Abstract
Background
Bovine theileriosis results from infection with obligate intracellular protozoa of the genus Theileria. The phylogenetic relationships between two isolates of Theileria annulata, and 36 Theileria spp., as well as 6 outgroup including Babesia spp. and coccidian protozoa were analyzed using the 18S rRNA gene sequence.
Methods
The target DNA segment was amplified by PCR. The PCR product was used for direct sequencing. The length of the 18S rRNA gene of all Theileria spp. involved in this study was around 1,400 bp.
Results
A phylogenetic tree was inferred based on the 18S rRNA gene sequence of the Iran and Iraq isolates, and other species of Theileria available in GenBank. In the constructed tree, Theileria annulata (Iran vaccine strain) was closely related to other T. annulata from Europe, Asia, as well as T. lestoquardi, T. parva and T. taurotragi all in one clade.
Conclusion
Phylogenetic analyses based on small subunit ribosomal RNA gene suggested that the percent identity of the sequence of Iran vaccine strain was completely the same as Iraq sequence (100% identical), but the similarity of Iran vaccine strain with other T. annulata reported from China, Spain and Italy determined the 97.9 to 99.9% identity.
Keywords: Theileria, Vaccine strain, Phylogenetic Analysis, PCR, 18S rRNA gene
Introduction
Bovine theileriosis results from infection with obligate intracellular protozoa of the genus Theileria. The two most important species in cattle and water buffalo are T. parva, as the agent for causes east coast fever, and T. annulata, which causes tropical theileriosis. Theileria parva occurs in 13 countries in sub-Saharan Africa causing East Coast fever (ECF), Corridor disease, and January disease. Theileria annulata, the cause of tropical theileriosis, occurs in large parts of the Mediterranean coast of north Africa, extending to northern Sudan, and southern Europe. South-eastern Europe, the near and Middle East, India, China and central Asia are also affected (1–3).
Theileria is classified based on microscopic observations, ultrastructural features, lifecycle, geographic region and vertebrate and non-vertebrate host (4). Clinical classification categorizes Theileria to malignant, moderate and benign species. Nonetheless, the exact taxonomic Theileria spp. have been difficult to establish and the subject of considerable debate (5–7). There is a number of factors makes complexity of assigning taxonomic positions, including similar morphology among this group of parasites regardless of vertebrate host, incomplete lifecycle data, serologic tests that are not specific enough to discriminate individual species in the presence of mixed infections, and the difficulty of obtaining pure isolates for studies when the circulating parasitemia may be very low (8–10).
The advances in molecular biology and specially sequence data analysis allowed the researchers to identify and characterize the hemoparasites species in particular Theileria group. Ribosomal RNA is the most abundant constituent of nucleic acids in any non-viral organism with the eukaryotic RNA transcription unit consisting of the large and small subunit (18S rRNA) and the 5.8S rRNA gene (11). The 18S rRNA gene is increasingly accepted as a widely used marker for characterization, taxonomic classification, and phylogenetic analysis and this gene has been sequenced from a variety of different organisms, resulting in a large database for sequence comparisons (6, 10, 12–22). The conserved function and structure of the 18S rRNA molecule allow sequences to be aligned, even among divergent species. However, the molecule also possesses phylogenetically informative variable regions that are useful for determining relationships among species (23).
In this study, we decided to determine the phylogenetic position of T. annulata infected cell line S15 Iran vaccine strain and T. annulata Iraq field isolate with 36 different Theileria spp. and six out-group protozoan parasites using 18S rRNA gene sequences comparison.
Materials and Methods
Parasites
Two T. annulata strain/isolates were used in this study. Theileria annulata infected cell line (S15 Iran vaccine strain, Tehran, Vasfenard) and Iraq T.annulata field isolate were provided from Protozoology and Vaccine Production Department of Razi Institute and Duhok Province, Kurdistan of Iraq respectively.
DNA isolation
Proteinase K and further phenol chloroform purification were performed for DNA extraction (24). Briefly, after treating the cells with lyses buffer, followed by centrifugation, proteinase K and SDS solution was added to the pellet, and then was incubated until most of the cellular protein was degraded. The digest was deproteinized by phenol/chloroform/isoamyl alcohol extraction, recovered by ethanol precipitation, then was dried and resolved in TE buffer. DNA concentration was determined either by agarose gel electrophoresis and spectrophotometry (A260) and measuring the ratio of A260/A280. Moreover, quality of the isolated DNA was evaluated by agarose gel electrophoresis.
PCR Primer Design
The specific primers were designed based on T. annulata 18S ribosomal RNA gene sequence (accession # EU083801) (by CinnaGen, Iran). Two primer pairs were designed in order to span the major hyper variable regions along the 18S ribosomal RNA gene sequence (Gene Runner program, Version 3.05). The first two primers, F1 (5’ GGC GGC GTT TAT TAG ACC 3’) and R1 (5’ TCA ATT CCT TTA AGT TTC AGC C 3’) were used to amplify bases between 186-1093 and the second primers, F2 (5'CAG ATA CCG TCG TAG TCC 3’) and R2 (5’ CCT TGT TAC GAC TTC TCC 3’) were applied to amplify bases between 945-1714 of T. annulata 18S ribosomal RNA gene sequence (EU083801) and these two primers sets covered the majority length of 18S ribosomal RNA gene sequence with 127 bp overlapping.
Polymerase Chain Reaction
PCR was performed in a final reaction volume of 20 µl containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.1% Triton X-100, 200 µM (each) deoxynucleoside triphosphate, 0.5 U of Taq DNA polymerase (CinnaGen, Iran), 10 pmol of each primers, and 2 µl of DNA template. The reactions were performed in an automatic DNA thermal cycler (Techne, Germany) with the first incubation at 94 oC for 3 min and were pursued by 35 cycles. Each cycle consisted of a denaturing step of 20 seconds at 95 oC, an annealing step of 45 seconds at 56 oC, and an extension step of 50 seconds at 72 oC, followed by final extension step of 10 min at 72 oC.
PCR product detection and sequencing
Amplified PCR products were separated by electrophoresis on a 2% agarose gel, stained with ethidium bromide, and visualized by UV transillumination. PCR products were cleaned and extracted from agarose gel and were submitted for bidirectional DNA sequencing by using chain termination method (MWG, Germany). The provided sequences from F1R1 and F2R2, first were merged and offered to be aligned for multiple sequence alignment and phylogenetic study of Theileria spp. and outgroups as well.
Sequence alignment and phylogenetic analysis
The DNA sequences of 18S rRNA gene obtained from two studied T. annulata samples and 42 sequences of 18S rRNA gene sequences including 36 Theileria spp., 4 Babesia spp. and two coccidian protozoa were accessed from GenBank. The sequences were aligned by Clustal W multiple alignments program (25). The alignment was manually edited in BioEdit and truncated to the size of the smallest sequence (∼1,400bp). Phylogenetic tree was constructed by using DNADist Neighbor-Joining method (version 3.6a2.1), sequence identity matrix of all sequences were computed as well (BioEdit phylogeny package, Version 7.0.1).
Nucleotide sequence accession numbers
The 18S rRNA gene sequences of the T. annulata S15 Iran vaccine strain and T. annulata field isolate from Iraq, Duhok have been submitted to GenBank and can be retrieved under accession numbers of HM628581, and HM628582 respectively. All 42 small subunit ribosomal RNA gene sequences were used for this phylogenetic study were listed in Table 1.
Table 1.
The sequence identity values between T. annulata Iran S15 Vaccine Strain and 43 small subunit of Ribosomal RNA gene sequences belong to different Theileria species and four Babesia spp. as well as two outgroup coccidians, Toxoplasma and Isospora. Different Theileria species are categorized in six clades (Q, Y, O, M, B and A) in addition to two outgroup clades for Babesia spp. and Coccidian spp.
| Sequence | Accession number | Clade * | Identity percent ** |
|---|---|---|---|
| T.equi | ab515315 | 90.1 | |
| T.equi | ay534882 | Q | 91.5 |
| T.equi | eu888906 | 88.2 | |
| T.youngi | af245279 | 91.1 | |
| T.bicornis | af499604 | Y | 92.7 |
| T.velifera | af097993 | 95.3 | |
| T.capreoli | ay726011 | 96.4 | |
| T.ovis | ay260171 | 97.3 | |
| T.ovis | ay260172 | 97.3 | |
| T.ovis | eu622911 | O | 97.1 |
| T.ovis | ay533144 | 97.0 | |
| T.ovis | fj603460 | 97.4 | |
| T.mutans | af078815 | 94.2 | |
| T.mutans | fj213586 | M | 92.9 |
| T.buffeli | dq287959 | 95.7 | |
| T.buffeli | ef126184 | 96.1 | |
| T.buffeli | af236094 | 95.9 | |
| T.buffeli | dq104611 | 95.9 | |
| T.buffeli | z15106 | 95.9 | |
| T.buffeli | af236097 | B | 95.9 |
| T.buffeli | fj426360 | 96.6 | |
| T.sergenti | gu143088 | 92.1 | |
| T.sergenti | fj225392 | 95.5 | |
| T.sergenti | eu083803 | 95.8 | |
| T.separata | ay260175 | 95.4 | |
| T.orientalis | ab520958 | 95.9 | |
| T.sinensis | eu274472 | 96.0 | |
| T.parva | l02366 | 98.5 | |
| T.taurotragi | l19082 | 97.9 | |
| T.annulata | eu073963 | 99.8 | |
| T.annulata | eu083799 | 99.4 | |
| T.annulata | eu083800 | 99.8 | |
| T.annulata | eu083801 | A | 99.9 |
| T.annulata | fj426369 | 99.2 | |
| T.annulata | dq287944 | 99.6 | |
| T.annulata , Iran Vaccine Strain | hm628581 | id | |
| T.annulata Iraq | hm628582 | 100.0 | |
| T.lestoquardi | AF081135 | 99.5 | |
| B.ovis | ay150058 | 85.3 | |
| B.motasi | ay533147 | Babesia | 87.0 |
| B.divergens | AY572456 | 88.7 | |
| B.cabali | AY534883 | 87.6 | |
| Toxoplasma gondii | L37415 | Coccidian | 84.4 |
| Isospora suis | U97523 | 83.3 |
The analyzed sequences were grouped in eight major clades, based on 18S rRNA gene sequences
Nucleotide identities are given in percentage
Results
The expected amplicons with sizes of 770 and 908 base pairs (bp) were observed in all of the examined samples (Fig. 1). The sequences of the 18S rRNA gene of the T. annulata Iran vaccine strain and Iraq (Duhok isolate) were determined from the overlapping flanking sequences of two generated PCR fragments. The sequencing of the PCR products yielded 1424 and 1413 base pair length for two Iran and Iraq samples respectively. The sequences were then subjected to phylogenetic analysis by using the BioEdit programme. The sequence identity matrix was also determined and showed 88.2-100% homology between T. annulata 18S rRNA sequence of S15 Iran vaccine strain and Theileria spp. from all over the world (Table 1). The sequence of Iran vaccine strain showed completely same as Iraq sequence (100% identical). But the similarity of Iran vaccine strain with other T. annulata reported 18S rRNA gene sequences from China, Spain and Italy determined the 97.9 to 99.9% identity. The closest similarity between T. annulata Iran vaccine strain and other Theileria species was belonged to T. lestoquardi Iran vaccine strain (AF081135) 99.5% similarity. There were selected 6 outgroup 18S rRNA gene sequences from Babesia ovis, B. motasi, B. cabali, B. divergens, Toxoplasma gondii and Isospora suis those exhibited the most difference among the comparison (88.3-88.7% similarity).
Fig. 1.
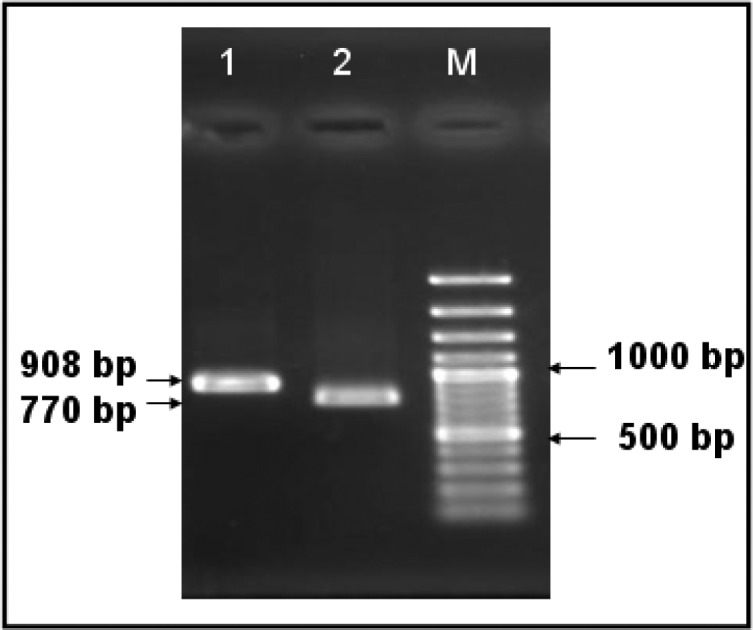
Gel agarose (2%) electrophoresis of amplified 18S rRNA gene sequence of Theileria annulata. Two fragments of 18S rRNA gene sequences were amplified for Theileria annulata. Lane 1; fragment of 908 bp, lane 2; fragment of 770bp and, M; 100 bp DNA ladder as size marker
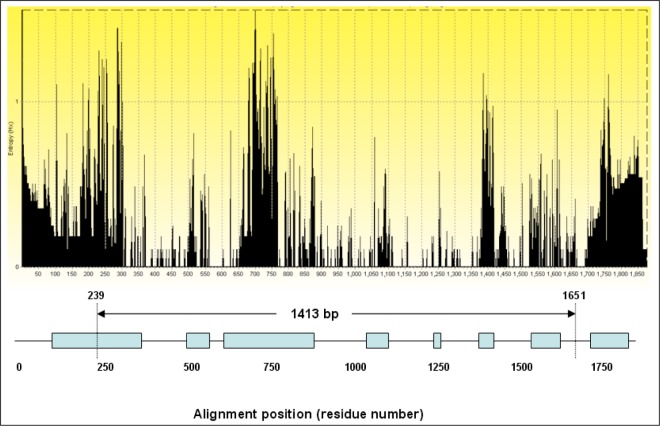
Entropy plot
The alignment was manually edited in BioEdit software and truncated to the size of the smallest sequence (1,413 bp). After resizing the aligned 44 sequences, in order to have the correct comparison, the designed entropy plot was plotted and this design showed the amplified 1413 bp length of the 18S rRNA gene sequence in this study, spans most of the hypervariable regions exist along the alignment (Fig. 2).
Fig. 2.
Plotted entropy shows the 4 major hypervariable regions along the 18S rRNA gene sequence. Two studied T.annulata sequences had 1413bp length. The plot shows the fragment of Theileria annulata have an enough length to cover the majority of hypervariable regions. The boxes are the hypervariable regions along the gene sequence and the arrows demonstrate the region was sequenced and applied for phylogenetic analysis
Sequence alignment and phylogenetic analysis
The phylogenetic tree was constructed based on the T. annulata Iran S15 vaccine strain, Iraq isolate, 36 Theileria spp. and 6 out-group sequences including Babesia spp., Toxoplasma gondii and Isospora suis sequences (Table 1).
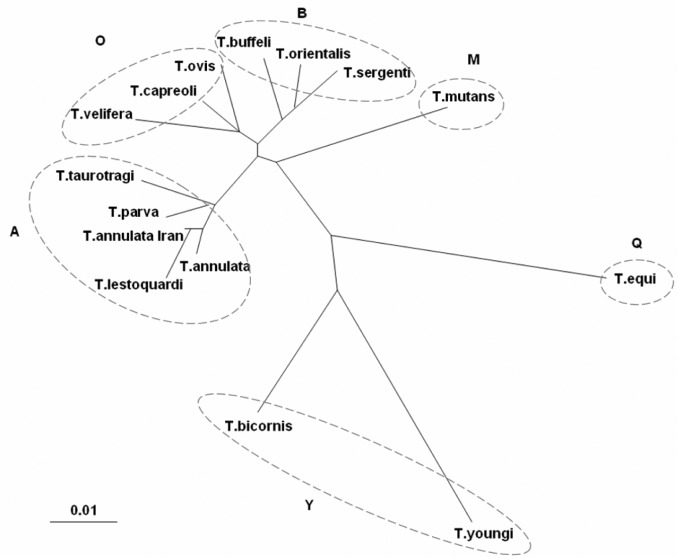
Theileria sequences was divided into six clades in the constructed tree, including “Q” clade; consists T. equi, “Y” clade; includes T. youngi and T. bicornis, “O” clade; contains T. ovis, T. capreoli and T. velifera, “M” clade; includes T. mutans, “B” clade; consists T. buffeli, T. sergenti, T. sinensis, T.separata and T. orientalis, “A” clade; includes T. annulata, T. parva, T. lestoquardi and T. taurotragi, and two more clades for outgroup sequences; “Babesia” clade; consists B. ovis, B. divergens, B. motasi and B. cabali, and “Coccidian” clade; contains Toxoplasma gondii and Isospora suis (Table 1 and Fig. 3).
Fig. 3.
Unrooted phylogenetic tree is constructed using Theileria spp. 18S rRNA gene sequences. Theileria spp. sequences were grouped in six major clades (detailed description is in the text). Scale bar represents nucleotide substitutions per position
There are eight T. annulata 18S rRNA gene sequences in clade “A”, identity percent of T. annulata Iran vaccine strain and other T. annulata in this clade are 99.2% to 100%.
The identity percent between T. annulata Iran strain and T. lestoquardi was 99.5% and this similarity rate was more than the identity percent between T. annulata Iran and Italy or China, Xinjiang isolates.
Discussion
Based on available literature, this is probably the first phylogenetic analysis, molecular characterization of T. annulata infected cell line of Iran vaccine strain, and a field isolate from clinically infected cattle in Iraq, Duhok by using 18S rRNA gene sequence.
One of the most important points in Good Manufacturing Practice (GMP) is to characterize the local vaccine seed in Razi Vaccine and Serum Research Institute, Karaj, Iran. Therefore, we decided to classify the T. annulata Iran vaccine strain cell line. Hence, we focused on to establish the phylogenetic relationships of Iran T. annulata vaccine strain with other Theileria species using 18S rRNA gene sequences.
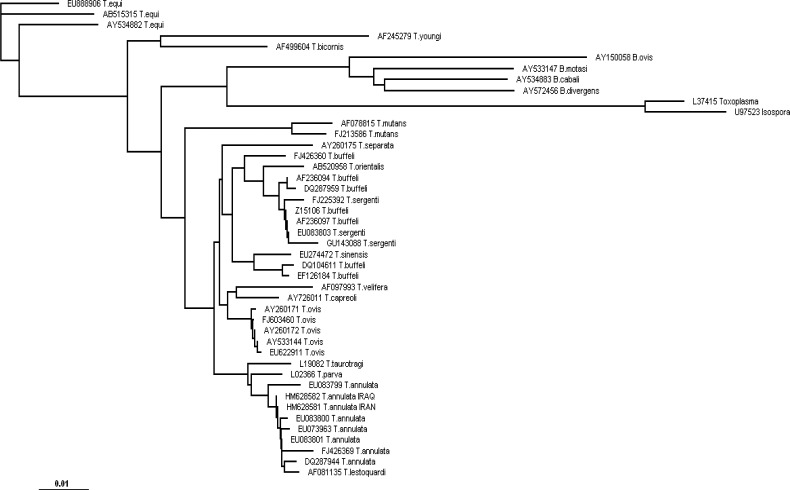
Basically, there are three steps in all phylogenetic analysis; multiple alignment of the sequences, distance calculation and tree construction. Using taxa (the outgroup) that are known to fall outside of the group of interest (the ingroup) is the way to root tree. In this study, we applied Babesia species and two coccidian parasites to root Theileria species.
In addition, Theileria species are host and vector specific (26, 27) but in some countries more than one species can infect animals, which causes a problem in diagnosis and epidemiology. Although recent molecular studies suggest that the genus Theileria including T. annulata, T. parva and T. lestoquardi are very similar in features of microscopic characteristics; these species are phylogenetically distinct and can be differentiated by accurate molecular techniques (28, 29). Although, Theileria spp. are tick-transmitted and the parasite is fully corresponded to the specific hosts, and this association makes a geographical distribution of the Theileria species around the world, but the phylogenetic analysis clearly shows close relationship of different species, T. annulata, T. lestoquardi, T. parva and T. taurotragi in constructed tree all are within clade “A” (Fig. 4).
Fig. 4.
Rooted phylogenetic tree of gene sequences of Theileria annulata S15 Iran vaccine strain, Iraq isolate and other Theileria spp. also outgroup 18S rRNA gene sequences existing in GenBank (Outgroup sequences are the least related to the group of taxa that we are studying). The outgroups are considered for rooting a tree. Scale bar represents nucleotide substitutions per position
In this study, two fragments of 18S rRNA gene sequences of T. annulata 770bp and 908bp were amplified for two examined samples. Nucleotide sequence identity data demonstrated that the Iran S15 vaccine strain (GenBank Accession Nr: HM628581) has nucleotide homology of 99.9% with China, Neimeng (EU083801), 99.8% with China, Ningxia (EU083800), 99.8% with China, Yining (EU073963), 99.2% with Italy (FJ426369), 99.6% with Spain (DQ287944), 99.4% with China, Xinjiang (EU083799) and 100% with Iraq, Duhok isolate (HM628582). From this perspective, the S15 live attenuated vaccine strain has a great homology with other T. annulata isolates; Iraq in neighborhood and China, Spain and Italy far from Iran. But the effectiveness of this vaccine for use in these countries as a valuable vaccine is an objective of future studies.
The minimum identity belongs to T. annulata Italy isolate and maximum similarity was for Iraq isolate. The interesting finding is that all pathogenic Theileria species including T. annulata, T. parva and T. lestoquardi are in “A” clade (Table 1).
Furthermore, T. lestoquardi and T. annulata share an extremely high similarity (99.5%) in their 18S rRNA gene sequences. This finding and the recent occurrence of T. lestoquardi infection in bovine cells might be a good explanation for very high closeness of these two Theileria species.
In the present study, another sequence identity analysis was performed by using a smaller size of 18S rRNA gene sequence of 793 nucleotides. The T. annulata field isolate from Golestan Province of Iran (HM535613) showed the 100% homology with the T. annulata Iran S15 vaccine strain, Iraq Duhok and Turkey isolates (data not shown).
Molecular phylogeny was used to classify Theileria spp. obtained in the Hubei province of China; the results clearly determined that the Theileria spp. from ruminants found in Hubei belonged to the benign group of Theileria spp. (30).
Three Theileria genotypes were phylogenetically analyzed by Nagore et al. in Spain that, sharing 96.7%–97.0% similarity between their 18S rRNA gene sequences: T. ovis, Theileria sp. OT1, and Theileria sp. OT3 (29).
Yin et al. in China suggested that the infective Theileria for small ruminants are comprised of two species with the same morphology, vector and life cycle. Regarding to the classical taxonomy, they should be one species, but the phylogenetic tree showed several Theileria spp. including China1 and 2 that were isolated from sheep and goats (31).
Nijhof et al. showed the cause of deaths in wild ruminants in South Africa that was previously attributed to the genus Cytauxzoon, but phylogenetic analysis has clarified the Theileria is the cause of mortality (32).
In conclusion, according to the data presented here, there are high homologies between T. annulata Iran 18S rRNA gene sequence strain with other T. annulata from above mentioned countries, in particular Iraq and Turkey. Therefore, if this phylogenetic data correlate with immunological response of susceptible cattle, it might be a new sight to find an efficient vaccine to control and prevention of Tropical Theileriosis through molecular epidemiological methods.
Acknowledgment
The author would like to thank Dr. Adel Talib Al-Saeed from College of Veterinary Medicine, Duhok University, for providing the Theileria annulata field isolate from Duhok in Kurdistan Region of Iraq, Dr. Farahi for the kind supply of the T. annulata field isolate from Golestan province of Iran, and all personnel of Parasite Research and Vaccine Production Department of Razi Institute. The author declares that there is no conflict of interests.
References
- 1.Hashemi-Fesharki R. Control of Theileria annulata in Iran. Parasitol Today. 1988;4(2):36–40. doi: 10.1016/0169-4758(88)90062-2. [DOI] [PubMed] [Google Scholar]
- 2.Pipano E, Shkap V. Vaccination against tropical theileriosis. Ann N Y Acad Sci. 2000;916:484–500. doi: 10.1111/j.1749-6632.2000.tb05328.x. [DOI] [PubMed] [Google Scholar]
- 3.Mohammad Al-Saeed AT, Omer LT, Abdo J, Habibi G, Salih DA, Seitzer U, Ahmed J. Epidemiological studies on tropical theileriosis (Theileria annulata infection of cattle) in Kurdistan Region, Iraq. Parasitol Res. 2010;106(2):403–7. doi: 10.1007/s00436-009-1675-7. [DOI] [PubMed] [Google Scholar]
- 4.Allsopp MT, Cavalier-Smith T, De Waal DT, Allsopp BA. Phylogeny and evolution of the piroplasms. Parasitology. 1994;108(Pt 2):147–52. doi: 10.1017/s0031182000068232. [DOI] [PubMed] [Google Scholar]
- 5.Kim SJ, Tsuji M, Kubota S, Wei Q, Lee JM, Onuma M. Sequence analysis of the major piroplasm surface protein gene of benign bovine Theileria parasites in East Asia. Int J Parasitol. 1998;28:1219–27. doi: 10.1016/s0020-7519(98)00095-2. [DOI] [PubMed] [Google Scholar]
- 6.Gubbels MJ, Yin H, Bai Q, Liu G, Nijman IJ, Jongejan F. The phylogenetic position of the Theileria buffeli group in relation to other Theileria species. Parasitol Res. 2002;88:S28–S32. doi: 10.1007/s00436-001-0566-3. [DOI] [PubMed] [Google Scholar]
- 7.Kawazu S, Sugimoto C, Kamio T, Fujisaki K. Analysis of the genes encoding immunodominant piroplasm surface proteins of Theileria sergenti and Theileria buffeli by nucleotide sequencing and polymerase chain reaction. Mol Biochem Parasitol. 1992;56:169–176. doi: 10.1016/0166-6851(92)90164-f. [DOI] [PubMed] [Google Scholar]
- 8.d'Oliveira C, van der Weide M A, Habela P, Jacquiet F. Jongejan. Detection of Theileria annulata in blood samples of carrier cattle by PCR. J Clin Microbiol. 1995;33(10):2665–2669. doi: 10.1128/jcm.33.10.2665-2669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda J, Bakheit MA, Liu Z, Yin H, Mu Y, Guo S, Beyer D, Oliva A, Ahmed JS, Seitzer U. Development of a recombinant indirect ELISA for the diagnosis of Theileria sp. (China) infection in small ruminants. Parasitol Res. 2006;98(6):561–567. doi: 10.1007/s00436-005-0105-8. [DOI] [PubMed] [Google Scholar]
- 10.Chae J, Allsopp BA, Waghela SD, Park J, Kakuda T, Sugimoto C, Allsopp MTEP, Wagner GG, Holman PJ. A study of the systematics of Theileria spp. based upon small subunit ribosomal RNA gene sequences. Parasitol Res. 1999;85:877–883. doi: 10.1007/s004360050651. [DOI] [PubMed] [Google Scholar]
- 11.Waters AP, McCuthan TF. Ribosomal RNA: Natures own polymerase-amplified target for diagnosis. Parasitol Today. 1990;6(2):56–9. doi: 10.1016/0169-4758(90)90071-b. [DOI] [PubMed] [Google Scholar]
- 12.Ellis J, Hefford C, Baverstock PR, Dalrymple BP, Johnson AM. Ribosomal DNA sequence comparison of Babesia and Theileria . Mol Biochem Parasitol. 1992;54:87–95. doi: 10.1016/0166-6851(92)90097-4. [DOI] [PubMed] [Google Scholar]
- 13.Allsopp BA, Baylis HA, Allsopp MT, Cavalier-Smith T, Bishop RP, Carrington DM, Sohanpal B, Spooner P. Discrimination between six species of Theileria using oligonucleotide probes which detect small subunit ribosomal RNA sequences. Parasitology. 1993;107(Pt 2):157–65. doi: 10.1017/s0031182000067263. [DOI] [PubMed] [Google Scholar]
- 14.Kakuda T, Shiki M, Kubota S, Sugimoto C, Brown WC, Kosum C, Nopporn S, Onuma M. Phylogeny of benign Theileria species from cattle in Thailand, China and the USA based on the major piroplasm surface protein and small ribosomal RNA genes. Int J Parasitol. 1998;28:1261–7. doi: 10.1016/s0020-7519(98)00113-1. [DOI] [PubMed] [Google Scholar]
- 15.Katzer F, McKellar S, Kirvar E, Shiels B. Phylogenetic analysis of Theileria and Babesia equi in relation to the establishment of parasite populations within novel host species and the development of diagnostic tests. Mol Biochem Parasitol. 1998;95:33–44. doi: 10.1016/s0166-6851(98)00085-1. [DOI] [PubMed] [Google Scholar]
- 16.Chae J, Lee J, Kwon O, Holman PJ, Waghela SD, Wagner GG. Nucleotide sequence heterogeneity in the small subunit ribosomal RNA gene variable (V4) region among and within geographic isolates of Theileria from cattle, elk and white-tailed deer. Vet Parasitol. 1998;75:41–52. doi: 10.1016/s0304-4017(97)00183-0. [DOI] [PubMed] [Google Scholar]
- 17.Chae J, Levy M, Hunt JR, Schlater J, Snider G, Waghela SD, Holman PJ, Wagner GG. Theileria sp. infection a ssociated with bovine fatalities in 76 the United States confirmed by small subunit rRNA gene analyses of blood and tick samples. J Clin Microbiol. 1999;37:3037–3040. doi: 10.1128/jcm.37.9.3037-3040.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chae J, Waghela SD, Craig TM, Kocan AA, Wagner GG, Holman PJ. Two Theileria cervi SSU rRNA gene sequence types found in isolates from white-tailed deer and elk in North America. J Wildlife Dis. 1999;35:458–465. doi: 10.7589/0090-3558-35.3.458. [DOI] [PubMed] [Google Scholar]
- 19.Chansiri K, Kawazu S, Kamio T, Terada Y, Fujisaki T, Philippe H, Sarataphan N. Molecular phylogenetic studies on Theileria parasites based on small subunit ribosomal RNA gene sequences. Vet Parasitol. 1999;83:99–105. doi: 10.1016/s0304-4017(99)00052-7. [DOI] [PubMed] [Google Scholar]
- 20.Stockham SL, Kjiemtrup AM, Conrad DA, Schmidt Scott MA, Robinson TW, Tyler JW, Johnson GC, Carson CA, Cuddihee P. Theileriosis in a Missouri beef herd caused by Theileria buffeli: case report, herd investigation, ultrastructure, phlyogenetic analysis and experimental transmission. Vet Pathol. 2000;37:11–21. doi: 10.1354/vp.37-1-11. [DOI] [PubMed] [Google Scholar]
- 21.Schnittger L, Yin H, Jianxun L, Ludwig W, Shayan P, Rahbari S, Voss-Holtmann A, Ahmed JS. Ribosomal small-subunit RNA gene-sequence analysis of Theileria lestoquardi and a Theileria species highly pathogenic for small ruminants in China. Parasitol Res. 2000;86:352–358. doi: 10.1007/s004360050680. [DOI] [PubMed] [Google Scholar]
- 22.Gubbels MJ, Hong Y, Weide M, Qi B, Nijman IJ, Guangyuan L, Jongejan F. Molecular characterization of the Theileria buffeli / orientalis group. Int J Parasitol. 2000;30:943–952. doi: 10.1016/s0020-7519(00)00074-6. [DOI] [PubMed] [Google Scholar]
- 23.Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66(4):411–53. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Vol. 3. Cold Spring Harbor; NY: Cold Spring Harbor Laboratory; 1989. Molecular Cloning: A Laboratory Manual; p. B.16. [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop R, Musoke A, Morzaria S, Gardner M, Nene V. Theileria: intracellular protozoan parasites of wild and domestic ruminants transmitted by ixodid ticks. Parsitology. 2004;129(Suppl):S271–83. doi: 10.1017/s0031182003004748. S.P. [DOI] [PubMed] [Google Scholar]
- 27.Morzaria. Identification of Theileria species and characterization of Theileria parva stocks. http://www.fao.org/wairdocs/ILRI/x5549E/x5549e0T.htm.
- 28.Bakheit MA, Scholzen T, Ahmed JS, Seitzer U. Molecular characterization of a Theileria lestoquardi gene encoding for immunogenic protein splice variants. Parasitol Res. 2006;100(1):161–170. doi: 10.1007/s00436-006-0255-3. [DOI] [PubMed] [Google Scholar]
- 29.Nagorea Daniel, García-Sanmartína Josune, García-Péreza Ana L, Justea Ramón A, Hurtado Ana. Identification, genetic diversity and prevalence of Theileria and Babesia species in a sheep population from Northern Spain. Int J Parasitol. 2004;34(9):1059–1067. doi: 10.1016/j.ijpara.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Zhou YQ, He GS, Oosthuizen MC, Zhou DN, Zhao JL. Molecular phylogenetic studies on Theileria spp. isolates (China) based on small subunit ribosomal RNA gene sequences. Trop Anim Health Prod. 2010;42(1):109–14. doi: 10.1007/s11250-009-9392-x. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Luo JX, Schnittger L, Lu BY, Beyer D, Ma ML, Guan GQ, Bai Q, Lu CP, Ahmed J. Phylogenetic analysis of Theileria species transmitted by Haemaphysalis qinghaiensis. Parasitol Res. 2004;2:36–42. doi: 10.1007/s00436-003-0900-z. [DOI] [PubMed] [Google Scholar]
- 32.Nijhof AM, Pillay V, Steyl J, Prozesky L, Stoltsz WH, Lawrence JA, Penzhorn BL, Jongejan F. Molecular characterization of Theileria species associated with mortality in four species of African antelopes. J Clin Microbiol. 2005;43(12):5907–11. doi: 10.1128/JCM.43.12.5907-5911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]