Abstract
Background
Polarization of tissue is achieved by asymmetric distribution of proteins and organelles within individual cells. However, existing quantitative assays to measure this asymmetry in an automated and unbiased manner suffer from significant limitations.
Results
Here, we report a new way to assess protein and organelle localization in tissue based on correlative fluorescence analysis. As a proof of principle, we successfully characterized planar cell polarity dependent asymmetry in developing Drosophila melanogaster tissues on the single cell level using fluorescence cross-correlation.
Conclusions
Systematic modulation of signal strength and distribution show that fluorescence cross-correlation reliably detects asymmetry over a broad parameter space. The novel method described here produces robust, rapid and unbiased measurement of biometrical properties of cell components in live tissue that is readily applicable in other model systems.
Keywords: Fluorescent microscopy, asymmetric protein and organelle localization, image analysis, cross-correlation, PCP, planar cell polarity
INTRODUCTION
While it is well established that asymmetric protein localization within cells is essential for development, currently available methods to assess asymmetry of proteins and cell structures in tissues have limitations. Existing methods to study protein/organelle asymmetry can be grouped into two categories. The first is based on the creation of a vector and calculation of the length/angle between the protein/organelle of interest to a reference point (Park, Mitchell et al. 2008; Hirota, Meunier et al. 2010; Mirzadeh, Han et al. 2010; Happe, de Heer et al. 2012). Although proven to be useful in some cases, this approach is not generally applicable for a number of reasons. An aspect often not considered is the difficulty of finding a reliable cell-internal reference. Variance in the localization of the reference point within individual cells and lack of spatial separation from the structure of interest are the main contributors to measurement errors associated with this approach. A second limitation of the method is the distribution of the protein of interest. In order to create a vector, it is necessary to identify the center of the respective structures (eg. cell nucleus, cilia). Reduction of the protein/organelle of interest to a single point of origin is a correct approximation for spherical distributions. However, the method is conceptually more difficult for structures with multiple centers or for non-spherical protein-distributions (eg. where the center of mass may be located outside of the effective structure). Furthermore, correct automated assignment of contributions to individual cells can be problematic. A last concern with this approach is the variability associated with non-uniform fluorescence intensities within the structures of interest. The identification of the center of mass often involves binarization of the image. Consequently, gating parameters that define the area of interest can substantially influence the localization of the center or mass.
The second category of methods to assess asymmetry is based on comparison of fluorescent intensities along a given line (Greenan, Brangwynne et al. 2010; Stephens, Haase et al. 2011; Omelchenko and Hall 2012). While this approach does not suffer from the geometric constraints of vector-based methods, it is associated with other limitations. This manual form of measuring asymmetry is useful to visualize protein asymmetry but only sparsely samples the data and yields few data points with considerable variability depending on specific placement of the line. Reduction of protein localization or reference sites to a single line is thus not suitable for rapid, quantitative analysis of multiple parameters over a large set of samples.
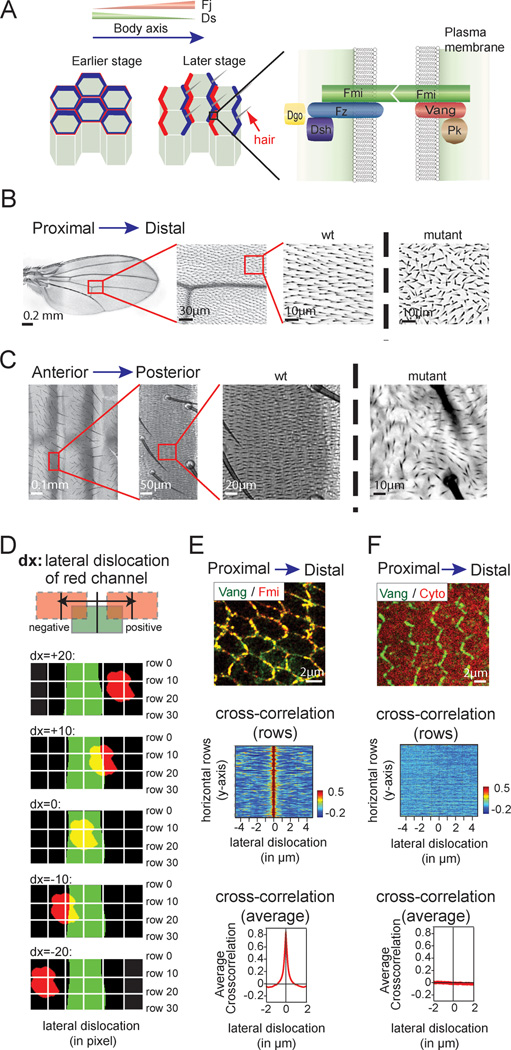
Here we aimed to develop an unbiased, automated and easily implemented assay to characterize the asymmetric subcellular protein and organelle localization within a tissue. To demonstrate the efficacy of the method, we took advantage of unique features of planar cell polarity (PCP) that is evident in a variety of epithelial tissues in both vertebrates and invertebrates (Zallen 2007; Vladar, Antic et al. 2009; Bayly and Axelrod 2011). In PCP, cells in epithelial tissue are polarized orthogonal to the apical–basal axis by the PCP signaling pathway (Fig. 1A). This planar polarization is controlled by the PCP machinery consisting of six core polarity proteins that acquire asymmetric distributions to distinguish proximal and distal sides of cells. Starting from a symmetric apical distribution in pupal wing cells, three of the six proteins, Frizzled (Fz), Dishevelled (Dsh) and Diego (Dgo) become highly enriched at the distal domain of the cell cortex, at the level of the adherens junctions, while Van Gogh (Vang) and Prickle (Pk) localize to the proximal side. Flamingo (Fmi) localizes to both proximal and distal sides (Fig. 1A). The orientation of cell asymmetry with respect to the tissue axis depends on a global signal. This is achieved at least in part through opposing expression gradients of the global PCP proteins Dachsous (Ds; an atypical cadherin) and the Golgi kinase Four-jointed (Fj), and transduced by the atypical cadherin Fat (Ft) (Axelrod 2009); Thomas and Strutt 2012). PCP is required for planar polarization of numerous epithelia in Drosophila, including the wing (Fig. 1B) and abdominal epithelia (Fig. 1C). In these tissues, polarization triggers asymmetric polymerization of the actin cytoskeleton to form hairs at the distal end of each wing cell and posterior end of each abdominal cell, respectively. Failure of the PCP mechanism results in incorrect orientation of hairs that is readily recognized visually (Fig. 1B, C; right images).
Fig. 1.
Planar cell polarity is a model for asymmetric protein localization in polarized tissue. A Schematic model of planar cell polarity (PCP) in Drosophila. B PCP is responsible for hair orientation in the Drosophila wing. Each cell in the wing produces a single hair pointing distally in a PCP-dependent manner (left and middle panels). Loss of PCP causes loss of orientation of wing hairs. For illustration, a magnified section of a dsh1 mutant wing is shown to the right. C Hair orientation in the Drosophila abdomen. Each cell on the abdomen makes hairs that point posteriorly in a PCP dependent manner (left and middle). The disruption of polarity in a dsh1 mutant fly is shown in the right panel. D Principle of the cross-correlation function. Corresponding fluorescence intensities of the two channels are compared pixel by pixel for each row to measure the cross-correlation. This is repeated upon lateral shift of the two channels in the horizontal direction (bottom). A detailed description can be found in Fig. S1. E Analysis of two PCP proteins shows a rapid drop in cross-correlation upon lateral shift. Drosophila wing expressing fluorescently labeled Vang (green) and Fmi (red) was imaged at 24h APF. As illustrated in the image, both proteins co-localize at the plasma membrane. Correspondingly, the cross-correlation is very high. The correlation drops rapidly upon lateral displacement. This is depicted in the heat-map, where individual cross-correlation scores for each row are color-coded and positioned according to the lateral dislocation (middle). An average score of all rows is show in the graph at the bottom. F No cross-correlation is observed for a PCP protein and a cytosolic marker. Vang (green) and a cytosolic fluorescent protein (red) were expressed in a Drosophila wing at 24h APF. No co-localization is visible (middle), and no cross-correlation is detected (bottom). Scale bars (B), 0.2mm; (C), 0.1mm; (E, F), 2 µm.
RESULTS
To assess PCP dependent asymmetric protein localization in Drosophila tissue, we took advantage of correlative fluorescent comparison (cross-correlation) that has previously been successfully used to assess co-localization of fluorescently tagged proteins (Bolte and Cordelieres 2006). Cross-correlation compares the respective fluorescent intensities of two channels as a function of lateral displacement (Fig. 1D, Fig. S1A). For proteins with overlapping localization, the correlation score rapidly drops as lateral shift disrupts co-localization of the corresponding channels (Fig. 1E), while no such drop is observed in the absence of co-localization (Fig. 1F).
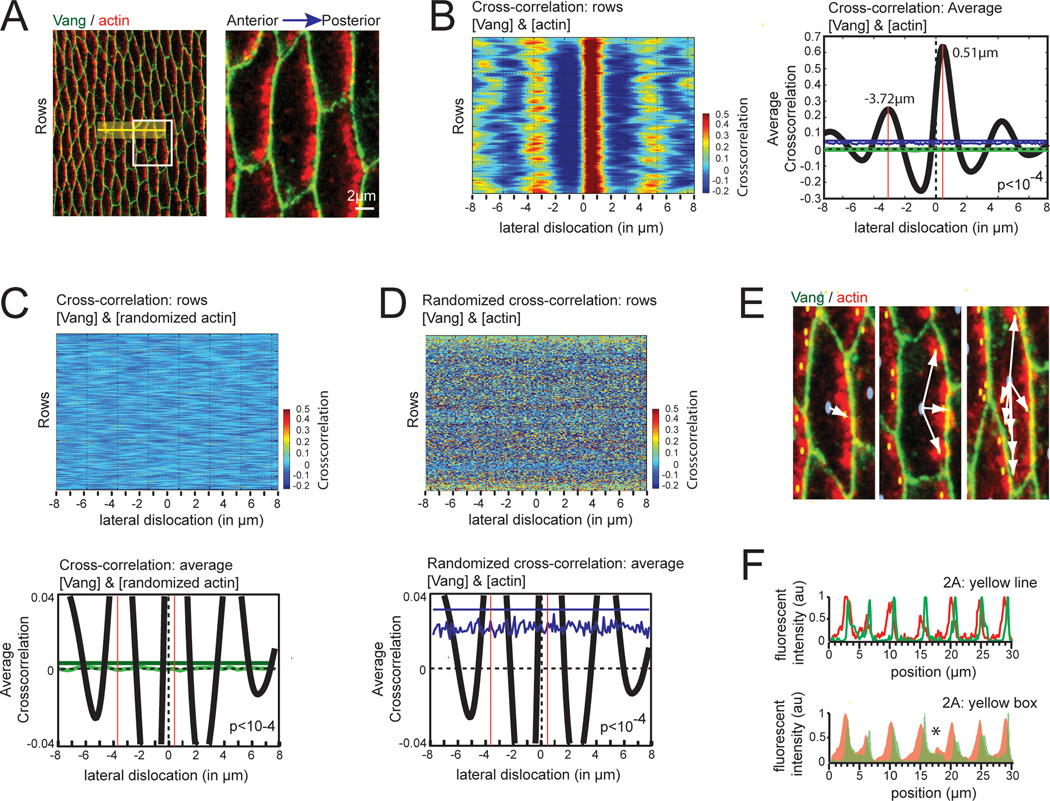
We hypothesized that comparison of fluorescent intensities of a protein with asymmetric cytosolic localization to a membrane reference would allow us to assess polarization in tissue on the single cell level across a cellular ensemble without specifically recognizing individual structures. To test this, we determined the correlative scores in fluorescence intensity between the membrane-associated PCP core protein Vang (green) and the accumulating actin (red) that precedes hair formation at the posterior side of Drosophila abdominal cells (Fig. 2A). As expected, lateral displacement of the actin signal in the posterior direction led to a substantial increase in the cross-correlation score, reflective of an artificial overlap of membrane and actin fluorescence at the posterior site of the cells caused by the shift (Fig. 2B). Likewise, displacement of actin in the anterior direction led to an increase in the cross-correlation score.
Fig. 2.
Cross-correlation, a novel method to detect asymmetric protein localization in tissue. A Asymmetric localization of actin in Drosophila epithelial cells. Abdominal cells expressing the membrane-anchored PCP core protein Vang (green) stained for actin (red) were imaged at 46 hrs APF. For visualization, a magnified section is shown to the right (white box). Note that actin polarizes to the posterior side of individual cells. The yellow box and line depicted in the image are used in Fig. 2F for the linescan. B Lateral displacement causes an increase in the cross-correlation score. The left panel shows a heat-map of the correlation scores for gray values of Vang and actin within individual rows of the image (ie. Fig. 2A) as a function of the lateral shift. To the right, the average value for all rows (black line) as well as the mean distance to the anterior and posterior boundary of the cell (red lines) are shown. Note that the correlation score peaks upon a lateral shift, reflective of an increased co-localization. Multiple peaks are visible as a consequence of periodic tissue structure. The blue and green lines in the graph are randomization controls discussed in Figs. 2C,D. C No cross-correlation upon randomization of the actin channel. Heatmap depicting the correlation score of Vang and the randomized actin channel of the image shown in Fig. 2A (top panel) and the graph of the average values for all rows (bottom panel, green line) are illustrated. As a reference, the average value for all rows prior to actin-randomization has been included (bottom panel, black line). Note that no correlation is visible (bottom panel, green jagged line). The cut-off was set at p<10−4 (ie. 4 standard deviations). The probability of generating an amplitude above this threshold (green line, 0.003786) by chance is p<10−4. A detailed description of the used randomization can be found in Fig. S1B. D No cross-correlation upon randomization of the heat map. The heatmap from Fig. 2B depicting the correlation score of Vang and the actin channel was randomized (top panel), and the average values for all rows plotted (bottom panel, blue line). Again, the average value for all rows prior to heatmap-randomization was included as a reference (bottom panel, black line). Note that no correlation is visible (bottom panel, blue jagged line). The cut-off (blue line, 0.03879) was set at p<10−4 (ie. 4 standard deviations). A detailed description of the randomization protocol can be found in Fig. S1C. E Vector based analysis is not suitable for analysis of actin staining. The center of individual cells (blue) and of the actin staining (yellow) is shown as individual, colored circles. Only some cells had one center of mass (left) while the majority showed 2 or more centers of mass for actin due to non-uniform actin distribution (middle, right). Note that in some cases multiple connected actin patches shared one center of mass, which was located outside of the fluorescent signal (middle). F Linescan depicting asymmetric localization of actin in cells. Fluorescent intensities along the yellow line depicted in Fig. 2A for Vang and actin is shown in the top panel. Note, that fluorescent intensities of Vang peak before actin, indicative of the lateral shift. Integrated fluorescent intensities for the yellow box shown in Fig. 2A is shown in the bottom panel. The asterisk depicts a region where the membrane within the box is curved, as well as oriented in an angle to the axis along which the gray values were averaged (A), 2 µm.
To analyze whether these relative peaks in cross-correlation were significant, we performed two separate controls. First, we randomized the position of individual pixels within each row of the asymmetric channel (actin) (Fig. S1B). Although the randomized and the original image of actin share the same distribution of gray values within individual rows, no cross-correlation was visible for Vang and the randomized actin signal (Fig. 2C, top panel). We then took the average cross-correlation of the randomized image and determined the standard deviation. We suggest setting a threshold of 4 standard deviations (4σ) above the mean value to provide a high likelihood of significance. Assuming a normal distribution, the probability to reach by chance an average cross-correlation score that is above this threshold is less than 0.01% (p<10−4). Notably, the threshold was more than an order of magnitude below the amplitude of the signal (Fig. 2B,C, green line). As a second control, we took the cross-correlation analysis data obtained in Fig. 2B and randomized the position of individual pixels within each row of the heatmap (Fig. S1C). The standard deviation was bigger compared to the previous control and the average correlation score was above zero, since the randomized heatmap is derived from cross-correlation values of the original heatmap (Fig. 2D, bottom panel). However, no cross-correlation was visible for the randomized heatmap (Fig. 2B,D), and the threshold (p<10−4) remained more than an order of magnitude below the amplitude of the signal (Fig. 2B,D, blue line). Together, for the example in Fig. 2A, these controls clearly show that the cross-correlation signal is statistically significant and thus can be used to characterize asymmetric protein distribution in tissue.
Once significance of the measurement is established, a simple measurement of the degree of asymmetry is possible. As illustrated in Fig. 2B, the distance in the anterior direction (3.72 µm) was substantially larger compared to the posterior displacement (0.51 µm), indicative of asymmetric protein localization. To determine the relative degree of asymmetry of the signal within the cell, we defined the center of the cell as 0% and the membrane as 100% dislocation. We find for the actin signal a 76% dislocation along this A/P axis towards the posterior end of the cell. Notably, this metric value for the degree of asymmetry of the signal towards the plasma membrane is independent of cell dimensions and may thus be suited for a comparison between samples.
Using the same image, we next tested currently used assays. As illustrated in Fig. 2E, the center of individual actin patches (yellow dots) could easily be detected, allowing the formation of a vector from the center of the cell (blue dots) to these sites (white arrows). However, the interpretation of these vectors was difficult due to the heterogeneity in actin immunostaining within individual cells (Fig. 2E). We next assessed the fluorescence distribution using a line scan (Fig. 2A, yellow line). As expected, we find that actin is asymmetrically distributed within the cell (Fig. 2F, top panel). This method illustrates the asymmetric distribution, and while an average distance of actin to the plasma membrane could be extracted from this assay, the stochastic noise component and small sample size would render the result highly sensitive to placement of the line. Increasing the width of the line (Fig. 2A, yellow box) led to some reduction of the noise (Fig. 2F, bottom panel). However, with increasing width it also blurred the boundaries where the membrane within the box was curved, or oriented in an angle to the axis along which the gray values were averaged (Fig. 2F, asterisk).
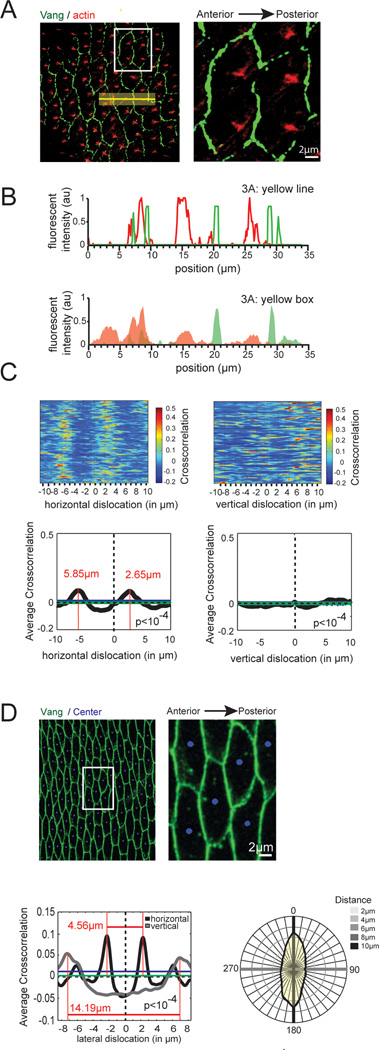
To test the robustness of our assay, we next explored hair formation at a later stage, in which actin asymmetry was less obvious (Fig. 3A). As in the previous case, the vector-based assay had limited value due to geometrical constrains. Neither did a line scan provide conclusive results (Fig. 3B). In contrast, cross-correlation clearly shows statistically significant asymmetry of the actin patches towards the posterior side with a median distance of 2.65 µm to the posterior and 5.85 µm to the anterior membrane, respectively (Fig. 3C, left panels). Using the same metrics as before, we find the signal to be 38% polarized along the A/P axis towards the posterior plasma membrane. We next translocated the two channels orthogonal to the A/P axis. One elongated peak was slightly above our significance threshold (p<10−4) when images were shifted in this direction, suggesting that the actin patches have a subtly asymmetric distribution along this axis as well, at least in the region analyzed here (Fig. 3C, right panels),. Together these results clearly indicate that the correlative fluorescent intensity method that we here developed is suitable to assess protein localization during PCP-dependent Drosophila abdomen development.
Fig. 3.
Cross-correlation detects asymmetry of low signal strength. A Asymmetric localization of hairs in the abdomen. Cells expressing fluorescently tagged Vang (green) stained for actin (red) were imaged at 50 hrs APF. For visualization, a magnified section is shown to the right (white box). The yellow box and line depicted in the image are used in Fig. 3B for the linescan. Note that individual cells contain multiple hairs that are located toward the posterior side of the cell. B No asymmetry can be detected for a line (top panel) or an integrated area (bottom panel) comparing the fluorescent intensities of Vang (green) and actin (red) along a given axis. C Cross-correlation detects asymmetric localization of hairs along the horizontal but not the vertical axis in the abdomen. Heat map (top panels) as well as average values (bottom panels, black line) and significance thresholds obtained from randomization controls (bottom panels, blue and green lines) are shown. Cross-correlation detects subtle, but significant (p<10−4 for green line = 0.005955, for blue line = 0.0182) asymmetric actin enrichment at the posterior site. No significant (p<10−4 for green line = 0.00683, for blue line = 0.01593) pattern is detected upon vertical dislocation (right). D Radial analysis of cells detects average cell shape in a tissue. Vang protein (green) and the center of cells (blue) were used for the radial analysis. For visualization, a magnified section is shown to the right (white box). Lateral displacement of the center of gravity in horizontal (black line) and vertical (gray line) direction identified the median width (4.45µm) and length (14.19µm) of all cells within the image (bottom, left). All peaks were significant (horizontal randomization, p<10−4 for green line = 0.006414, for blue line = 0.01179; vertical randomization, p<10−4 for green line = 0.005725, for blue line = 0.008076). In yellow, the average shape and size of all cells identified by radial displacement in all polar directions is shown (bottom, right). Scale bars (A, D), 2 µm.
It is sometimes of interest to monitor cell shape during development (Classen, Anderson et al. 2005; Aigouy, Farhadifar et al. 2010). We therefore asked whether cross-correlation may also be used to investigate cell shape. For this, we performed a cross-correlation analysis of a membrane marker (green) and the geometric center of individual cells (blue) in all directions (ie. radial displacement). As shown in Fig. 3D, assembly of individual values of the mean peak distance (peak amplitude was p<10−4) obtained with this assay into a radial graph, led to the average cell shape within the tissue that was analyzed (Fig. 3D, yellow area). Information on changes in cell shape and size may be of particular interest for tissue with characteristic patterns (eg. Drosophila eye or wing), or where elongation along a given axis and changes in cell size are expected (e.g. during pupal wing development, 0 to 30 hAPF).
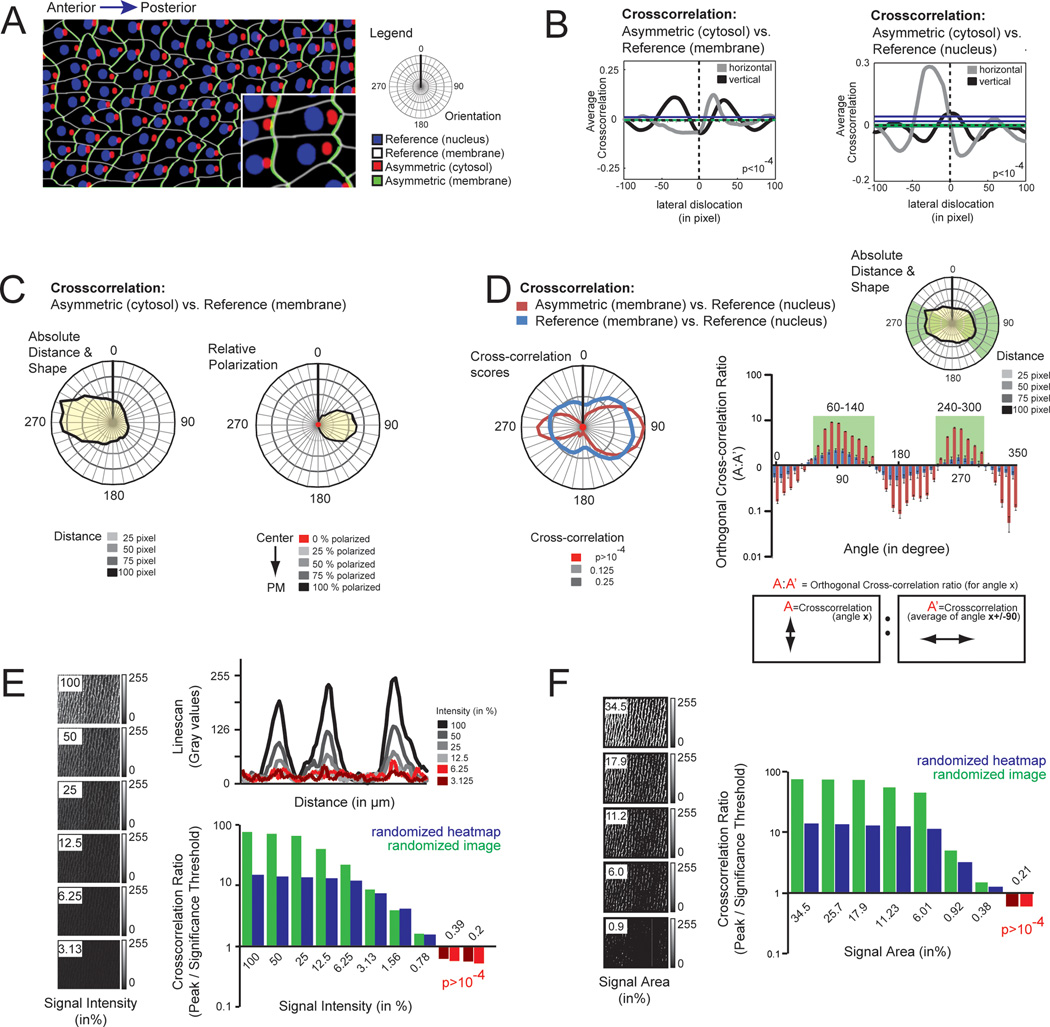
To further investigate the possibilities of the radial analysis, we then created a hypothetical tissue composed of irregularly shaped cells that contain a nuclear (blue) and a membrane (white) anchored reference as well as an asymmetric cytosolic (red) and membrane-associated asymmetric signal (green) (Fig. 4A). Cross-correlation between the asymmetric cytosolic signal (red) and a membrane-associated (white) or with the nuclear reference (blue) yielded comparable results, indicating that any cellular structure with relatively constant localization can be used as a reference (Fig. 4B). By extending the analysis to sampled angles of dislocation, we obtained the median cell shape as well as the average position of the asymmetric signal within cells (Fig. 4C, left panel). As before, we defined the center of the cell as 0% and the membrane as 100% dislocation, and measured the position of the asymmetric signal on this scale for each angle, and find the maximal asymmetry at 90 degrees (Fig. 4C, right panel).
Fig. 4.
Cross-correlation can detect asymmetry independent of tissue shape and cell-internal reference localization. A Model tissue composed of irregular cell shapes. Each cell contained a membrane-anchored (white) and a nuclear (blue) reference, as well as an asymmetric cytosolic (red), and asymmetric membrane-anchored (green) signal. For orientation, an A/P axis along the 270-90 angle was added. B Cross-correlation detects asymmetry of the asymmetric cytosolic signal using both, the membrane (left graph) and the nuclear (right graph) reference. The asymmetrically distributed signal was shifted laterally in horizontal (gray line) and vertical (black line) direction. Note that horizontal displacement led to an increase in the cross-correlation score using both the membrane-anchored (left graph) and the nuclear reference (right graph), while no asymmetry was detected in the vertical direction. Significance thresholds: left, gray (p<10−4 for green line = 0.006375, for blue line = 0.01742); left, black (p<10−4 for green line = 0.00505, for blue line = 0.02035); right, gray (p<10−4 for green line = 0.007259, for blue line = 0.04173); right, black (p<10−4 for green line = 0.007547, for blue line = 0.02233). C Radial displacement can be used to detect the asymmetry axis of the cell. Radial analysis of the asymmetric cytosolic component was compared to the membrane-associated reference. Plot of average distance and angle show the average cell shape (left graph, yellow). Position of the asymmetric signal within the cell is reflected in the reduced distance to the membrane. Analysis of the relative asymmetry in percent is shown in the right graph. D Comparative radial displacement can be used to detect asymmetry of membrane-associated proteins. Radial analysis of the cross-correlation scores for of the asymmetric membrane signal vs. the nuclear reference (red) and cross-correlation scores of the membrane-associated reference vs. the nuclear reference (blue) are shown to the right. Note the increase in cross-correlation values along the A/P axis for the cross-correlation of the asymmetric membrane vs. nuclear reference (red) compared to the control cross-correlation (blue). The plot to the right depicts the ratio of a cross-correlation of a given angle divided by the mean cross-correlation in the perpendicular direction (ie. orthogonal cross-correlation ratio). This ratio detects relative changes in the distribution (ie. circularity) of radial cross-correlation scores. Note that the asymmetric membrane-associated signal is significantly increased above the reference for angles between 60–140 and 240–300 degrees (green box), consistent with accumulation along the A/P axis (green sector). Error bars (p<10−4) were calculated from the randomized heatmap of the radial cross-correlation scores of the asymmetric signal vs. the nuclear reference. E Cross-correlation detects asymmetry of dim objects. Starting with the original image from Fig. 2D, the intensity was divided by half with each frame leading to a series of 8 images with progressively reducing signal strength (left panels). To simulate the stochastic noise component, a randomized noise mask was added to each frame before analysis (Fig. S2B). Analysis shows the ratio of the maximal cross-correlation amplitude of the individual images normalized to the significance threshold (p<10−4) of the same image upon randomization of the image (green) or randomization of the heatmap (blue), respectively. Intensities below the significance threshold are shown in red. Note that signal/noise ratio >0.75 (the average gray value of this noise component is 3% of the maximal intensity) are reliably detected. F Cross-correlation is suitable to detect asymmetry of small structures. To assess the minimal area required to reliably detect asymmetry using cross-correlation, we progressively reduced the actin area (left panels). Analysis shows the ratio of the maximal cross-correlation amplitude of the individual images normalized to the significance threshold (p<10−4) of the same image upon randomization of the image (green) or randomization of the heatmap (blue), respectively. Intensities below the significance threshold are shown in red. Note that a signal of >0.5% of the total area is sufficient to detect asymmetry using cross-correlation.
As many asymmetric proteins locate at or close to the plasma membrane (Mellman and Nelson 2008), we next tested whether cross-correlation may be used to detect asymmetry of membrane-associated proteins. For this, we made a radial analysis of the cross-correlation for the asymmetric membrane attached signal (green) and the nuclear reference (blue) and compared it to a control cross-correlation derived from the membrane-localized reference (white) and the nuclear reference (blue). Compared to the control, the cross-correlation scores of the polarized signal show greater elongation along the A/P polarity axis, indicating that the axis of asymmetric membrane-anchored signal can be detected using this approach (Fig. 4D, left). To quantify the asymmetry, we compared the two cross-correlations, as a significant asymmetric enrichment of the protein of interest should produce a greater cross-correlation compared to the membrane control. Specifically, we calculated orthogonal cross-correlation ratios, by dividing the cross-correlation along a given axis by the average cross-correlation score in the perpendicular direction (Fig. 4D, right). For the control, changes in this ratio are the consequence of asymmetry in localization of the nuclear reference and the cell shape. Values for the asymmetric signal that are above this value likely reflect relative enrichment within the plasma membrane. Notably, angles where the ratio was significantly above the reference line clustered to sectors where the membrane protein is enriched (Fig. 4D, green boxes and sectors), and angles where the ratio is significantly below the reference correspond to sectors where the membrane protein is depleted. However, we would like to note that application of the orthogonal cross-correlation ratio should be handled with great care. As the amplitude of the cross-correlation score is affected by signal strength, noise levels and signal distribution, only membrane-associated signals with comparable signal/noise ratio should be used for this comparative analysis. Furthermore, any form of image processing (eg. background subtraction, contrast enhancement) should be avoided as this will change the shape of the radial cross-correlation.
Finally, we determined the robustness of the cross-correlation assay by artificially changing individual parameters. Using the example from Fig. 2A, we generated a series of images where the signal intensity of one channel was continuously diminished (each frame had 50% intensity compared to the previous sample). To all images, a constant noise component (ie. random gray values with a Gaussian distribution centered around 10) was added to maintain the background constant (Fig. 4E, Fig. S2). As expected, the amplitude of the cross-correlation was reduced when signal approached the noise level. Intriguingly, we were able to detect significant asymmetry (p<10−4) for gray values that were as little as 75% above the average noise level (Fig. 4E). This indicates that cross-correlation can be used to analyze low intensity signals. Next, we characterized how the area occupied by the signal affects sensitivity. As non-uniform fluorescent intensities within the signal area will affect the analysis, the signal was first binarized and then successively reduced. Again a noise component was added to the analysis. We find that asymmetry can be reliably detected for signals that cover less than 0.5% of the total area (Fig. 4F), although non-uniform and/or reduced signal strength of a biological sample will likely require a larger area.
As an added note, we would like to stress that the absolute value of the cross-correlation integrates signal intensity, signal area and noise into one value. This restricts a direct interpretation of cross-correlation scores. For that reason, our study uses a comparison of the position and amplitude of cross-correlation of original and randomized images to obtain statistically significant measurements of relative signal strength. Our examples show that this approach can be reliably used to interrogate dependency of two signals within a cell (direction, amplitude and angle).
DISCUSSION
Here we used spatial cross-correlation to assess asymmetric subcellular protein localization within a tissue. Methodical sampling shows that it can detect statistically significant differences over a wide array of patterns and a broad parameter space (Figs. 4E,F). In addition we show that this method can also be used to measure changes in cell size and shape (Figs. 3D, 4C). Importantly, the method uses information from variable fluorescence intensity and does not depend on specific geometry of protein distribution, providing an increased sensitivity and robustness over existing approaches.
A similar method that defines a “magnitude and axis of nematic order for a single cell” and applies it over a field within an image has been described (Aigouy et al 2010). The output of the method yields data that are similar to a subset of those that our method produces as shown in Fig 4D. One advantage of the Aigouy method is that it maps varying orientation of polarity across an image. However, the Aigouy method was only shown to be suitable for measuring asymmetry of plasma membrane associated proteins, but not for intracellular proteins, and no statistical analysis of significance is incorporated. Our method, which does not rely on recognition of any specific structures, is much more general in its ability to detect asymmetry.
We have described an approach for determining statistical significance of the method presented here. In addition, we have made suggestions for metrics to “quantify” the degree of asymmetry. However, we recognize that specific metrics may be more or less relevant to particular users’ requirements. For example, one may be interested in how eccentrically a signal is placed within a cell, while another may wish to incorporate the amount of signal from that object in a weighted measure of eccentricity. For a plasma membrane signal, one may be interested in how focal the asymmetric protein is accumulated, while another may be interested in the proportion in a given half of the cell. We therefore caution that users may wish to derive their own measures from the data produced by the analysis.
This study focused on the use of cross-correlation to study polarized protein localization within tissue. However, it is not limited to assessing asymmetry in tissues but may be applicable to study directionality of dynamic processes within single cells. For example, comparing the fluorescence of an individual vesicle at two time points may be used to assess its direction and speed. Here, the cross-correlation score would remain high only when lateral displacement occurred along the microtubule axis.
Finally we would like to emphasize that this method is available as a plugin for ImageJ (Bolte and Cordelieres 2006) and as a function in Matlab (Fig. S2A). Analysis with this method is thus not only suitable to make statistical statements about polarization in a variety of experimental setups, but is also fast, inexpensive, and easily accessible.
EXPERIMENTAL PROCEDURES
Fly lines
Following fly lines and mutant alleles were used: OreR, dsh1, actinP-Vang::EYFP.
Immunohistochemistry
Drosophila pupal wing were prepared for imaging as previously described (Axelrod 2001. Pupal abdomens were dissected as follows: pupae were placed on double-sticky tape on their side and cut in half along a sagittal plane. The cuticle together with epidermis was transferred in PBS and washed by gently pipetting PBS solution toward the abdominal epidermis to remove the body fat and organs. The dissected tissue was then fixed in 4% paraformaldehyde for 15 min at room temperature, followed by blocking in 2% BSA (Sigma) in PBS with added 0.1% Triton X-100 (Thermo Scientific) at room temperature for 1 hour and stained with Alexa 660 conjugated phalloidin (Invitrogen) at room temperature for 30 min. The primary antibody against Fmi was anti-Fmi mAb (#74, DSHB), and the secondary antibody was Alexa-Fluor 546-goat anti-mouse (1:200, Invitrogen).
Image acquisition and analysis
Images were acquired on a Leica TCS SP5 AOBS confocal microscope using a 100x Objective and processed with LAS AF (Leica). Low magnification images of the wing and abdomen in Figs. 1B, C were composed from several separate images using the stitch function in ImageJ/Fiji. The image in Fig. 4F was modified using the erode function in ImageJ/Fiji. Center of individual cells in Fig. 3D was determined with “Center of mass” function in ImageJ/Fiji.
The cross-correlation function
The whole image is used for the cross-correlation analysis. In detail, within a horizontal row for each pixel the gray values of the two channels are measured and one correlation score is determined. This is done for all horizontal rows. Next, the correlation-score is determined upon lateral displacement by one pixel for each horizontal row individually (Fig. S1A). These data are summarized in the heatmaps. Note that each point in the heatmap reflects one correlation score: the y-axis in the heatmap represent the individual horizontal rows of the original image, and the x-axis shows the correlation score of individual horizontal rows upon lateral displacement of the two channels within the row by n pixels.
ImageJ and Matlab Script
Several cross-correlation Plugins for ImageJ are publicly available and can be found on the ImageJ homepage (http://rsb.info.nih.gov/ij/plugins/index.html). We tested JACoP (http://rsb.info.nih.gov/ij/plugins/track/jacop.html), although better plugins may be available. Documentation and further reading can be found on the ImageJ Wiki site (http://imagejdocu.tudor.lu/). The Matlab script used in this manuscript and illustrated in Figs. S1 and S2 can be fund on our homepage (http://www.stanford.edu/group/axelrodlab/). It contains a documented code with instructions how to use all statistical analyses (including both randomizations) that were used in this manuscript. For updates please check the homepage.
Radial Cross-Correlation
A circular section of the image was analyzed to prevent artifacts induced by the rotation. For the radial cross-correlation analysis in Figs 3D and 4C,D, a circular image was rotated in 10 degree steps.
Supplementary Material
Key findings.
We present a novel technique to identify asymmetric protein and organelle distribution on the single cell level across the entire tissue.
This method can analyze subcellular localization (ie. mean distance and direction) from the structure of interest to any cellular reference within the tissue.
Systematic sampling shows that this approach yields significant results over a wide parameter space making it suitable to assess asymmetry in a variety of tissues and potentially to study dynamic, polarized processes on the single cell level.
This method, which is based on cross-correlation of fluorescent intensities, is freely accessible and can be directly used without the need for additional image-processing.
ACKNOWLEDGMENTS
M.M was supported by grants from AXA and Stanford Deans Postdoctoral fellowship. M.G. was supported by grants from the Swiss National Science Foundation, Novartis Jubilaeumsstiftung and Stanford Deans Postdoctoral fellowships. J.D.A. acknowledges funding from the NIH.
Footnotes
AUTHOR CONTRIBUTIONS
M.M. performed the experiments and M.G. made the analysis. M.M, J.D.A. and M.G. discussed all results and wrote the manuscript.
REFERENCES
- Aigouy B, Farhadifar R, et al. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell. 2010;142(5):773–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
- Axelrod JD. Unipolar membrane association of Dishevelled mediates Frizzled planar cell polarity signaling. Genes & development. 2001;15(10):1182–1187. doi: 10.1101/gad.890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod JD. Progress and challenges in understanding planar cell polarity signaling. Seminars in cell & developmental biology. 2009;20(8):964–971. doi: 10.1016/j.semcdb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Bayly R, Axelrod JD. Pointing in the right direction: new developments in the field of planar cell polarity. Nature reviews. Genetics. 2011;12(6):385–391. doi: 10.1038/nrg2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Classen AK, Anderson KI, et al. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell. 2005;9(6):805–817. doi: 10.1016/j.devcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, et al. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Current biology : CB. 2010;20(4):353–358. doi: 10.1016/j.cub.2009.12.050. [DOI] [PubMed] [Google Scholar]
- Happe H, de Heer E, et al. Morphometric analysis of centrosome position in tissues. Methods in molecular biology. 2012;839:249–255. doi: 10.1007/978-1-61779-510-7_20. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Meunier A, et al. Planar polarity of multiciliated ependymal cells involves the anterior migration of basal bodies regulated by non-muscle myosin II. Development. 2010;137(18):3037–3046. doi: 10.1242/dev.050120. [DOI] [PubMed] [Google Scholar]
- Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nature reviews. Molecular cell biology. 2008;9(11):833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Han YG, et al. Cilia organize ependymal planar polarity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(7):2600–2610. doi: 10.1523/JNEUROSCI.3744-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T, Hall A. Myosin-IXA Regulates Collective Epithelial Cell Migration by Targeting RhoGAP Activity to Cell-Cell Junctions. Curr Biol. 2012 doi: 10.1016/j.cub.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, et al. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nature genetics. 2008;40(7):871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Haase J, et al. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. The Journal of cell biology. 2011;193(7):1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Strutt D. The roles of the cadherins Fat and Dachsous in planar polarity specification in Drosophila. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241(1):27–39. doi: 10.1002/dvdy.22736. [DOI] [PubMed] [Google Scholar]
- Vladar EK, Antic D, et al. Planar cell polarity signaling: the developing cell's compass. Cold Spring Harbor perspectives in biology. 2009;1(3):a002964. doi: 10.1101/cshperspect.a002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA. Planar polarity and tissue morphogenesis. Cell. 2007;129(6):1051–1063. doi: 10.1016/j.cell.2007.05.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.