Abstract
Background
Infectivity of herbivores with Trichostrongylus nematodes is widespread in many countries, having a major economic impact on breeding, survivability, and productivity of domestic livestock. This study was carried out on Trichostrongylus species isolated from domestic livestock in order to develop an easy-to-perform method for species identification.
Methods
Trichostrongylus isolates were collected from sheep, goat, cattle, and buffaloes in Khuzestan Province, southwest Iran. Primary species identification was carried out based on morphological characterization of male worms. PCR amplification of ITS2-rDNA region was performed on genomic DNA and the products were sequenced. Phylogenetic analysis of the nucleotide sequence data was conducted employing Bayesian Inference approach. Consequently, a restriction fragment length polymorphism (RFLP) profile was designed to differentiate Trichostrongylus species.
Results
A consensus sequence of 238 nucleotides was deposited in the GenBank for Iranian isolates of Trichostrongylus species including T. colubriformis, T. capricola, T. probolurus and T. vitrinus. The designated RFLP using restriction enzyme TasI could readily differentiate among species having different ITS2 sequence. The molecular analysis was in concordance with morphological findings.
Conclusion
Phylogenetic analysis indicated a close relationship among the sequences obtained in this study and reference sequence of relevant species. ITS2-RFLP with TasI is recommended for molecular differentiation of common Trichostrongylus species.
Keywords: Trichostrongylus, ITS2, Iran, Livestock
Introduction
Infection of digestive tract of herbivores with Trichostrongylus nematodes are distributed throughout the world and are of major veterinary and economic importance because of their high prevalence, pathological outcomes of infection and impacts on animal products. Usually, herbivores can be infected by several species of Trichostrongylus nematodes, each with a different pathological consequence on the animal (1, 2). Therefore, precise differentiation of Trichostrongylus species is important for operative control programs of the parasite. Although conventional morphological methods for identification of Trichostrongylus spp. are quite reliable in Trichostrongylus males, they are laborious and cannot be used for recognition of females and eggs (3, 4). Until date, many studies have reported DNA-based methods as an effective approach to differentiate Trichostrongylus species (4–7). In this case, rDNA-ITS2 has been indicated as a useful and reliable region for species identification within Trichostrongylus species (6–8).
Iran had long been considered as a major focus of human and animal trichostrongylid infection in the world (9–11). Different species had been prevalent in ruminants throughout the country (9, 12, 13), among those the occurrence of seven species in human is documented (9) with highest prevalence in nomadic tribes (11). Therefore, molecular studies of Trichostrongylus species will be a useful approach to have a perspective on trichostrongylid nematodes of the country compared with those from other parts of the world.
The aim of this study was to compare sequences of ITS2 region of ribosomal DNA in Trichostrongylus species isolated from some domestic livestock in Iran with those published from reference Trichostrongylus sequences of other countries, and to establish an easy and reliable method to distinguish Trichostrongylus species by a PCR-restriction fragment length polymorphism (RFLP) approach.
Materials and Methods
Parasite
Worms were collected from abomasums and small intestines of sheep, goats, cattle and buffalos from different abattoirs of Khuzestan Province, southwest of Iran. The samples were washed extensively in distilled water and preserved in ethanol 70%. The species of male nematodes were identified based on morphological characterizations, using a light microscope and nematodes taxonomic identification keys (14, 15).
DNA extraction and Polymerase Chain Reaction
DNA was obtained from individual male Trichostrongylus nematodes. An isolate refers to a single male worm was obtained from any different hosts. Briefly, each individual single worm was crushed between two microscopic slides for 1 min with 300µl lysis buffer (NaCl 0.1M, EDTA 0.01M, Tris-HCl 0.1M, Triton X-100 2%, SDS 2%). The lysate from each sample was treated by 30 µg/ml proteinase K in 56 °C for an hour. Then, the DNA was extracted by conventional phenol/chloroform extraction and ethanol precipitation (16). Finally, the DNA pellet was eluted in 20 µl deionized distilled water and inserted at -20 °C until more analysis.
ITS2 fragment of ribosomal DNA was PCR-amplified, using forward (NC1: 5-ACGTCTGGTTCAGGGTTGTT-3) and reverse (NC2: 5-TTAGTTTCTTTTCCTCCGCT-3) primer pair (17). The PCR reactions were performed in a 25 µl volume containing, 12.5 µl of 2× premix (Ampliqon, Skovlunde, Denmark), 25 pmol of each primer, 10.5 DDW and 1 µl of extracted DNA in a thermocycler (Corbett Research, Sydney, Australia). The PCR program was one cycle of 95 °C for 6 min followed by 35 cycles of 94 °C for 45 seconds, 60 °C for 90 seconds and 72 °C for 60 seconds with a final extension of 72 °C for 5 min. Samples with 1µl DDW instead of templates were used in each run as negative controls.
Two microliters of each amplification product was conducted to 1.5% agarose gel electrophoresis in TBE buffer (90 mM Tris, 90 mM boric acid and 2 mM EDTA) at 100 V for 1 h. The gels included 0.5 µg/ml ethidium bromide (Roche, Mannheim, Germany) for staining. A 100-bp DNA ladder (Fermentas, Vilnius, Lithuania) was run to estimate the size of DNA in gels. The bands were visualized and photographed under UV light employing a transilluminator (UVItec, UK).
Sequencing and phylogenetic analysis
Three or four PCR products of each Trichostrongylus species from different livestock hosts were subjected to sequencing, using above-mentioned primers. The results were analyzed by ClustalX (18) and DNASIS (Hitachi, Tokyo, Japan) softwares and were compared with relevant sequences related to sequences deposited in GenBank, using BLAST (http://www.ncbi.nlm.nih.gov/). The levels of sequence difference (D), were obtained, using pairwise comparison of a consensus representative sequence containing 238 nucleotides from different Trichostrongylus species found in the current study, via the formula of D=1-(M/L) (19), in which M is the number of alignment positions at which the two sequences had a common base, and L is the total number of alignment positions over which the two sequences are compared. For better understanding of relationship among isolates from Iran and other countries, all the different Trichostrongylus sequences described here conducted to a phylogenetic reconstruction along with reference sequences of Trichostrongylus species deposited in the GenBank from previous studies (6, 8, 20, 21), and an ITS2 sequence of T. colubriformis in the GenBank from Iran (AN: HQ389232). The analysis performed by Bayesian Inference approach employing MrBayes software v.3.1.2 (http://mrbayes.csit.fsu.edu/index.php). Trichostrongylus tenuis, the prevalent species in birds, was used as the outgroup. The tree was run several times to obtain the most frequent topology.
RFLP
The sequences were subjected to in-silico cutting with almost all known restriction enzymes using DNASIS software. Consequently, TasI has been considered to differentiate Trichostrongylus species. Five microliters of PCR product, 0.5 µl (5 units) of TasI enzyme (Fermentas, Vilnius, Lithuania), 1.5 µl of 10X supplied buffer and 8 µl double distilled water, were incubated at 65 °C for 3 h. Restriction fragments were separated on 2.2% agarose gel in TBE buffer, stained with ethidium bromide and photographed.
Results
Among 107 individual male worms of Trichostrongylus collected in the present study, four species were identified morphologically. Trichostrongylus colubriformis was the most identified species. All extracted DNA samples successfully yielded an expected 320 bp PCR product. No PCR amplification was seen in negative controls.
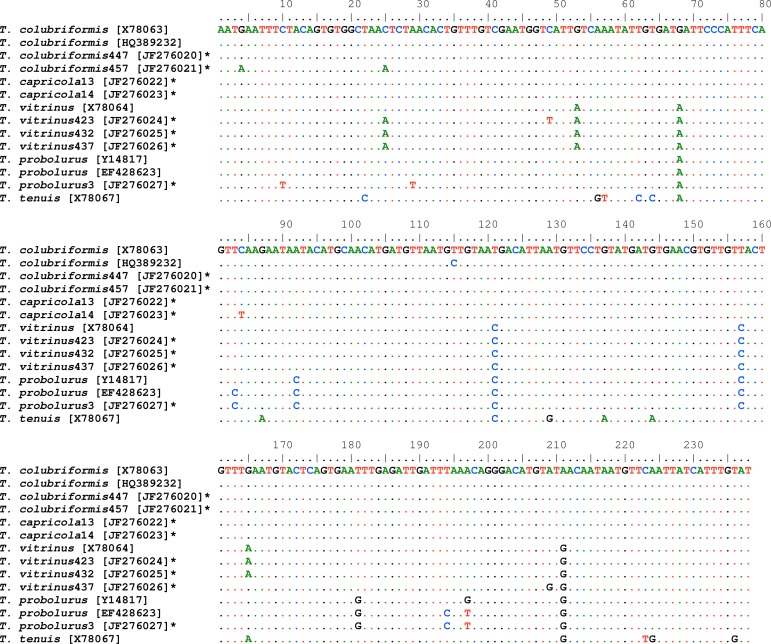
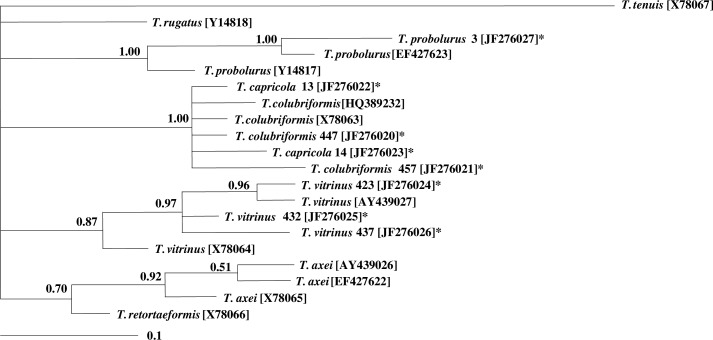
After sequencing, eight ITS2-sequences inferred from Trichostrongylus isolates, including two T. colubriformis, two T. capricola, three T. vitrinus and one T. probolurus. A consensus sequence containing 238 nucleotides for each individual sequence was deposited in GenBank (accession numbers: JF276020 to JF276027). Figure 1 shows multiple alignment of the sequences. Based on pairwise comparison, the differences among all of the different sequences of ITS2 ranged 0.5–3.4% (data not shown). The consensus phylogenetic tree indicated four major groups with high statistical supports (PP: 0.76 to 1.00). All Trichostrongylus species obtained in this study were in a cluster with the relevant reference sequences from previous studies (Fig. 2).
Fig. 1.
Alignments of the ITS2 sequences representing for all Trichostrongylus isolates in the present study with key reference sequences for Trichostrongylus species from previous studies (6, 8, 20–22). The accession numbers of individual sequences are given in square parentheses. Sequences with the bold asterisk inferred from this study. T. tenuis was used as the outgroup
Fig. 2.
Genetic relationships of Trichostrongylus isolates from Iran and reference sequences for different species of Trichostrongylus selected from previous studies (6, 8, 20, 21). The titles with asterisk inferred from the present study. The relationships were inferred based on phylogenetic analysis of ITS2 sequence data using Bayesian Inference. Nodal support is given as a pp value. The scale bar indicates distance.
The expected fragments after digesting the ITS2-PCR products of different species, by TasI restriction enzyme, are showed in Table 1.
Table 1.
The obtained fragments after in silico cutting of ITS2 region in common Trichostrongylus species. Relevant GenBank accession numbers was included
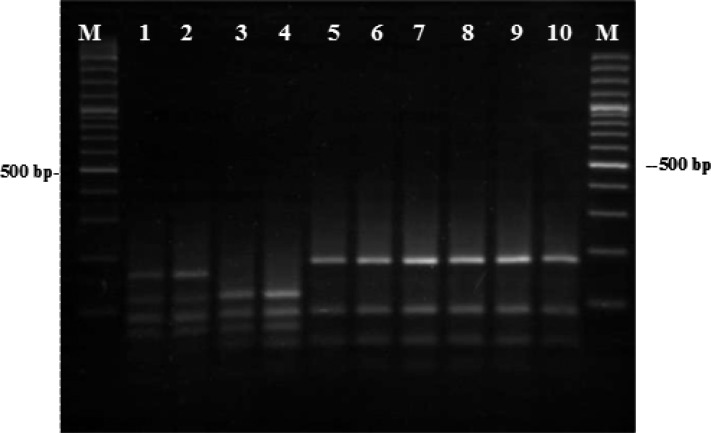
After RFLP analysis, the restriction enzyme TasI produced three different patterns as expected based on in-silico analysis; one relevant to both T. colubriformis and T. capricola, one to T. vitrinus and another to T. probolurus (Fig. 3). In RFLP-gels, detected Trichostrongylus species were in concordance with morphology and sequences results. Not any intra-specific variation was seen in the RFLP patterns of the species with multiple isolates.
Fig. 3.
The pattern of PCR products after digestion with TasI: lanes 1–2 are T. probolurus, lanes 3–4 are T. vitrinus, lanes 5-7 and 8-10 are T. capricola and T. colubriformis, respectively. Lanes M, 100-bp DNA ladder
Discussion
Molecular characterization of ITS2-rDNA region in four Trichostongylus species including T. colubriformis, T. capricola, T. probolurus and T. vitrinus collected from southwest Iran, were studied. All these species have been previously reported both in human (9–11, 23–24) and domestic livestock (9) in Iran. Furthermore, a recent study indicated that T. colubriformis was the main zoonotic species causing helminth infection in a rural village in Laos (24). In the present study, these species were studied using sequence analysis of ITS2-rDNA region and the sequence data for T. capricola, T. vitrinus and T. probolurus are submitted to the GenBank for the first time from Iran. Recently, a complete ITS1-5.8S-ITS2 rDNA sequence of T. colubriformis was deposited in the GenBank from Iran (HQ389232). This sequence has one nucleotide difference in ITS2 sequence with reference sequence of T. colubriformis (X78063) and also one of T. colubriformis ITS2 sequences from this study [JF276020]. The mentioned sequence was placed in one group along with other T. colubriformis and T. capricola sequences, having maximal statistical support (pp: 1.00) in the phylogenetic tree. Surprisingly, the ITS2-rDNA sequences of T. capricola isolates indicated 100% (JF276022) and 99.6% (JF276023) homology with reference sequence for T. colubriformis (X78063). Probably it can be explained with high similarity of these two species in morphological characters. As there is not any previous data for ITS2-rDNA sequence of T. capricola in the the GenBank to compare with the findings of the present study, the phylogenetic tree yields an overview of relationship of this species with other species. Two T. capricola sequences (JF276022 and JF276023) were placed in one cluster along with two T. colubriformis sequences from the present study (JF2760220 and JF276021), as well as the reference sequences for T. colubriformis (X78063), having maximum statistical support (pp= 1.00). The three T. vitrinus sequences (JF2760224- JF276026) indicated more relationship to T. vitrinus from UK (AY439027) (pp= 0.89) rather than those from Australia (X78064) (pp= 0.88). In addition, the sequence of T. probolurus was in a cluster with T. probolurus from Russia (EF427623), and these two were placed in an outer cluster with T. probolurus from Australia (Y14817), both clusters supported by maximum pp (1.00).
The PCR-RFLP approach has been used previously to distinguish among species of trichostrongylid nematodes, including T. colubriformis, T. axei, T. vitrinus, Haemonchus contortus, Teladorsagia circumsinata and Cooperia oncophora (7). RFLP analysis in current study could differentiate the Trichostrongylus species with different sequences. Due to high sequence similarity of T. colubriformis and T. capricola, these two species produced same RFLP pattern.
The present study represents preliminary sequence information of Trichostrongylus species from Iran. Few numbers of sequenced Trichostrongylus isolates and using one DNA locus (ITS2) to characterize the isolates were limitations in the current study. The T. capricola isolates could not be distinguished from T. colubriformis by ITS2-RFLP indicating the sequence is identical to T. colubriformis, whereas alignments of T. colubriformis sequences recently added to the GenBank (AB503241-52) from a village in Laos, with two T. capricola sequences (AF210006 and AF210030) originally from France, demonstrates that these two species have clear differences in 28s rDNA sequences (24, 25). As, trichostrongylid species are distributed all throughout the country, in different livestock (8), and by means of traditional methods the occurrence of seven species in human have been reported (9), utilization of molecular tools employing sequences of multiple genes will benefit understanding of the rate of zoonocity for each species, as well as obtaining a better perspective of Trichostongylus species in various host species in different geographical areas.
Acknowledgments
This study was financially supported by the National Institute of Health Research, Tehran University of Medical Sciences, Project number 86-10-02-241/429. The authors would like to thank the staff at molecular biology lab in Isfahan Station of National Institute of Health Research for their kind helps particularly Mrs. Niloofar Jalaizand and Mr. Reza Jafari. The authors declare that they have no conflicts of interest.
References
- 1.Holmes PH. Pathophysiology of nematode infections. In: Howell M. J, editor. Parasitology - Quo Vadit? Canberra: Australian Academy of Science; 1986. pp. 443–51. [Google Scholar]
- 2.Symons LEA. Sydney: Academic Press; 1989. Pathophysiology of endoparasitic infection compared with ectoparasitic infestation and microbial infection. [Google Scholar]
- 3.Georgi J, McCulloch C. Diagnostic morphometry: identification of helminth eggs by discriminant analysis of morphometric data. P Helm Soc Wash. 1989;56(1):44–57. [Google Scholar]
- 4.Gasser RB, Hoste H. Genetic markers for closely-related parasitic nematodes. Mol Cell probes. 1995;9(5):315–20. doi: 10.1016/s0890-8508(95)91588-5. [DOI] [PubMed] [Google Scholar]
- 5.Hoste H, Chilton NB, Beveridge I, Gasser RB. A comparison of the first internal transcribed spacer of ribosomal DNA in seven species of Trichostrongylus (Nematoda : Trichostrongylidae) Int J Parasitol. 1998;28(8):1251–60. doi: 10.1016/s0020-7519(98)00093-9. [DOI] [PubMed] [Google Scholar]
- 6.Kuznetsov D, Kuznetsova N. Sequences of the second internal transcribed spacer of ribosomal DNA for three species of Trichostrongylus (Nematoda: Trichostrongylidae) from sheep in Russia. Helminthologia. 2007;44(2):43–6. [Google Scholar]
- 7.Gasser RB, Chilton NB, Hoste H, Stevenson LA. Species identification of Trichostrongyle nematodes by PCR-linked RFLP. Int J Parasitol. 1994;24(2):291–3. doi: 10.1016/0020-7519(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 8.Hoste H, Chilton NB, Gasser RB, Beveridge I. Differences in the second internal transcribed spacer (Ribosomal DNA) between five species of Trichostrongylus (Nematoda: Trichostrongylidae) Int J Parasitol. 1995;25(1):75–80. doi: 10.1016/0020-7519(94)00085-3. [DOI] [PubMed] [Google Scholar]
- 9.Ghadirian E, Arfaa F. Present Status of Trichostrongyliasis in Iran. Am J Trop Med Hyg. 1975;24(6):935–41. doi: 10.4269/ajtmh.1975.24.935. [DOI] [PubMed] [Google Scholar]
- 10.Ghadirian E, Arfaa F, Sadighian A. Human Infection with Trichostrongylus Capricola in Iran. Am J Trop Med Hyg. 1974;23(5):1002–3. doi: 10.4269/ajtmh.1974.23.1002. [DOI] [PubMed] [Google Scholar]
- 11.Ghadirian E, Arfaa F, Arvanaghi A. Prevalence of intestinal helminthiasis among settled nomads and those with moving habits in southern Iran. Iran J Publ Health. 1974;3(3):147–52. [Google Scholar]
- 12.Borji H, Razmi G, Movassaghi AR, Naghibi AM, Maleki M. A study on gastrointestinal helminths of camels in Mashhad abattoir, Iran. Iran J Vet Res. 2010;11(2):174–9. [Google Scholar]
- 13.Eslami A, Meydani M, Maleki S, Zargarzadeh A. Gasterointestinal nematodes of wild sheep (ovis orientalis) from Iran. J Wildlife Dis. 1979;15(2):263–5. doi: 10.7589/0090-3558-15.2.263. [DOI] [PubMed] [Google Scholar]
- 14.Skrjabin KI. Essentials of nematodology; Helminthological Institute of the Academy of Sciences of the USSR; 1960. [Google Scholar]
- 15.Anderson R. 2nd edition. Wallingford, UK: CABI; 2000. Nematode parasites of vertebrates: Their development and transmission. [Google Scholar]
- 16.Sambrook J, Fritsch E. F, Maniatis T. New York, USA: Cold Spring Harbor Laboratory Press; 2004. Molecular Cloning: A laboratory manual. [Google Scholar]
- 17.Chilton NB. The use of nuclear ribosomal DNA markers for the identification of bursate nematodes (order Strongylida) and for the diagnosis of infections. Anim Health Res Rev. 2004;5(2):173–87. doi: 10.1079/ahr200497. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chilton NB, Gasser RB, Beveridge I. Differences in a ribosomal DNA sequence of morphologically indistinguishable species within the Hypodontus macropi complex (Nematoda: Strongyloidea) Int J Parasitol. 1995;25(5):647–51. doi: 10.1016/0020-7519(94)00171-j. [DOI] [PubMed] [Google Scholar]
- 20.Chilton NB, Hoste H, Newton LA, Beveridge I, Gasser RB. Common secondary structures for the second internal transcribed spacer pre-rRNA of two subfamilies of trichostrongylid nematodes. Int J Parasitol. 1998;28(11):1765–73. doi: 10.1016/s0020-7519(98)00129-5. [DOI] [PubMed] [Google Scholar]
- 21.Wimmer B, Craig BH, Pilkington JG, Pemberton JM. Non-invasive assessment of parasitic nematode species diversity in wild Soay sheep using molecular markers. Int J Parasitol. 2004;34(5):625–31. doi: 10.1016/j.ijpara.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 22.Hoste H, Gasser RB, Chilton NB, Mallet S, Beveridge I. Lack of intraspecific variation in the second Internal Transcribed Spacer (ITS-2) of Trichostrongylus colubriformis ribosomal DNA. Int J Parasitol. 1993;23(8):1069–71. doi: 10.1016/0020-7519(93)90128-l. [DOI] [PubMed] [Google Scholar]
- 23.Ghadirian E. Human Infection with Trichostrongylus lerouxi (Biocca, Chabaud, and Ghadirian, 1974) in Iran. Am J Trop Med Hyg. 1977;26(6):1212–3. doi: 10.4269/ajtmh.1977.26.1212. [DOI] [PubMed] [Google Scholar]
- 24.Sato M, Yoonuan T, Sanguankiat S, Nuamtanong S, Pongvongsa T, Phimmayoi I, Phanhanan V, Boupha B, Moji K, Waikagul J. Human Trichostrongylus colubriformis Infection in a Rural Village in Laos. Am J Trop Med Hyg. 2011;84(1):52–4. doi: 10.4269/ajtmh.2011.10-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouy de Bellocq J, Ferte H, Depaquit J, Justine J-L, Tillier A, Durette-Desset M-C. Phylogeny of the Trichostrongylina (Nematoda) Inferred from 28S rDNA Sequences. Molecular Phylogenetics and Evolution. 2001;19(3):430–42. doi: 10.1006/mpev.2001.0925. [DOI] [PubMed] [Google Scholar]