall, the apical localization of several markers in the sorted cells and the gradual polarization of these cells seem to be essential for the sorting process.
Signaling cues might also be important for the final sorting step. A possible candidate could be the PDGF signaling. As mentioned previously, Pdgfrα is one of the first markers to be expressed in the PrE precursor cells (along with Gata6) and PDGF signaling plays a crucial role for the expansion of PrE-committed cells in late blastocysts (Artus et al. 2010). Nevertheless, the sorting process in Pdgfrα mutant embryos seems to occur normally, even though the PrE epithelium consists of fewer cells compared to wild-type blastocysts (Artus et al. 2010). However, positional-induced signaling cues seem to have a more direct effect on the sorting process. To this end, Wnt9α, which is expressed on the surface of the blastocyst, was shown to facilitate repositioning of the Gata6-expressing cells (Meilhac et al. 2009). In conclusion, it is becoming widely accepted that the sorting process does not simply involve cell movement governed by the expression of lineage-specific transcription factors. Instead, several other parameters, such as gradual polarization, changes in cell adhesion properties, and signaling cues induced by position, all likely have a cumulative effect on this final sorting step, which culminates in lineage commitment and tissue segregation.
10.6 Making a Compromise: Is Lineage Allocation in the ICM a Stochastic Process with a Lineage Bias?
Many aspects regarding lineage allocation in the ICM remain puzzling. However, seemingly conflicting reports might actually be revealing complementary mechanisms governing the EPI versus PrE fate choice. The initial EPI- and PrE-specification at the onset of the salt-and-pepper distribution noticeably occurs in a spatially disorganized manner. This observation could point to a stochastic process underlying the second fate decision. Indeed, the stem cell population of the ICM might exhibit a similar dynamically heterogeneous state as their in vitro counterparts (Chambers et al. 2007; Kalmar et al. 2009; Singh et al. 2007). Moreover, heterogeneities in the levels of Cdx2 and Nanog expression have been observed at even earlier stages in the embryo, indicating that stochastic processes play a significant role during the first ICM versus TE fate decision as well (Dietrich and Hiiragi 2007). However, amongst this apparent randomness, the fate of an individual ICM cell could be biased; to this end, stochastic does not necessarily mean unbiased (Zernicka-Goetz and Huang 2010). Indeed, if lineage allocation were solely based on random and unbiased mechanisms, its phenomenally certain outcome would have been unpredictable. Moreover, the final cell sorting is clearly influenced by deterministic cues, such as positional signaling originating from the cavity surface, which stabilizes lineage commitment. Therefore, the early embryo provides an experimental model in order to investigate in vivo the crosstalk between stochastic and deterministic processes and how intercellular signaling pathways, such as the FGF pathway, influence these processes. Future experiments will likely elucidate the order of these events, and the mechanisms driving them during embryogenesis, which ultimately give rise to the remarkably structured blastocyst stage embryo.
Acknowledgements
We thank Jerome Artus, Silvia Munoz-Descalzo and Marilena Papaioannou for discussions and comments on this review. Work in our laboratory is supported by the HFSP, NIH, and NYSTEM.
References
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Artus J, Panthier JJ, Hadjantonakis AK. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 2010;137:3361–3372. doi: 10.1242/dev.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol. 2011;350:393–404. doi: 10.1016/j.ydbio.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Casanova J, Grabel L. Localization of endoderm-specific mRNAs in differentiating F9 embryoid bodies. Mech Dev. 1992;37:3–12. doi: 10.1016/0925-4773(92)90010-h. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Zernicka-Goetz M. Developmental control of the early mammalian embryo: competition among heterogeneous cells that biases cell fate. Curr Opin Genet Dev. 2010;20:485–491. doi: 10.1016/j.gde.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, Tortorice CG, Cardiff RD, Cross JC, Muller WJ, Pawson T. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95:793–803. doi: 10.1016/s0092-8674(00)81702-x. [DOI] [PubMed] [Google Scholar]
- Chisholm JC, Houliston E. Cytokeratin filament assembly in the preimplantation mouse embryo. Development. 1987;101:565–582. doi: 10.1242/dev.101.3.565. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Enders AC, Given RL, Schlafke S. Differentiation and migration of endoderm in the rat and mouse at implantation. Anat Rec. 1978;190:65–77. doi: 10.1002/ar.1091900107. [DOI] [PubMed] [Google Scholar]
- Fassler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Can developmentally significant spatial patterning of the egg be discounted in mammals? Hum Reprod Update. 1996;2:3–27. doi: 10.1093/humupd/2.1.3. [DOI] [PubMed] [Google Scholar]
- Gerbe F, Cox B, Rossant J, Chazaud C. Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst. Dev Biol. 2008;313:594–602. doi: 10.1016/j.ydbio.2007.10.048. [DOI] [PubMed] [Google Scholar]
- Grabarek JB, Żyżyńska K, Saiz N, Piliszek A, Frankenberg S, Nichols J, Hadjantonakis AK, Plusa B. Differential plasticity of epiblast and primitive endoderm precursors within the ICM of the early mouse embryo. Development. 2012;139:129–139. doi: 10.1242/dev.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Papaioannou VE. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J Exp Zool. 1977;200:365–376. doi: 10.1002/jez.1402000307. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Li L, Arman E, Ekblom P, Edgar D, Murray P, Lonai P. Distinct GATA6- and laminin-dependent mechanisms regulate endodermal and ectodermal embryonic stem cell fates. Development. 2004;131:5277–5286. doi: 10.1242/dev.01415. [DOI] [PubMed] [Google Scholar]
- Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc Natl Acad Sci USA. 1975;72:1441–1445. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec JF, Zernicka-Goetz M. Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Dev Biol. 2009;331:210–221. doi: 10.1016/j.ydbio.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Kemler R. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol. 2010;344:129–137. doi: 10.1016/j.ydbio.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci USA. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P, Edgar D. Regulation of the differentiation and behaviour of extra-embryonic endodermal cells by basement membranes. J Cell Sci. 2001;114:931–939. doi: 10.1242/jcs.114.5.931. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, McMahon AP, Eggan K. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Hadjantonakis AK. Cellular dynamics in the early mouse embryo: from axis formation to gastrulation. Curr Opin Genet Dev. 2010;20:420–427. doi: 10.1016/j.gde.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- Plusa B, Hadjantonakis AK, Gray D, Piotrowska-Nitsche K, Jedrusik A, Papaioannou VE, Glover DM, Zernicka-Goetz M. The first cleavage of the mouse zygote predicts the blastocyst axis. Nature. 2005;434:391–395. doi: 10.1038/nature03388. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Stephens LE, Sutherland AE, Klimanskaya IV, Andrieux A, Meneses J, Pedersen RA, Damsky CH. Deletion of beta 1 integrins in mice results in inner cell mass failure and periimplantation lethality. Genes Dev. 1995;9:1883–1895. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol. 2008;322:133–144. doi: 10.1016/j.ydbio.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18:155–180. [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Wilder PJ, Kelly D, Brigman K, Peterson CL, Nowling T, Gao QS, McComb RD, Capecchi MR, Rizzino A. Inactivation of the FGF-4 gene in embryonic stem cells alters the growth and/or the survival of their early differentiated progeny. Dev Biol. 1997;192:614–629. doi: 10.1006/dbio.1997.8777. [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Yang DH, Smith ER, Roland IH, Sheng Z, He J, Martin WD, Hamilton TC, Lambeth JD, Xu XX. Disabled-2 is essential for endodermal cell positioning and structure formation during mouse embryogenesis. Dev Biol. 2002;251:27–44. doi: 10.1006/dbio.2002.0810. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Huang S. Stochasticity versus determinism in development: a false dichotomy? Nat Rev Genet. 2010;11:743–744. doi: 10.1038/nrg2886. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]