Abstract
The molecular basis for recognition by human P2Y1 receptors of the novel, competitive antagonist 2′-deoxy-N6-methyladenosine 3′,5′-bisphosphate (MRS 2179) was probed using site-directed mutagenesis and molecular modeling. The potency of this antagonist was measured in mutant receptors in which key residues in the transmembrane helical domains (TMs) 3, 5, 6, and 7 were replaced by Ala or other amino acids. The capacity of MRS 2179 to block stimulation of phospholipase C promoted by 2-methylthioadenosine 5′-diphosphate (2-MeSADP) was lost in P2Y1 receptors having F226A, K280A, or Q307A mutations, indicating that these residues are critical for the binding of the antagonist molecule. Mutation of the residues His132, Thr222, and Tyr136 had an intermediate effect on the capacity of MRS 2179 to block the P2Y1 receptor. These positions therefore appear to have a modulatory role in recognition of this antagonist. F131A, H277A, T221A, R310K, or S317A mutant receptors exhibited an apparent affinity for MRS 2179 that was similar to that observed with the wild-type receptor. Thus, Phe131, Thr221, His277, and Ser317 are not essential for antagonist recognition. A computer-generated model of the human P2Y1 receptor was built and analyzed to help interpret these results. The model was derived through primary sequence comparison, secondary structure prediction, and three-dimensional homology building, using rhodopsin as a template, and was consistent with data obtained from mutagenesis studies. We have introduced a “cross-docking” procedure to obtain energetically refined 3D structures of the ligand–receptor complexes. Cross-docking simulates the reorganization of the native receptor structure induced by a ligand. A putative nucleotide binding site was localized and used to predict which residues are likely to be in proximity to agonists and antagonists. According to our model TM6 and TM7 are close to the adenine ring, TM3 and TM6 are close to the ribose moiety, and TM3, TM6, and TM7 are near the triphosphate chain.
Introduction
The physiological actions of extracellular ATP1,2 have been comprehensively described and include contractile regulation of the heart, as well as vascular and visceral smooth muscle, excitatory and inhibitory effects on neurons of the central and peripheral nervous system, and activation of neuroendocrine secretion. These effects are apparently mediated by a family of G-protein-coupled receptors (P2Y) and ligand-gated ion channels (P2X).3 Seven subtypes of P2X receptors and at least six distinct subtypes of P2Y receptors have been cloned. P2Y1 receptors were the first in the series to be cloned4 and were shown to have the seven transmembrane helical domain (TM) structure common for the superfamily of rhodopsin-like G-protein-coupled receptors.5 Like other P2Y receptors, P2Y1 receptors activate phospholipase C (PLC), which generates inositol phosphates and diacylglycerol from phosphatidyl-inositol(4,5)bis-phosphate.6
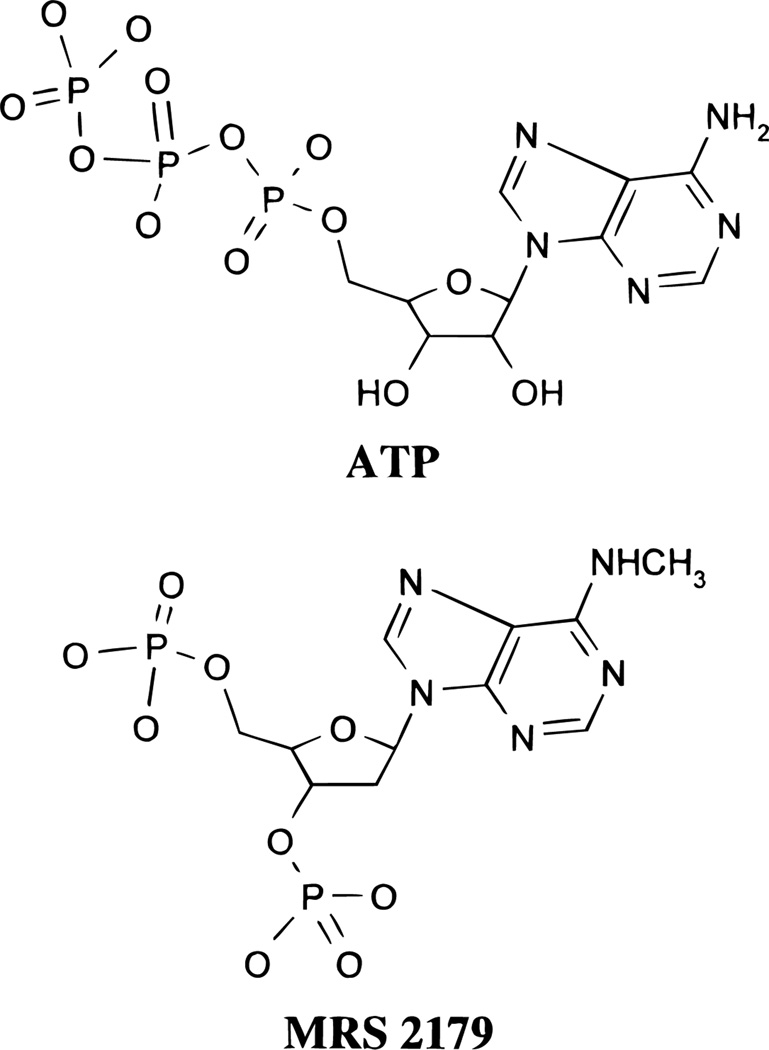
Detailed structural studies of P2Y1 receptors have been limited by the lack of selective P2Y1 antagonists.7 Recently, we have synthesized a new, selective antagonist for P2Y1 receptors: 2′-deoxy-N6-methyladenosine 3′,5′-bisphosphate (MRS 2179),8 the chemical structure of which is shown in Figure 1. A Ki of approximately 100 nM was determined for MRS 2179 acting as an antagonist at the PLC-coupled P2Y1 receptor in turkey erythrocyte membranes.8 Due to its potency and structural similarity to agonists, we chose MRS 2179 as a ligand to probe the P2Y1 receptor binding pocket.
Figure 1.
Chemical structures of ATP and MRS 2179 (charges not shown).
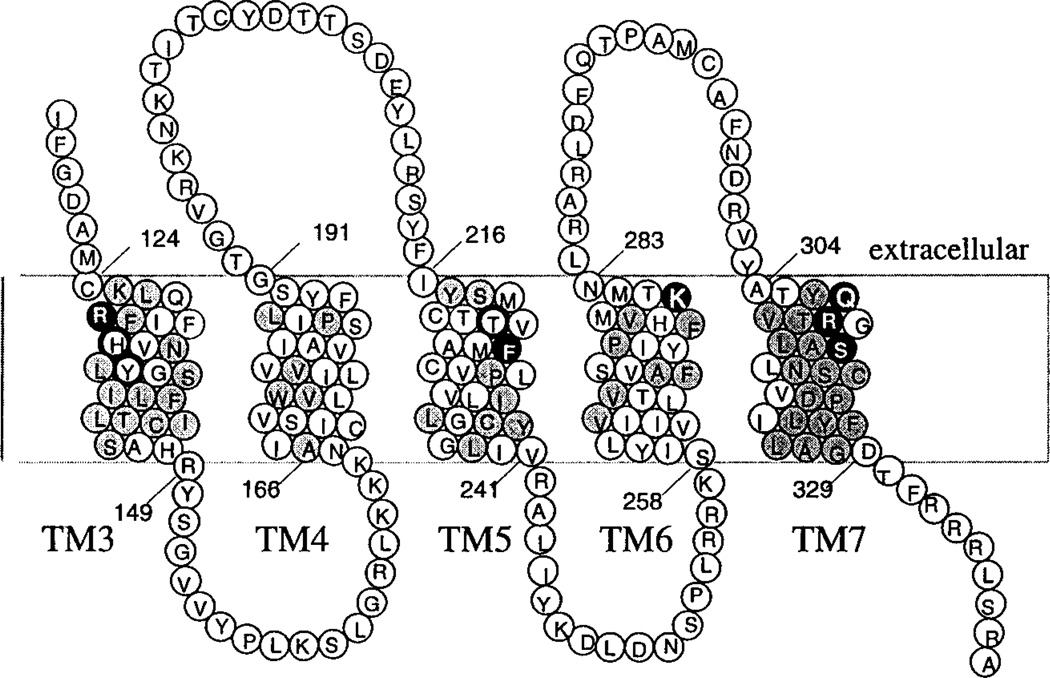
Site-directed mutagenesis of human P2Y receptors was utilized for localizing agonist recognition elements in two previous studies.9,10 Amino acid residues in TM3, TM5, TM6, and TM7 were found to be involved in nucleotide binding to P2Y1 receptors.10 The potent P2Y1 receptor agonist 2-methylthioadenosine 5′-triphosphate (2-MeSATP) had no activity in cells expressing the R128A, R310A, and S314A mutant receptors (see Figure 2), and a markedly reduced potency of 2-MeSATP was observed with the K280A and Q307A mutant receptors. These and additional residues in the same transmembrane regions were selected to probe the influence of individual side chains on the molecular recognition of the antagonist MRS 2179 in the present study. Site-directed mutagenesis studies have been used in combination with molecular modeling to predict the environment of agonist and antagonist recognition pockets of the P2Y1 receptor.
Figure 2.
Partial topology of the human P2Y1 receptor showing residues proposed to be involved in recognition of the antagonist MRS 2179. Mutation sites in which replacement by alanine significantly impedes the ability of MRS 2179 to block activation of the receptor are highlighted. Gray shading indicates residues that are conserved between P2Y1 and P2Y2 receptors. Solid highlighted circles indicate a 1–3-fold change in the EC50 for 2-MeSADP in the presence of 1 µM MRS2179; circles with thick outline, a 3–6-fold change; solid nonhighlighted circles, a 6–12-fold change. Residues R128, R310, and S314 are highlighted; however the effects of antagonist at these Ala mutant receptors could not be evaluated experimentally due to lack of activation by agonist. The effects of non-Ala substitutions on the potency of MRS 2179 are S314T, moderate reduction; R310K, identical to wild-type receptors.
Results
Functional Characterization of Mutant P2Y1 Receptors
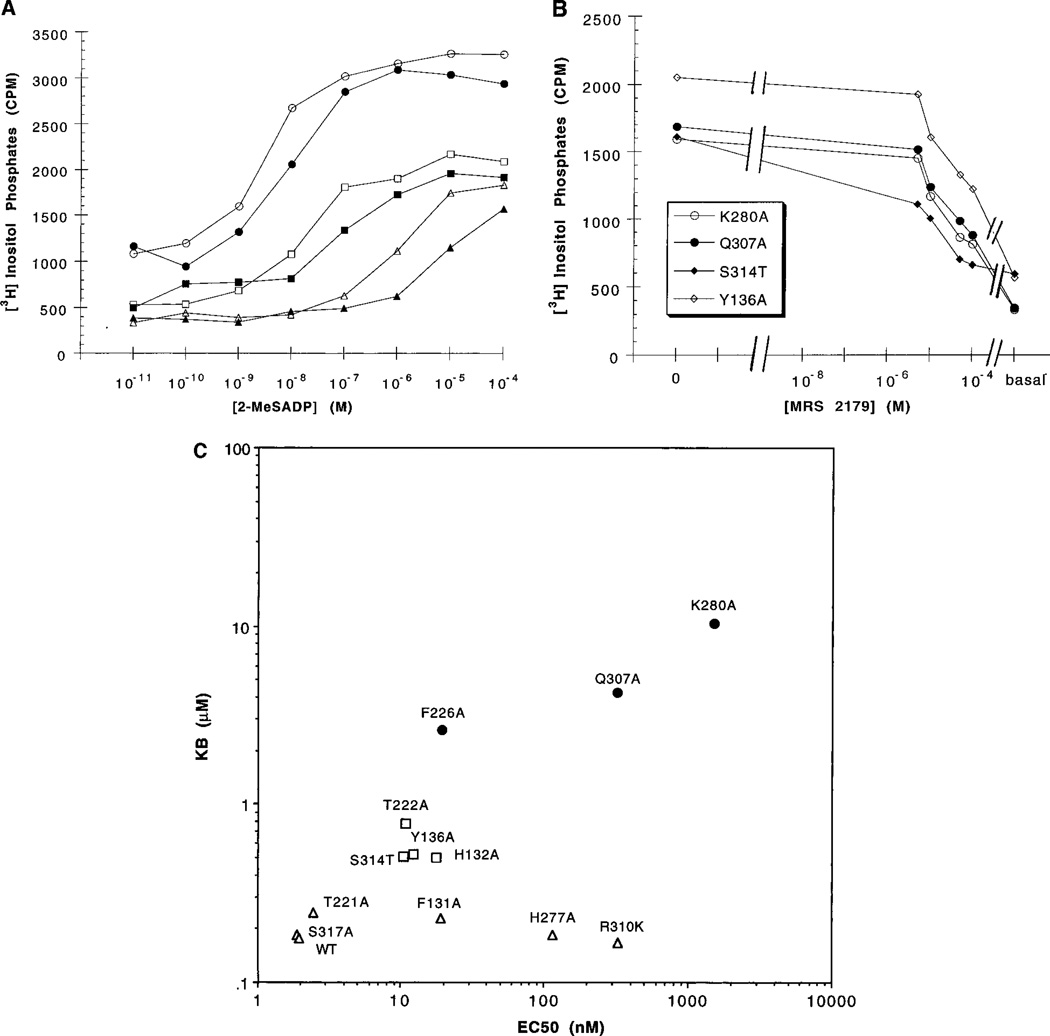
Site-directed mutagenesis was carried out by standard methodology10–16 using oligonucleotide primers. Wild-type and mutant HA-tagged human P2Y1 receptors were expressed in Cos-7 cells and shown to be present in the plasma membrane by ELISA measurements, as shown previously.10 The agonist 2-MeSADP, previously shown to be 5-fold more potent than 2-MeSATP at this receptor,10 displayed an EC50 value for stimulation of PLC17 in wild-type receptors of 1.94 nM (Table 1). Recently, we have synthesized 2′-deoxy-N6-methyladenosine 3′,5′-bisphosphate, MRS 2179, as a selective and competitive P2Y1 antagonist.8 The potency of this nucleotide antagonist was measured in the wild-type (Figure 3A) and mutant (Figure 3A,B) human P2Y1 receptors. In wild-type receptors the antagonist MRS 2179 at a concentration of 1 µM resulted in a 10-fold right shift of the concentration–response curve of 2-MeSADP. This shift corresponded to a KB value for MRS 2179 of 0.177 µM. Thus, the potency of MRS 2179 as a competitive antagonist at the human P2Y1 receptor was similar to its reported potency at the turkey P2Y1 receptor.8
Table 1.
Antagonism of the Activation of Phospholipase C in Mutant Human P2Y1 Receptorsa
| EC50 (2-MeSADP, nM) |
KBe,f (MRS 2179, µM) |
||||
|---|---|---|---|---|---|
| constructb | residuec | n | aloned | +MRS 2179 (1 ±M) | |
| WT | 4 | 1.94 ± 0.80 | 12.9 ± 2.6** | 0.177 | |
| F131A | 3.32 | 3 | 19.2 ± 0.7 | 103 ± 20* | 0.229 |
| H132A | 3.33 | 4 | 17.7 ± 4.7 | 53.0 ± 9.1** | 0.501 |
| Y136A | 3.37 | 4 | 12.3 ± 3.7 | 36.0 ± 8.3** | 0.519 |
| T221A | 5.42 | 3 | 2.45 ± 0.60 | 12.4 ± 1.1** | 0.246 |
| T222A | 5.43 | 2 | 10.9 ± 1.1 | 25.0 ± 0.2** | 0.773 |
| F226A | 5.47 | 3 | 19.5 ± 1.9 | 27.0 ± 4.7 | >1 |
| H277A | 6.52 | 6 | 115 ± 29 | 738 ± 4** | 0.185 |
| K280A | 6.55 | 6 | 1520 ± 310 | 1580 ± 110** | 10.4g |
| Q307A | 7.36 | 7 | 323 ± 53 | 378 ± 46** | 4.25g |
| R310K | 7.39 | 4 | 324 ± 41 | 2270 ± 840** | 0.166 |
| S314T | 7.43 | 5 | 10.4 ± 1.3 | 30.9 ± 5.1** | 0.507 |
| S317A | 7.46 | 4 | 1.88 ± 0.35 | 12.1 ± 0.5** | 0.184 |
Data are presented as means ± SD of two to seven independent experiments, each performed in duplicate.
All constructs contain the HA-epitope tag sequence at the N-terminus and a hexa-His tag at the C-terminus.10
Using the sequence identifier as defined in ref 10.
Agonist at the highest concentrations caused only a small increase or no change in inositol phosphates in the following mutants: R128A (2.29); R310A (7.39); R310S/S314R (7.39/7.43); S314A (7.43).
Calculated according to the Schild equation: KB = [antagonist]/((EC50 with antagonist/EC50 agonist alone) − 1).
EC50 values, as follow, for the stimulation of PLC by 2-MeSADP at each of the following mutant receptors, in the presence of 50 µM MRS 2179: 188 ± 18 nM (Y136A); 8.82 ± 3.51 µM (K280A); 4.12 ± 0.35 µM (Q307A); or in the presence of 20 µM MRS 2179: 106 nm (S314T).
KB values for MRS 2179 were determined using increases in EC50 for 2-MeSADP in the presence of 50 µM MRS 2179 (see footnote f).
Statistical significance: *p < 0.05;
p < 0.005.
Figure 3.
Stimulation of phospholipase C by 2-MeSADP in human P2Y1 receptors. Transfected COS-7 cells were incubated for 30 min at 37 °C (see the Experimental Procedures for details). Data are presented as absolute accumulations of tritiated inositol phosphates above basal levels in the absence of 2-MeSADP for representative experiments. EC50 values (average of two to four independent experiments, each carried out in duplicate) are given in Table 1. (A) Concentration–response curves for stimulation of PLC in the absence (open symbols) or presence (closed symbols) of MRS 2179 in COS-7 cells transiently expressing wild-type HA-tagged-hP2Y1 (○) or mutant hP2Y1 receptors (K280A, △; F131A, □) Concentrations of MRS 2179 used were K280A, 50 µM; wild-type and F131A, 1 µM. (B) Concentration–response curves for antagonism by MRS 2179 of agonist-induced stimulation of PLC in four mutant receptors. Concentrations of 2-MeSADP used were: K280A, 5 µM (○); Q307A, 2.5 µM (●); S314T, 100 nM (◆); Y136A, 1 µM (◇). (C) Correlation of agonist (EC50, nM) vs antagonist (KB, µM) potencies at mutant hP2Y1 receptors (mutations having little or no effect on antagonist affinity, △; mutations having an intemediate effect on antagonist affinity; □, mutations having the largest effect on antagonist affinity, ●). An estimated KB value for F226A of 2.6 µM has been used.
The effects of point mutations in the P2Y1 receptor on antagonist recognition were examined and correlated with effects on agonist potency (Figure 3C). These mutant receptors can be divided into three groups based on the effects observed on antagonist potency. For most mutations there is a linear relationship between EC50 for agonist vs KB for antagonist. Thus, the effect of most mutations on agonist potency is proportional to the effect on antagonist affinity. However, F131A, H277A, and R310K mutant receptors show essentially the same antagonist affinity as wild-type receptors, while agonist affinity is markedly reduced.
Residues Essential for Antagonist Recognition
Although 2-MeSADP is a potent activator of the F226A mutant receptor,10 1 µM MRS 2179 had no significant effect (p = 0.06) on the potency of 2-MeSADP in the stimulation of PLC (<2-fold) in cells expressing this mutant receptor. Thus, amino acid residue Phe226 is apparently more critical for recognition of the antagonist than for agonist. At K280A and Q307A mutant receptors, 1 µM MRS 2179 was also ineffective as an antagonist of PLC activation. For the K280A, Q307A, and Y136A mutant receptors, additional concentration–response curves for 2-MeSADP were measured at a much higher concentration of MRS 2179 (50 µM). This concentration was selected based on the concentration effect relationships for the antagonist at fixed concentrations of 2-MeSADP (Figure 3B) at these three and at the S314T mutant receptors. Approximate IC50 values (µM) for antagonism of PLC activation by MRS 2179 were 5 (S314T), 42 (Y136A), 30 (K280A), and 42 (Q307A). In these experiments, the concentration of agonist was 3.3- (K280A), 7.7- (Q307A), 9.6- (S314T), and 81- (Y136A) fold greater than the EC50 value at each mutant receptor in the absence of MRS 2179. It was not feasible to measure the effect of MRS 2179 at the R128A, R310A, and S314A mutant receptors, because the agonist did not fully activate these receptors.10
Mutations Modulatory to Antagonist Recognition
At the H132A, Y136A, T222A, and S314T mutant receptors, MRS 2179 (1 µM) produced an intermediate shift, i.e. a 2- to 3-fold reduction in agonist potency, of the concentration response curve of 2-MeSADP. Thus, the His132, Tyr136, and Thr222 residues located in TM3 and TM5 appear to have a modulatory role in recognition of this antagonist. Steric requirements are present at Ser314, since the Thr substitution reduced the affinity of the antagonist.
Mutations That Do Not Affect Antagonist Recognition
The shift in EC50 of 2-MeSADP promoted by 1 µM MRS 2179 at the F131A, T221A, H277A, R310K, and S317A mutant receptors was nearly identical to that observed in wild-type receptors (roughly an order of magnitude reduction in agonist potency). Thus, the residues Phe131, Thr221, His277, and Ser317 are not essential for recognition of MRS 2179, and at amino acid residue Arg310, a side chain of similar positive charge (Lys) could be substituted with no effect on antagonist potency.
Model of the Agonist Recognition Site
On the basis of the structure of rhodopsin which has sequence homology to GPCRs, we have derived a human P2Y1 receptor model using the Sybyl program18 and other computational methods,19–28 and docked ATP, the natural agonist, in the hypothetical binding site, in a fashion that is consistent with all available pharmacological data.
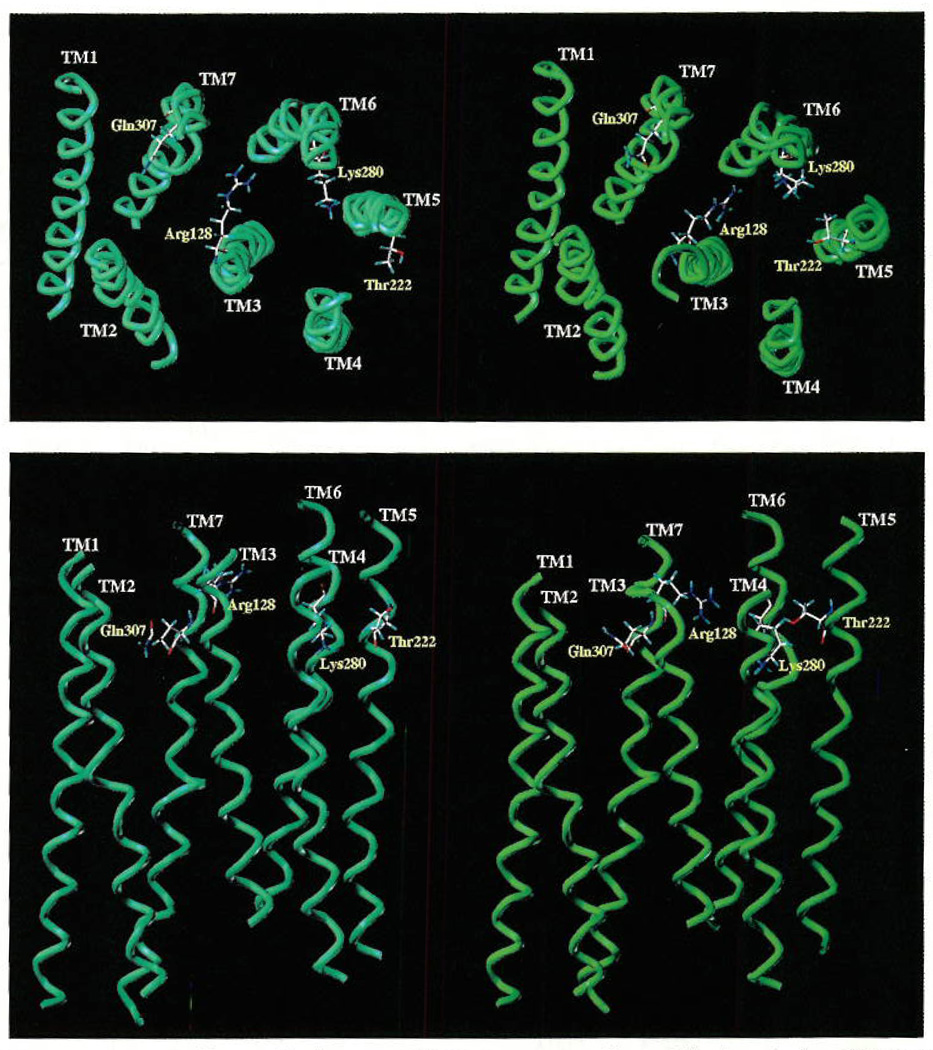
Although a three-dimensional rhodopsin-like model of the chick P2Y1 receptor was published previously,5 our description of ligand/P2Y1 receptor interactions has been improved by including additional computational steps to explore the reorganization of the native receptor structure induced by the ATP coordination (cross-docking). Figure 4 shows the 3D structural models of the human P2Y1 receptor before and after the application of cross-docking with ATP. Several geometric parameters were unaffected by cross-docking: the total length of the membrane-spanning region is about 40 Å; the interhelical distance between the pairs of adjacent helical axes is about 10 Å, consistent with a common interhelical contact distance;25 the interhelical angles, measured between the principal axes of adjacent helices, are between −150° and 170° for antiparallel and between 10° and 25° for the parallel helices (typical of a 3–4 type helix–helix contact associated with optimal interactions between nearly parallel aligned helices).25 Each helix maintained almost the same position and tilting found in the published rhodopsin 2D electron density map.26,27 TM5 in the ATP-bound cross-docked model has been rotated clockwise 60° about its transmembrane axis with respect to the ligand-free receptor model. Consequently, the position of Thr222 is shifted inside the helical bundle. This residue seems to be moderately important in the coordination of the γ-phosphate of ATP, as demonstrated by site-directed mutagenesis.10 Moreover, in the cross-docked model TM3, TM4, TM6, and TM7 were rotated clockwise 5°, 15°, 10°, and 5°, respectively, about its transmembrane axis with respect to the ligand-free receptor model. The energy of cross-docked ATP–receptor complex structure is about 65 kcal/mol lower with respect to the original one.
Figure 4.
Stereoview of human P2Y1 receptor transmembrane helical bundle model viewed along the helical axes from the extracellular end (A, top) and perpendicular to the helical axes (B, bottom), before (left) and after (right) the “cross-docking” procedure for the P2Y1–ATP complex (see the Experimental Procedures for details). The docked ATP molecule is not shown. Side chains of some amino acids important for ligand recognition are highlighted.
As in the earlier modeling study of van Rhee et al.,5 ATP was present in the anti conformation (χ, the torsion angle of the glycosidic bond C9–N9–C1′-O4′ was −3.8°), consistent with the typical conformation based on crystallographic data for protein-bound nucleotides. The ring puckering, defined by the dihedral angle C1′–C2′–C3′–C4′ was 18.8°, resulting in a 2′-exo, 3′-endo (3T2, North) conformation of the tetrahydrofuran ring.
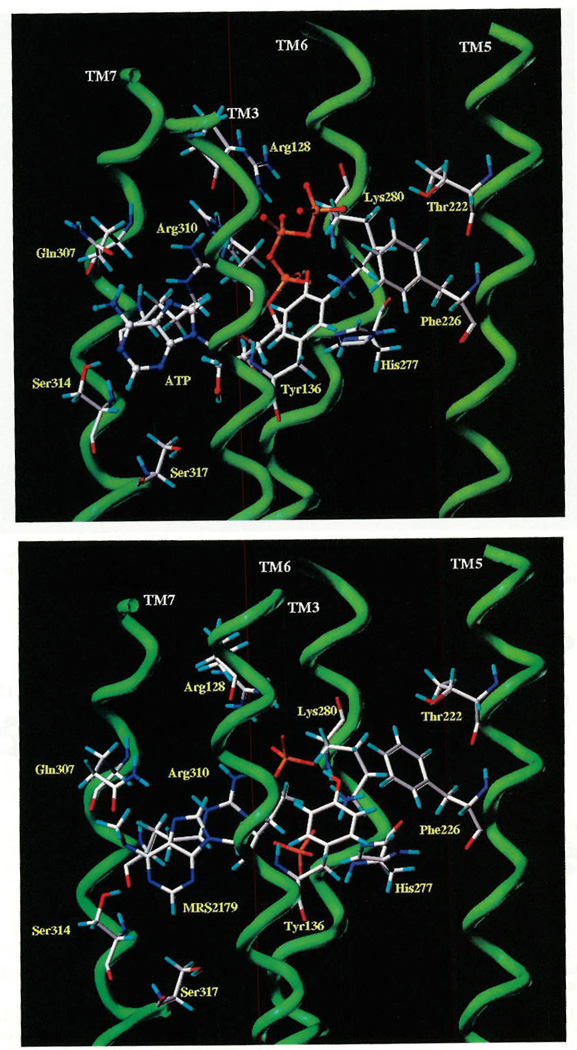
Figure 5A represents the final helical bundle with ATP docked into the putative ligand binding cavity. The putative orientation of bound nucleotide is slightly different from that predicted in the previous modeling study,5 based on new specific interactions proposed as a result of our site-directed mutagenesis studies.10 As shown in Figure 5A, the recognition of ATP seems to occur in the upper region of the helical bundle. The adenine moiety of the ATP molecule is most favorably oriented perpendicular to the plane of the lipid bilayer, with the γ-phosphate of the triphosphate chain in proximity to the exofacial portion of TM5 and the adenosine moiety near the midpoints of TM6 and TM7. From Figure 5A it is possible to distinguish three different parts of the transmembrane domain responsible for ATP recognition: TM6 and TM7 are close to the adenine ring; TM3 and TM6 are close to the ribose moiety; and TM3, TM6, and TM7 are near the triphosphate chain.
Figure 5.
(A, Top) Side view of the P2Y1–ATP complex model. The side chains of the important residues in proximity to the docked ATP molecule are highlighted and labeled. Residues in proximity (≤5 Å) to the docked ATP molecule: adenine Q307, R310, S314; ribose H132, H277, S317; triphosphate R128, Y136, T222, K280, R310. (B, Bottom) Side view of P2Y1–MRS 2179 complex model. The side chains of the important residues in proximity to the docked MRS 2179 molecule are highlighted and labeled. Residues in proximity (≤5 Å) to the docked MRS2179 molecule: adenine Q307, R310, S314; ribose H132, H277; bisphosphate R128, Y136, K280, R310 (R128 and K280 present different conformations with respect to the ATP binding model).
There appear to be three favorable interactions between the adenine moiety of ATP and the receptor. The side chain of Gln307(TM7) is within hydrogen-bonding distance of the N6 atom at 2.7 Å, and the side chain of Ser314(TM7) is positioned 2.0 Å from the N1 atom of the purine ring. Arg310(TM7) is within ionic coupling range to both N7 and N9 atoms of the adenine moiety. Three residues are tentatively implicated in the coordination of the ribose structure. The side chain of His132(TM3) and O4′ are separated by 3.2 Å, His277-(TM6) and O3′ are separated by 3.6 Å, and the Ser317-(TM7) is within hydrogen-bonding distance of O2′ at 3.9 Å. The triphosphate side chain appears to be coordinated by Arg128 (TM3; 1.8 Å, O3γ and 1.7 Å, O2β), Tyr136 (TM3; 3.6 Å, O2α), Thr222 (TM5; 4.1 Å, O2γ), Lys280 (TM6; 1.7 Å, O3α), Arg310 (TM7; 2.6 Å, O3β).
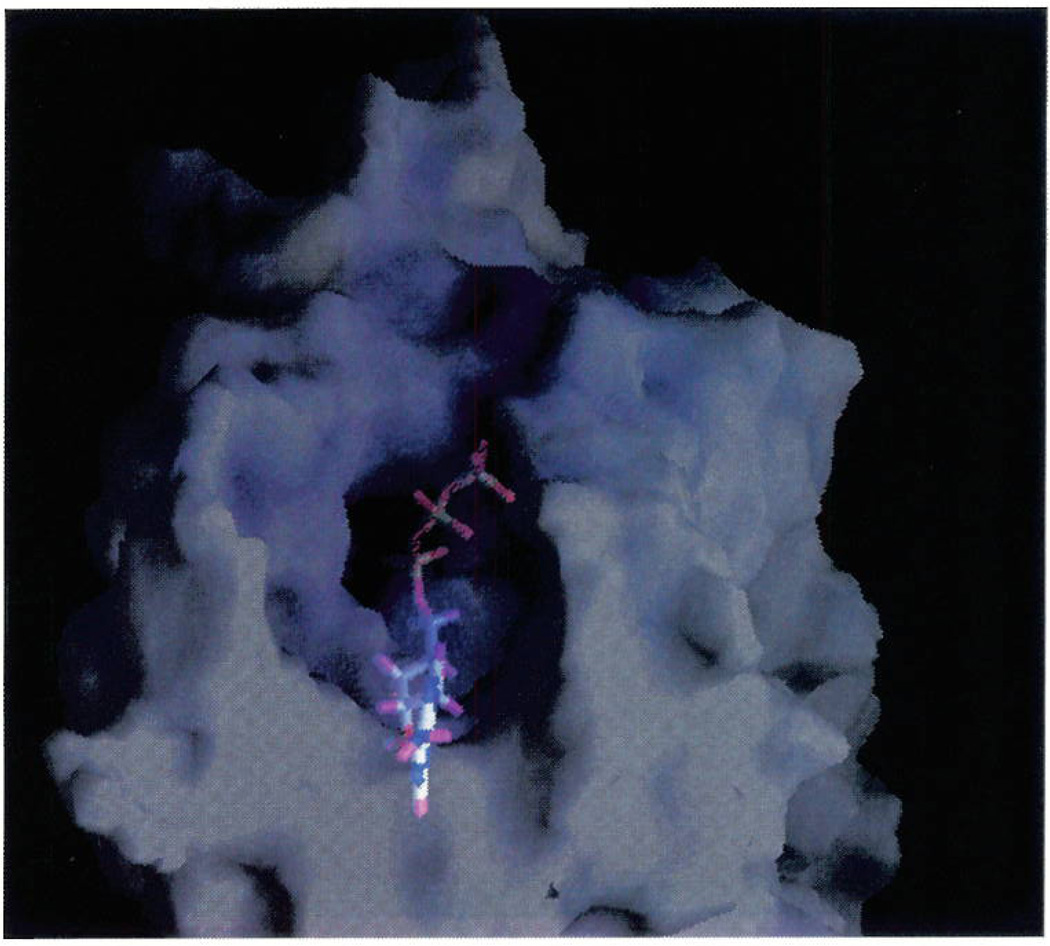
The presence of three important basic residues (Arg128, Lys280, and Arg310) near the extracellular environment seems to indicate that these amino acids are essential to the coordination of the triphosphate moiety. Figure 6 shows the molecular surface of the P2Y1 receptor model color coded by electrostatic potential as calculated using GRASP25 with Amber all-atoms atomic charges. The view is from the extracellular side directly into the binding site, and the structure of ATP is highlighted. It is interesting to note the depth of the pocket and its positive electrostatic nature (blue color). The positive electrostatic potential of the binding cavity overlaps very well with the negative potential generated by the negative charges of the triphosphate moiety of ATP.
Figure 6.
Surface potential of P2Y1–ATP complex model, displayed with GRASP.25 The molecular surface is color coded by electrostatic potential. Potentials less than −20 kT are red, those greater than 20 kT are blue, and neutral potentials (0 kT) are white. The ATP binding site is clearly distinguishable as a region of intense positive potential.
We also docked 2-MeSATP into the helical bundle. 2-MeSATP is 350-fold more potent than ATP in activating P2Y1 receptors.2,30 We have tried to rationalize the higher potency of 2-MeSATP vs ATP by comparing the geometry of the two structures docked within the binding cavity. The methylthio derivative has almost the same position as the natural ligand, although the coordination environment is slightly different. The presence of the bulky methylthio substituent at the 2-position shifts the molecule further away from TM7 compared to ATP, as demonstrated by the increased distance of 3.4 Å between the side chain of the Ser314-(TM7) and the N1 atom of the adenine ring. At the same time, more favorable hydrogen-bonding interactions are possible between the side chains of Gln307(TM7) and Ser314(TM7) with the N6 atom (distance 2.0 and 2.6 Å respectively) of the 2-MeSATP. Also the interactions between Arg128 (TM3; 1.7 Å, O3γ and 1.6 Å, O2β) and Thr222 (TM5; 3.1 Å, O2γ) with the triphosphate chain seem to be more geometrically favorable with respect to those observed for ATP.
Model of the Antagonist Recognition Site
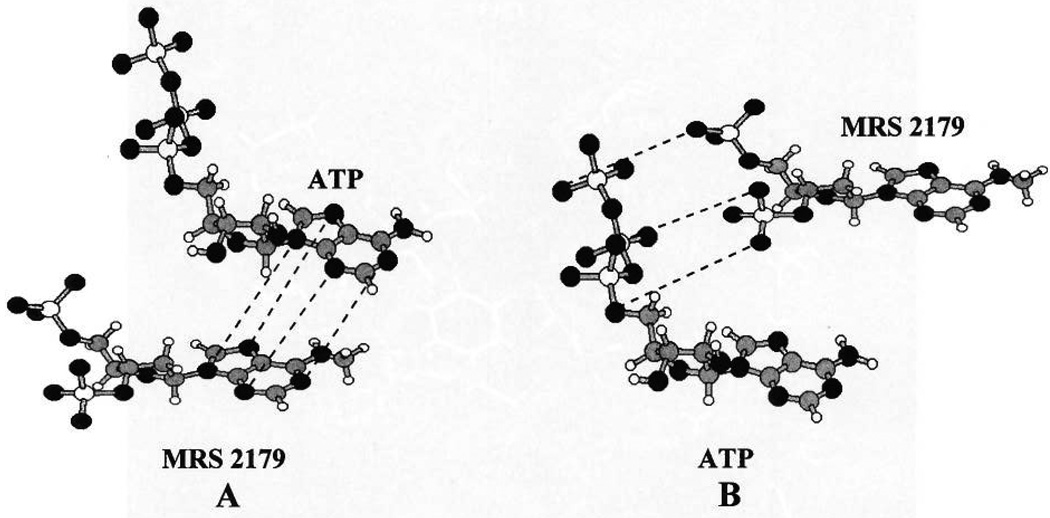
The possible modes of recognition of the 3′,5′-bisphosphate MRS 2179 in the P2Y1 binding pocket was examined. There are two possible ways of overlapping ATP and MRS 2179: one in which all the adenosine ring atoms are perfectly overlayed and another in which ATP α-and γ-phosphorus atoms are overlayed to 3′- and 5′-phosphorus atoms of MRS 2179 (see Figure 7). Ligand/receptor complexes with the two different arrangements of the MRS 2179 structure were constructed and optimized as previously described. The complex corresponding to the adenosine overlay appeared more stable by approximately 40 kcal/mol. Consistent with their structural similarity, the cross-docking procedure demonstrated that the receptor architecture found for ATP recognition was energetically appropriate also for MRS 2179. Figure 5B represents the lowest energy docked complex of MRS 2179 in the proposed ligand binding cavity. In the MRS 2179 cross-docked model, TM3, TM4, TM5, TM6, and TM7 were rotated clockwise 10°, 15°, 55°, 5°, and 5°, respectively, about their transmembrane axes with respect to the ligand-free receptor model. The energy of cross-docked MRS 2179–receptor complex structure is about 50 kcal/mol lower with respect to the original one. MRS 2179 was present in the anti conformation (χ, the torsion angle of the glycosidic bond C9–N9–C1′-O4′ was −6.5°), consistent with the typical conformation based on crystallographic data for protein-bound nucleotides. The ring puckering, defined by the dihedral angle C1′–C2′–C3′–C4′ was 22.3°, resulting in a 2′-exo, 3′-endo (3T2, North) conformation of the tetrahydrofuran ring.
Figure 7.
Possible superpositions of ATP and MRS 2179. (A) Superposition of all adenosine ring atoms or (B) superposition of ATP α- and γ-phosphate atoms with 3′- and 5′-phosphate of MRS 2179.
In this model, the side chain of Gln307(TM7) is within hydrogen-bonding distance of the N6 atom at 1.7 Å, and the side chain of Ser314(TM7) is positioned at 1.9 Å from the N1 atom and at 2.7 Å from the N6 of the purine ring. Another three amino acids are important for the coordination of the phosphate groups in this antagonist: Arg128(TM3), Tyr136(TM3), and Lys280(TM6). As shown in Figure 5B, Lys280 may interact directly with both 3′- and 5′-phosphates (1.6 Å, O3′ and 1.6 Å, O5′), whereas Arg128(TM3) is within ionic coupling range to both the O2 and O3 atoms of the 5′-phosphate. Tyr136 is also within hydrogen-bonding distance of the 5′-phosphate at 3.8 Å. Thr222 is positioned at 4.8 Å from the 5′-phosphate group of MRS 2179.
Discussion
Fundamental understanding of the molecular details of ligand/GPCR interactions remains very rudimentary. How an agonist binds and transforms a resting GPCR into its active form and the microscopic basis of binding site blockade by an antagonist are generally still unclear. In the absence of high-resolution structural knowledge of GPCRs, such questions can be addressed only by building models, which are tested through pharmacological and biochemical studies. Structural models can be used to describe the interatomic interactions between a ligand and its receptor. We have extended our P2Y1 receptor model to describe the possible manner in which the nucleotide coordination information is transmitted through the receptor. We have sought to improve the description of ligand/receptor interactions by introduction of a simple means to simulate the reorganization of the native receptor structure accompanying ligand recognition. Our results illustrate that cross-docking can be used to predict local structural changes induced by a ligand in a receptor binding site. As shown in Figure 4, the presence of a bound nucleotide favors a simultaneous adjustment in the orientation of TM3, TM5, TM6, and TM7. The most pronounced change upon docking of ATP, i.e. rotation of TM5, is probably promoted by electrostatic interactions between the phosphate groups and the basic amino acids of the binding cavity. Rotations and translations of the TM domains are crucial factors in the ligand recognition process in different GPCRs, as recently proposed by Gouldson et al.31 Consequently, our approach to docking is designed to mimic the natural domain movement within the receptors.
It appears that the (tri)phosphate moiety of ATP and other nucleotide derivatives are the key structure responsible for the binding and activation of the P2Y1 receptor. The negatively charged triphosphate chain is likely coordinated to basic amino acids in the TMs, as proposed previously,5,9,10 and may also be bound to the cationic side chains of the basic amino acids of the extracellular loops possibly oriented toward the receptor cavity. Our modeling study suggests that Arg128(TM3), Thr222(TM5), Lys280 (TM6), and Arg310(TM7) are likely candidates for this counterion function. In particular, Arg128(TM3) tentatively coordinates the α- and β-phosphate, Thr222(TM5) the γ-phosphate, Lys280-(TM6) the α- and β-phosphate, and Arg310(TM7) the β-phosphate. Consistent with this model, the R128A and R310A mutant receptors are not activated by agonists, and a markedly reduced response is observed with the K280A mutant.10 Moreover, T221A and T222A mutant receptors exhibit much larger reductions in triphosphate, rather than di- or monophosphate potency.10 This result may be indicative of a greater role of these TM5 residues in γ-phosphate recognition. Thr222(TM5) and Lys280(TM6) in the human P2Y1 receptor are equivalent to Asn253 and Phe182 in the human A2A receptor, both of which were implicated by Kim et al. in adenosine binding.29 Our model also reveals a high degree of coordination of the β-phosphate and, probably, its crucial role in the binding process. This observation is supported by the fact that ADP is more potent than ATP for activation of the human P2Y1 receptor and that AMP is a very weak agonist for this receptor.10
Gln307(TM7) and Ser314(TM7) are positioned, in our model, in the vicinity of the N6 amine of the adenine moiety. We speculate that these two amino acids may be involved in the recognition of the nucleotide base in the agonist structure. The important role of the exo-cyclic NH6 of the adenine moiety, putatively through Gln307 as a hydrogen-bond acceptor, has been demonstrated using a doubly alkylated N6 derivative of ATP, for which no agonist activity was observed.5 Moreover, a markedly reduced response of 2-MeSADP was observed for the Q307A mutant receptor compared with the wild-type receptor.10
A hydrogen bond between the N1 atom of the adenine ring and Ser314(TM7) is proposed in our model. The S314A mutant receptor indeed exhibits low affinity for agonist ligands.10 Arg310(TM7) can also interact with both N7 and N9 atoms of the adenine ring. It is to be noted that residues Gln307, Arg310, and Ser314 of the human P2Y1 receptor align with Tyr271, Ile274, and His278 in the human A2A receptor sequence, which have been shown by mutagenesis to be involved in ligand binding.29 Previous studies of A2A receptors have shown that TM7 is the critical region for the interaction of the adenine moiety in agonist recognition.29
As previously reported, substitution at the 2-position of the adenine ring of the agonist derivatives is tolerated and in some cases favored for P2Y receptor agonists.30 For example, 2-MeSADP is more potent than ADP, and 2-MeSATP is more potent than ATP. We suggest that the presence of the bulky methylthio substituent at the 2-position shifts the position of the agonist inside the binding cavity farther away from TM7, and this change enhances the electrostatic interaction between the basic amino acids, Arg128 and Lys280, and the negative charges of the phosphate side chain.
Tyr136(TM3) can also interact with the α-phosphate group of the ATP molecule. We suggest that this interaction is a “secondary” interaction. In fact, alanine replacement of Phe131(TM3), His132(TM3), Tyr136-(TM3), Phe226(TM5) or His277(TM6) resulted in mutant receptors that exhibited 7–18-fold reductions in the potency of 2-MeSATP compared with the wild-type receptor.10 These five residues in TM3, TM5, and TM6 are all close to the ATP binding site and seem to play a modulatory role in ligand binding to the P2Y1 receptor.
From the present modeling study, the antagonist MRS 2179 can overlap the ATP structure by superimposition of all adenine ring atoms (Figure 7). If this hypothesis is correct, all amino acids important in the recognition of the adenine moiety of the ATP molecule should also be important for MRS 2179 binding. Accordingly, the antagonist at 1 µM did not affect the potency of 2-MeSADP in the Q307A and S314T mutant receptors, indicating that these residues are important for the binding of the antagonists. This is additional evidence that the TM7 region may be very important for the accommodation of the adenine moiety for both agonists and antagonists.
We speculate that hydrogen bonding occurs between the N6 position of the adenine moiety and the NH2 of Gln307. The apparent affinity of antagonists related to MRS 2179 is dramatically dependent on the size of the N6 substituent.8 For example, the N6 propyl group completely abolished antagonist properties, and double alkylation of the N6 amino group resulted in a 300-fold decrease in apparent affinity of MRS 2179. It is possible to rationalize these results from our model by proposing an unfavorable steric interaction between the large N6 substituent and the side chain of Gln307.
Ser314(TM7) in the P2Y1 receptor–MRS 2179 complex can interact with both the N6 and N1 atoms of the adenine moiety, but the N1 position seems to be preferred. This is in accordance with the experimental evidence that the N1-methyl analogue of 2′-deoxyadenosine 3′,5′-bisphosphate is a weak antagonist (13-fold loss of potency vs the corresponding adenosine bisphosphate).8 The proximity between Ser314 and the adenine moiety is also supported by the fact that the S314T mutation reduces the antagonist affinity.
Previously reported structure–activity relationships for adenosine bisphosphates at P2Y1 receptors8 suggest that either 2′- or 3′-deoxy modifications are completely tolerated in the case of antagonists, while 2′- and 3′-deoxyATP are less potent (6.9- and 27-fold, respectively) than ATP. For nucleotide antagonists, phosphate groups at either 2′- or 3′-positions are nearly equivalent and also equally essential as a 5′-phosphate group for antagonist properties. Our model suggests that two amino acids are important for the coordination of the phosphate groups: Arg128(TM3) and Lys280(TM6). In particular, Lys280 can interact directly with both 3′- and 5′-phosphates, and Arg128 is directly involved in the coordination of the 5′-phosphates. The high flexibility of the side chain of Lys280, and consequently the possibility that the amino group can also interact with the 2′-phosphate, may be the reason for the antagonist activity of both 2′,5′- and 3′,5′-bisphosphates.
Thr222 seems to be too far from the 5′-phosphate group (4.8 Å) to participate in a direct, strong hydrogen-bonding interaction with the ligand, but only one water molecule would be sufficient to bridge the two groups. Phe226(TM5) is also located in proximity to the 5′-phosphate group. Since a hydrophobic interaction occurring between the receptor and this portion of the nucleotide is untenable, we speculate that this amino acid has an important function in delineating the binding site cavity. It is likely that Arg128(TM3) and Arg310(TM7) are very important for the binding of MRS 2179, just as they are for agonist. However, the lack of responsiveness of these Ala mutant receptors to 2-Me-SADP made it impossible to measure antagonist-binding properties.
Tyr136(TM3) seems to be modulatory for antagonist potency, just as for agonist potency. In our model, Tyr136 can interact with the 5′-phosphate group of MRS 2179.
Alanine replacement of Phe131(TM3), His132(TM3), His277(TM6), or Ser317(TM7) resulted in mutant receptors that exhibited apparent antagonist affinities similar to those observed wisth the wild-type receptor. Thus, these residues are not essential for antagonist recognition. Mutation of Ser317 is inconsequential to activation;10 however, the aromatic residues Phe131, His132, and His277 are modulatory for agonist action.10 We speculate that coordination of residues in TM3 and TM6 by the nucleotide may be related to conformational changes leading to receptor activation. The differential effects on antagonist affinity and agonist potency in F131A, H277A, and R310K mutant receptors (Figure 3C) do not appear to be due to reduced intrinsic activity in these mutant receptors, since maximal efficacy noted for three different agonists in these mutant receptors reached levels similar to that type of wild-type receptors.10
Taken together, these results suggest that the adenosine and α-phosphate moieties are critical for the binding of both agonists and antagonists to the P2Y1 receptor and that the presence of β- and γ-phosphates are probably essential for the agonist activity of ATP. MRS 2179 and its related molecules are the only clearly competitive antagonists at the P2Y1 receptor. Other known antagonists such as PPADS, suramin, etc. likely have different binding requirements at the receptor.8 Further studies may differentiate these requirements and prove helpful for antagonist drug development.
Conclusions
Binding of the adenine nucleotide antagonist MRS 2179 to the human P2Y1 receptor involves many of the same amino acid residues that are important for recognition of agonist ATP derivatives: Lys280 and Gln307 are very important, and His132, Tyr136, and Thr222 are modulatory. Furthermore, residue S317 is not involved in binding of either agonists or the antagonist MRS 2179. However, differences between agonist and antagonist recognition were observed. In particular, Phe226 in TM5 is more important for the antagonist MRS 2179 than for agonists, and residues Phe131, Thr221, and His277 are less important for the antagonist than for agonists. This has allowed us to construct rhodopsin-based molecular models of the P2Y1–MRS 2179 and P2Y1–ATP complexes, allowing for minor adjustments of the helical positions upon nucleotide binding. The binding modes for these two ligands are very similar, especially in the adenine region; however a rotation of the antagonist molecule allows direct interaction of both 5′- and 3′-monophosphate groups with the same positively charged residues of the receptor that are putatively involved in recognition of the 5′-triphosphate moiety.
Experimental Procedures
Molecular Biological Methods
Single amino acids of the human P2Y1 receptor cDNA (pcDNA3P2Y1) were mutated as previously described.10 Briefly, the coding region of pCDNA3P2Y1 was subcloned into the pCD-PS expression vector,12 yielding pCDP2Y1. All mutations were introduced into pCDP2Y1 using standard polymerase chain reaction (PCR) techniques.10,13 Oligonucleotides (Bioserve Biotechnologies, Laurel, MD) were designed and used to generate a PCR fragment, which was then used to replace the corresponding wild-type P2Y1 sequence. The accuracy of all PCR-derived sequences was confirmed by dideoxy sequencing of the mutant plasmids.14 In addition to single amino acid replacement within the TMs, the N-terminus of the receptor was appended with an epitope tag, consisting of a 9-amino acid sequence derived from the influenza virus hemagglutinin (HA) protein (TAC CCA TAC GAC GTG CCA GAC TAC GCG; peptide sequence: YPYDVPDYA) which was inserted after the first Met residue.10,11 A HexaHis tag15 was also included at the C-terminus immediately after the Leu residue resulting in a construct potentially suitable for affinity chromatography using a chelated nickel column.
Mutant receptors were transiently expressed in COS-7 cells transfected approximately 24 h after seeding with plasmid DNA (4 µg of DNA/dish) using the DEAE-dextran method.16 Cells were grown for an additional 48 h at 37 °C in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum.
Pharmacological and Immunological Methods
Inositol phosphate determination was used as the measure of activity of the ligands at the mutant receptors. Stimulation by the agonist 2-methylthioadenosine 5′-diphosphate (2-Me-SADP) (RBI, Natick, MA) and its antagonism by MRS 2179 (synthesized as the ammonium salt, as described8) were measured in the wild-type and mutant receptors. The assay was carried out according to the general approach of Harden et al.6 using [3H]-myo-inositol (American Radiolabeled Chemicals, St. Louis, MO) in the presence of 10 mM LiCl for 30 min at 37 °C, 5% CO2. The [3H]inositol monophosphate fraction was isolated by anion exchange chromatography17 and eluted with 4.5 mL of 0.1 M formic acid/0.2 M ammonium formate. Pharmacological parameters were analyzed using the Kaleida-Graph program (Abelbeck Software, version 3.01). Statistical analysis was performed using the unpaired t-test (Prism, GraphPad, San Diego, CA).
Indirect cellular ELISA measurements were employed to demonstrate that the wild-type and mutant receptors were expressed in the plasma membrane with the proper orientation of N-terminus. Approximately 72 h after transfection, cells were fixed in 4% formaldehyde, incubated with HA-specific monoclonal antibody (12CA5, Boehringer-Mannheim, Indianapolis, IN), and then washed and incubated with a 1:2000 dilution of a peroxidase-conjugated goat anti-mouse IgG antibody (Sigma, St. Louis, MO) for 1 h at 37 °C.10,29 Color reaction took place in the presence of hydrogen peroxide and o-phenylenediamine (each 2.5 mM in 0.1 M phosphate–citrate buffer, pH 5.0) and was measured bichromatically in the BioKinetics reader (EL 312, Bio Tek Instruments, Inc., Winooski, VT) at 490 and 630 nm (baseline).
Computational Methods
The human P2Y1 receptor model was built and optimized using Sybyl 6.318 and Macro-model 5.019 modeling packages, respectively, based on the approach described by van Rhee et al.5 All calculations were performed on a Silicon Graphics Indigo2 R8000 workstation. Briefly, transmembrane domains were identified with the aid of Kyte–Doolittle hydrophobicity20 and Emini20 surface probability parameters. Transmembrane helices were built from the sequences and minimized individually. The minimized helices were then grouped together to form a helical bundle that matching the overall characteristics of the electron density map of rhodopsin. The helical bundle was minimized using the Amber21 all-atoms force field until the rms value of the conjugate gradient (CG) was <0.1 kcal/mol per Å. A fixed dielectric constant = 4.0 was used throughout these calculations.
A model of adenosine 5′-triphosphate (ATP) was constructed from the crystallographic coordinates of ATP-phosphoglycerate kinase.22 2-MeSATP and MRS 2179 models were constructed from ATP using the “Sketch Molecule” module of Sybyl. The ligands were then fully minimized using AM123 of MOPAC24 and were rigidly docked into the helical bundle using graphical manipulation coupled to continuous energy monitoring (Dock module of Sybyl). When a final position was reached, consistent with a local energy minimum, the complexes of receptor and ligand were subjected to an additional CG minimization run of 300 steps. Partial atomic charges for the ligands were imported from the MOPAC output files.
We wanted to explore possible ligand-induced rearrangements of 7TM bundle by sampling 7TM conformations in the presence of the docked ligands. The docking process was repeated for each model agonist– and antagonist–receptor complex after manually adjusting the relative positions of TM3, TM4, TM5, TM6, and TM7 around the initial docked ligands (our cross-docking procedure). Cross-docking was carried out using the Dock module of Sybyl. Each helix was separated from the ligand–receptor complex structure, and its relative position was changed until a new lower energy geometry was obtained. These adjustments consisted of small translations and rotations of the principal axis of the helix with respect to its original position. When a new final position was reached, consistent with the lowest local energy minimum, the separated helix was merge again into the ligand–receptor complex. The hydropathy profile for the new oriented helix was checked using Kyte–Doolittle method.20 The new complex was subjected to an additional CG minimization run of 300 steps. This procedure was repeated for TM3, TM4, TM5, TM6, and TM7. The manual adjustments were followed by 25 ps of molecular dynamics (MD module of Macromodel) performed at a constant temperature of 300 K using time step of 0.001 ps and a dielectric constant = 4.0. This procedure was followed by another sequence of CG energy minimization to a gradient threshold of <0.1 kcal/mol per Å. Energy minimization of the complexes was performed using the Tripos (SYBYL) and AMBER all-atom (MacroModel) force field. The three-dimensional energy-minimized structures appear to be force-field-independent, particularly in the binding region, as judged by backbone superposition with SYBYL’s root-mean-square superposition procedure (rms = 0.014–0.22 Å).
Supplementary Material
Acknowledgment
S. Moro is grateful to Gilead Sciences (Foster City, CA) for a postdoctoral fellowship grant. The authors are grateful for helpful comments and encouragement provided by Dr. Robert Pearlstein of the Center for Molecular Modeling (DCRT), NIH. We thank Dr. Carsten Hoffmann and Dr. Ivar von Kügelgen of Molecular Recognition Section, NIDDK/NIH, for helpful discussion.
Abbreviations
- ATP
adenosine 5′-triphosphate
- DMEM
Dulbecco’s modified Eagle’s medium
- ELISA
enzyme-linked immunosorbent assay
- FBS
fetal bovine serum
- GPCR
G-protein-coupled receptor
- HA
hemagglutinin
- HBSS
Hank’s balanced salt solution
- 2-Me-SADP
2-methylthioadenosine 5′-diphosphate
- 2-Me-SATP
2-methylthioadenosine 5′-triphosphate
- MRS 2179
2′-deoxy-N6-methyladenosine 3, 5′-bisphosphate
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PLC
phospholipase C
- PPADS
pyridoxal-5′-phosphate-6-azophenyl-2, 4-disulfonate
- rms
root-mean-square
- TM
transmembrane helical domain
- Tris
tris-(hydroxymethyl)aminomethane
Footnotes
Supporting Information Available: Coordinate files for models of complexes of MRS 2179, ATP, and 2-MeSATP with the human P2Y1 receptor (63 pages). Ordering information is given on any current masthead page.
References
- 1.Burnstock G. In: Purinergic Approaches in Experimental Therapeutics. Jacobson KA, Jarvis M, editors. New York: Wiley; 1997. [Google Scholar]
- 2.Fredholm BB, Abbracchio MP, Burnstock G, Daly JW, Harden TK, Jacobson KA, Leff P, Williams M. Nomenclature and classification of purinoceptors. Pharmacol. Rev. 1994;46:143–156. [PMC free article] [PubMed] [Google Scholar]
- 3.Burnstock G, King BF. Numbering of cloned P2 purinoceptors. Drug Dev. Res. 1996;38:67–71. [Google Scholar]
- 4.Webb TE, Simon J, Krishek BJ, Bateson AN, Smart TG, King BF, Burnstock G, Barnard EA. Cloning and functional expression of a brain G protein-coupled ATP receptor. FEBS Lett. 1993;324:219–225. doi: 10.1016/0014-5793(93)81397-i. [DOI] [PubMed] [Google Scholar]
- 5.van Rhee AM, Fischer B, van Galen PJM, Jacobson KA. Modelling the P2Y purinoceptor using rhodopsin as template. Drug Design Discovery. 1995;13:133–154. [PMC free article] [PubMed] [Google Scholar]
- 6.Harden TK, Hawkins PT, Stephens L, Boyer JL, Downes P. Phosphoinositide hydrolysis by guanosine 5′-(gamma-thio) triphosphate-activated phospholipase C of turkey erythrocyte membranes. Biochem. J. 1988;252:583–593. doi: 10.1042/bj2520583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobson KA, Kim Y-C, Camaioni E, van Rhee AM. Structure activity relationships of P2 receptor agonists and antagonists. Chapter 4 in The P2 Nucleotide Receptors. In: Turner JT, Weisman G, Fedan J, editors. The Receptors. Clifton, NJ: Humana Press; 1997. pp. 81–107. [Google Scholar]
- 8.Camaioni E, Boyer JL, Mohanram A, Harden TK, Jacobson KA. Deoxyadenosine-bisphosphate derivatives as potent antagonists at P2Y1 receptors. J. Med. Chem. 1998;41:183–190. doi: 10.1021/jm970433l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erb L, Garrad R, Wang YJ, Quinn T, Turner JT, Weisman GA. Site-directed mutagenesis of P2U purinoceptors—positively charged amino acids in transmembrane helix-6 and helix-7 affect agonist potency and specificity. J. Biol. Chem. 1995;270:4185–4188. doi: 10.1074/jbc.270.9.4185. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Q, Guo D, Lee BX, van Rhee AM, Kim YC, Nicholas R, Schachter J, Harden TK, Jacobson KA. Mutational analysis of residues essential for ligand recognition at the human P2Y1 receptor. Mol. Pharmacol. 1997;52:499–507. doi: 10.1124/mol.52.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schachter JB, Li Q, Boyer JL, Nicholas RA, Harden TK. Second messenger cascade specificity and pharmacological selectivity of the human P2Y1-purinoceptor. Br. J. Pharmacol. 1996;118:167–173. doi: 10.1111/j.1476-5381.1996.tb15381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okayama H, Berg PA. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol. Cell. Biol. 1983;3:280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higuchi R. Using PCR to Engineer DNA. In: Ehrlich HA, editor. PCR Technology. New York: Stockton Press; 1989. pp. 61–70. [Google Scholar]
- 14.Sanger R, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robeva AS, Woodard R, Luthin DR, Taylor HE, Linden J. Double tagging recombinant A1- and A2A-adenosine receptors with hexahistidine and the FLAG epitope. Development of an efficient generic protein purification procedure. Biochem. Pharmacol. 1996;51:545–555. doi: 10.1016/0006-2952(95)02235-x. [DOI] [PubMed] [Google Scholar]
- 16.Cullen BR. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 17.Berridge MJ, Dawson RM, Downes CP, Heslop JP, Irvine RF. Changes in the levels of inositol phosphates after agonist-dependent hydrolysis of membrane phosphoinositides. Biochem J. 1983;212:473–482. doi: 10.1042/bj2120473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The program SYBYL 6.3 is available from TRIPOS Associates. St. Louis, MO: 1993. [Google Scholar]
- 19.Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. MacroModel-An Integrated Software System for Modeling Organic and Bioorganic Molecules using Molecular Mechanics. J. Comput. Chem. 1990;11:440–450. [Google Scholar]
- 20.Kyte J, Doolittle RF. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Weiner SJ, Kollman PA, Nguyen DT, Case DA. An all-atom force field for simulation of protein and nucleic acids. J. Comput. Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 22.Watson HC, Walker NP, Shaw PJ, Bryant TN, Wendell PL, Fothergill LA, Perkins RE, Conroy SC, Dobson MJ, Tuite MF. Sequence and structure of yeast phosphoglycerate kinase. EMBO J. 1982;1:1635–1640. doi: 10.1002/j.1460-2075.1982.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewar MJSE, Zoebisch G, Healy EF. AM1: A new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985;107:3902–3909. [Google Scholar]
- 24.MOPAC 6.0 available from Quantum Chemistry Program Exchange. [PubMed] [Google Scholar]
- 25.Chiota C, Levitt M, Richardson D. Helix to helix packing in proteins. J. Mol. Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- 26.Schertler GF, Villa C, Henderson R. Projection structure of rhodopsin. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 27.Urger VM, Hargrave PA, Baldwin JM, Schertler GFX. Arrangement of rhodopsin transmenbrane α-helices. Nature. 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls A. Grasp: Graphical Representation and Analysis of surface Properties. New York: Columbia University; 1992. [Google Scholar]
- 29.Kim J, Wess J, van Rhee MA, Schoneberg T, Jacobson KA. Site-directed mutagenesis identifies residues involved in ligand recognition in the human A2a adenosine receptor. J. Biol. Chem. 1995;270:13987–13997. doi: 10.1074/jbc.270.23.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer B, Boyer JL, Hoyle CHV, Ziganshin AU, Brizzolara AL, Knight GE, Zimmet J, Burnstock G, Harden TK, Jacobson KA. Identification of potent, selective P2Y-purinoceptor agonists–structure–activity-relationships for 2-thioether derivatives of adenosine 5′-triphosphate. J. Med. Chem. 1993;36:3937–3946. doi: 10.1021/jm00076a023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouldson PR, Snell CR, Reynolds CA. A new approach to docking in the β2-adrenergic receptor that exploits the domain structure of G-protein-coupled receptors. J. Med. Chem. 1997;40:3871–3886. doi: 10.1021/jm960647n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.