Abstract
Human papillomavirus (HPV) infection is associated with a wide spectrum of disease that ranges from self-limited skin warts to life-threatening cancers. Since HPV plays a necessary etiological role in cervical cancer, it is logical to use HPV as a marker for early detection of cervical cancer and precancer. Recent advances in technology enable the development of high-throughput HPV assays of different formats, including DNA-based, mRNA-based, high-risk group-specific and type-specific methods. The ultimate goal of these assays is to improve the accuracy and cost-effiectiveness of cervical screening programs. HPV testing has several potential advantages compared to cytology-based screening. However, since the cancer to transient infection ratio is always low in the general population, HPV test results are bound to have a low positive predictive value that may subject women to unnecessary follow-up investigations. The wide-spread administration of prophylactic HPV vaccine will substantially decrease the incidence of cancer and precancer. This poses a number of challenges to cytology-based screening, and the role of HPV testing is expected to increase. Finally, apart from technical and cost-effiectiveness considerations, one should also keep in mind the psycho-social impact of using sexually-transmitted agents as a marker for cancer screening.
Keywords: Human papillomavirus, warts, epidermodysplasia verruciformis, skin cancer, cervical cancer, vaginal cancer, vulvar cancer, oropharyngeal cancer, penile cancer, cervical intraepithelial neoplasia, cervical screening, triage, biomarker, co-test, colposcopy
Introduction
To date, more than 120 types of human papillomavirus (HPV) have been characterized1. HPV exclusively infects epithelial cells and is associated with a broad spectrum of clinical manifestations that range from self-limiting lesions to life-threatening disease2–13.
With the advances in molecular techniques over the last decade, our understanding of this family of ubiquitous viruses has improved tremendously, a number of previously unrecognized important disease associations have been confirmed, and detection assays for epidemiological study and for confirming diagnosis to assist patient management have been developed. Vaccines targeting two HPV types that show the strongest association with cervical cancer have been developed14. Assessment of vaccine cost-effectiveness and priority becomes an important task for public health policy makers. To this end, HPV detection and typing assays are indispensable in defining the fraction of disease attributed to the types covered by current vaccines.
Disease spectrum
Benign lesions
Non-genital skin infections
With the improvement in the sensitivity of detection assays and coverage for a variety of HPV types, it is now recognized that asymptomatic carriage of HPV on healthy skin is common and can persist over several years15. Persistence of HPV infection on non-genital skin does not seem to be related to age, sex, immunosuppressive treatment or history of warts16,17. Clinical manifestations of these lesions are characteristic. In most situations, diagnosis can be made clinically without HPV testing18–19. Testing for HPV in these lesions is mainly for epidemiological study or other research purposes.
HPV causes benign warts on the skin, which present as flat or firm non-itchy papules10. Commonly affected areas include hands (verrucae palmares) and feet (verrucae plantares), which are associated typically with HPV 1, 2, and 4. Skin warts, except those growing over press areas, are non-painful. Verrucae planae are flat skin-colour warts found on the face, hands and forearms, and are caused most commonly by HPV 3 and 7. Periungal warts found at the nail fold are often painful. Butcher's warts are found rarely on the hands of butchers who have repeated trauma that predisposes them to infection with, most commonly, HPV 7; although it is a well-known occupational disease, there is no evidence that the source of infection is animal papillomaviruses20.
Genitial infections
Genitial tract HPV infection is the most common sexually-transmitted infection. Infections are often subclinical. Clinical lesions (condylomata acuminata) are often multiple and appear as exophytic papillomas, flesh or brown in colour. HPV 6 and 11 are the types most commonly found in visible anogenital warts, but other types, including those typically found in non-genital skin, can also be detected, especially from anogenital areas without visible lesions3,21–25.
Oral infections
The oral mucosa is also susceptible to HPV infection. Such infection involving the larynx represents a rare but severe disease3,7,26–28. Laryngeal papillomatosis has two age-related incidence peaks; those with early childhood onset are acquired vertically from maternal condyloma, whereas the adult onset group is presumably acquired via orogenital contact. In both groups, HPV 6 and 11 are the most commonly found HPV types. The lesions, which develop mainly over vocal cords and trachea, present as hoarseness of voice and stridor. Lesions may extend to lungs, nose and oral cavity. Treatment is difficult as the lesions often recur; thus it is also known as recurrent respiratory papillomatosis. Heck's disease, another rare condition, presents as multiple papillomas over the lip and buccal mucosa29–31.
Malignant lesions
The hypothesis that HPV plays a role in the development of cervical cancer was proposed in the mid-1970s. Over the last 40 years, a strong body of evidence has accumulated to prove the etiological association between HPV infection and a number of human cancers in addition to cervical cancer11,32,33.
Epidermodysplasia verruciformis
Epidermodysplasia verruciformis (EV), a genetically-inherited chronic skin condition, presents as disseminated flat warts. Recent studies have shown that some EV patients have mutations in the EVER1 and EVER2 genes34–36. Certain HPV types belonging to the beta genus (HPV 5, 8, 9, 12, 14, 15, 17, 19, 25, 36, 38, 47 and 50) are specifically linked to EV. These EV-associated HPV types are also commonly found in the general population. Some immunosuppressed individuals who do have EV may develop lesions caused by EV-associated HPV types. There is no evidence that EV patients are more susceptible to infection or disease manifestations caused by the alpha genus of HPV. About half of the EV patients develop squamous cell carcinoma in sun-exposed areas. Most malignant lesions are associated with HPV 5 and 8, though the pathogenic mechanism of these HPV types is still not yet clear8,37,38.
Non-melanoma skin cancers
In recent years, some evidence suggests that non-melanoma skin cancers including basal cell and squamous cell carcinoma may be linked to cutaneous HPV infection. The suspicion of a viral cause for cutaneous squamous cell carcinoma is based on the observation that its incidence increases dramatically in solid organ transplant recipients receiving long-term immunosuppressive therapy. While sun-exposure is a recognized risk factor, infection with cutaneous HPV (mainly the beta genus) seems to play a role in the development of non-melanoma skin cancers, especially squamous cell carcinoma. HPV types belonging either to beta species 1 or 2 have been detected from squamous cell carcinoma specimens, but unlike anogenital cancers, no predominant HPV types can be identified39,40. The etiological association between non-melanoma skin cancers and HPV infection is difficult to prove as the same spectrum of HPV types is prevalent among healthy subjects9,41. Furthermore, HPV may not be required in maintaining the cancer phenotype and it may therefore escape detection in tumour specimens42.
The mucosal group of HPV is associated with several malignant conditions. Bowenoid papulosis presents as multiple small flat pigmented papules on the external genitalia. Histologically, it is a carcinoma-in-situ, it can evolve to invasive carcinoma, and it is often associated with HPV 16. A subset of intraepithelial neoplasia and carcinoma of the penis, vulva and vagina is also associated with HPV infection, mainly HPV 1643.
Cancer of vagina
This uncommon cancer in women has an age-standardized rate of 0.3–0.7 per 100,000 worldwide44. The limited information about the role of HPV infection and occurrence of vaginal cancer is mostly based on analysis of a few HPV types using fixed tissues45. Epidemiological studies indicate that vaginal cancer resembles cervical cancer, and HPV DNA is detected in a majority of vaginal tumours and their precursor lesions. HPV is detected in 82–100% of vaginal intraepithelial neoplasia grade III, and 64–91% of vaginal cancers; as in cervical cancer, HPV16 is the most prevalent type found45,46.
Cancer of vulva
The age-standardized incidence rates of vulvar cancer lie between 0.5 and 1.5 per 100,000. The geographical pattern of vulvar cancer is different from cervical cancer and high rates are observed in several European populations (Scotland, Denmark, Spain, Italy), whereas the prevalence in sub-Saharan Africa, Southeast Asia, and Latin America is low. Distinct subtypes, such as the warty and basaloid types, have been recognized, but the majority of tumours are squamous cell carcinoma. Etiologically, because vulvar carcinomas are heterogeneous, the prevalence of HPV infection in invasive vulvar cancer cases varies47. Vulvar cancer with basaloid histopathology in young women is often associated with HPV. HPV 16, 31 and 33 are the most frequently-detected types in this type of vulvar cancer and its precursor lesions48. On the other hand, vulvar cancer with verrucus subtype and some cases of precancerous lesions of vulvar intraepithelial neoplasia are not associated with HPV infection49. In general, the HPV-positive and -negative groups of vulvar squamous cell carcinoma share a similar prognosis50.
Cancer of anus
The vast majority of anal cancers is associated with HPV infection. Cancers arising in the anal canal and tumors of the external skin (anal margin) are classified as skin cancers. The canal is lined in its upper part by colorectal-type mucosa, and in its lower third by squamous epithelium, with a specialized transitional zone in between. Therefore, cancers are predominantly squamous cell carcinoma, adenocarcinoma, or basaloid and cloacogenic carcinoma. In most populations, squamous cell carcinoma is twice as common in females as males. However the incidence is particularly high among men who have sex with men and the risk is increased further by infection with human immunodeficiency virus, cigarette smoking, anal intercourse, and more lifetime sexual partners51–53.
Cancer of penis
Globally, this rare cancer accounts for less than 0.5% of all cancers in men. The concordance of cervical and penile cancer in married couples and the geographical distribution of these cancers suggest that they share a common etiology54. Serological studies have confirmed the role of HPV 16 and HPV 18 in the etiology of penile cancer, and HPV DNA is detected in 40–50% of such cancers55–57.
Cervical cancer
Among the cancers for which a confirmed or probable etiological link with HPV infection has been established, cervical cancer has the strongest association and accounts for the largest share of disease burden2,12,58–60. In 2008, there were about 530,000 new cases of cervical cancer, and about half this number (275,000) died of the disease worldwide; the age-standardized incidences of new cases and mortality were 15 and 8 per 100,000, respectively61,62. Worldwide, cervical cancer ranked third among cancers in women, just following breast cancer (1.3 million new cases) and colorectal cancer (0.57 million new cases) in 2008. The incidence of cervical cancer varies widely and the developing world accounts for more than 85% of both incidence and mortality. The annual age-standardized incidence rates range from 56 per 100,000 (Guinea) to < 1 per 100,000, depending mainly on the availability of organized cervical screening programs. Overall, the lowest disease burden is recorded from Australia, New Zealand, North America and Western Europe, whereas highest burden is seen in Africa, South-Central Asia and South America62.
Oropharyngeal cancer
An increase in incidence of oropharyngeal squamous cell carcinoma, specifically those originated from the tonsil and tongue base, has been observed in some parts of the world63,64. A proportion of these lesions are associated with HPV infection65–68. The prevalence of HPV in these tumours varies geographically and reflects the variation in prevalence of oral HPV infection, which in turn mainly depends on the practice of oral sex. HPV-positive oropharnygeal squamous cell carcinomas differs from HPV-negative ones in several molecular aspects, reflecting that they are distinct entities. The molecular features observed in HPV-positive oropharyngeal cancers are consistent with the notion that HPV plays a role in the development of these tumours5,6,65,69.
Clinically important basic virology
Viral genome and key proteins
Papillomaviruses have a small double-stranded DNA genome of about 8 kb in length. The key functions and proteins encoded by different regions of the HPV genome that are relevant for designing detection assays are shown in Figure 1.70–72 The viral genome has eight open reading frames (ORF) encoding two structural (late) proteins L1 and L2; these form the viral capsid that is about 55 nm in diameter and protects the viral genome inside. L1 is the major structural protein which is conserved within a given HPV type. The fact that L1 protein is immunogenic and conserved within a given type makes it a prime target as antigens for serological assay as well as prophylactic vaccine development73. L1 proteins can reassemble themselves under appropriate in vitro conditions to form virus-like particles which are the main constituent of current prophylactic vaccines14,53,74–77. L2 is the minor capsid protein which can potentially elicit a broader spectrum of neutralizing antibodies against different types of HPV. The potential of using L2 protein as an additional component in future vaccines is being investigated78–81. The early proteins E5, E6 and E7 encoded by HPV contribute to tumour progression. The oncogenic activities of E6 and E7 are well-characterized. Both E6 and E7 have numerous cellular targets. E6 proteins encoded by high-risk HPV types primarily bind to the tumour suppressor protein p53, and the binding is mediated by the E6-associated proteins (E6-AP). Overexpression of E6, together with its interactions with other cellular proteins, results in the degradation of p53, anti-apoptosis, chromosomal destabilization, enhancement of foreign DNA integration and activation of telomerase82–89. The E7 proteins encoded by high-risk HPV types also demonstrate an important role in tumourigenesis. E7 binds to a large number of cellular proteins, most importantly the retinoblastoma protein (Rb) and the Rb-related pocket proteins. Such binding results in inactivation of Rb-related pocket proteins, activation of cyclins, inhibition of cyclin-dependent kinase inhibitors, and enhancement of foreign DNA integration and mutagenesis90–95. The expression of E6 and E7 is tightly controlled via a promoter located at the non-coding Long Control Region (LCR) of the viral genome. Other early proteins encoded by papillomaviruses are E1, E2 and E5. In contrast to E6 and E7, the oncogenic activities of E5 are much less well-defined. Recent studies indicate that E5 plays a role in escape from immune surveillance, upregulation of transcription factors and inhibition of apoptosis96–98. E2 is another important region of the papillomavirus genome99. E2 proteins form complexes with E1 to initiate viral replication. E2 also regulates the expression of E6 and E7, and can exert suppressive or activating effects depending on the abundance of E2. Disruption of E2 ORF as a result of integration of viral genome into the host genome allows an uncontrolled overexpression of viral oncoproteins E6 and E7, which is a hallmark in cervical cancer100–102.
Figure 1.
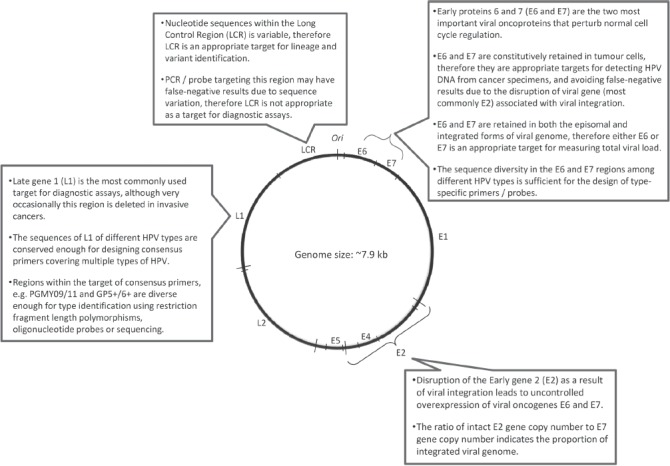
Key targets of human papillomavirus genome for detection.
Concept of HPV type
HPV are classified under the family Papillomaviridae which comprises members infecting humans, non-human primates, birds, and reptiles1,103–105. Unlike many other viruses, the concept of “strain” is not used for papil-lomaviruses as they cannot be grown by conventional cell culture methods. “Serotype” cannot be applied because of the lack of a robust antibody response following natural infection. Thus, HPV types refer to “genotypes”, which are classified based on sequence similarity. To date, more than 120 HPV types have been well-characterized1. The L1 ORF is a key region for classification of HPV types (Figure 1). Technically-speaking, the DNA sequence of the L1 region of an HPV type differs from all other types by more than 10%. When the differences in L1 ORF nucleotide sequences between two isolates are within 2–5%, they are regarded as subtypes; differences below 2% are referred as variants. The findings that sequence divergence of a given HPV type is limited and subtypes are rare indicate that HPV has probably gone through genetic drift that became amplified by founder effects and bottlenecks of evolution106–111. While the classification of HPV “types” is based on genome sequence, it has strong clinical implications112,113. Firstly, the phylogenetic grouping of HPV types reflects tissue tropism observed in clinical infections. HPV types belonging to the alpha genus (also known as super-group A) mainly infect the mucosal areas, whereas types in the beta genus (also known as supergroup B) mainly infect the cutaneous areas. Secondly, the protective immune response elicited by HPV infection is mainly type-specific. Similarly, the protection induced by the currently available prophylactic vaccines (Gardasil® quadrivalent with HPV 6, 11, 16, 18 from Merck & Co.; Cervarix® bivalent with HPV 16, 18 from GlaxoSmithKline Biologicals) is also mainly type-specific, although some degree of cross-protection, especially for the bivalent vaccine, has been observed114–118. The ability of HPV to induce cancer is also type-related. HPV types can be grouped into high-risk and low-risk based on their association with the development of cervical cancer. The risk associations identified by epidemiological and biochemical studies are similar. There is no dispute in grouping the commonly-found HPV types, but classification of rare types is still not clear119. In view of the current epidemiological, biochemical and phylogenetic data, the mucosal/anogenital HPV types can be grouped as “high-risk” (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59), “probable high-risk” (HPV 68), “possible high-risk” (HPV 26 and 73), and “low- or unknown risk” (HPV 6, 11, 30, 34, 40, 42, 43, 44, 53, 54, 55, 57, 61, 62, 66, 67, 69, 70, 82, 85 and 97)2,105,112,119,120.
Virus detection approaches
Because the replication cycle of papillomaviruses can be completed only in differentiated epithelial cells, isolation of viruses from clinical samples is difficult72. Growth of papillomaviruses has been accomplished only in a specialized primary human keratinocyte-derived cell culture system, organotypic “raft” culture, where epithelial cells are grown on semi-solid agar to allow differentiation of epithelial cells and hence productive replication of papillomaviruses121–123. Even with this technique, only a few types (mainly HPV 11 and 31) have been replicated successfully in laboratory conditions.
HPV is equipped with several immune evasion mechanisms. It multiplies in keratinocytes that have a short-life span, and thus the progeny viruses can be released in a natural way without inducing cell lysis that is seen with other non-enveloped viruses. Tis avoids triggering of infammation and immune responses associated with cellular damage. The lack of a viremic phase also minimizes stimulation to the systemic immune system. Furthermore, the virus actively downregulates the synthesis of interferon. As a result, although the immune system plays an important role in clearing infection and it is associated with a strong localized cellular immune response, natural infection with papillomaviruses does not result in a robust antibody response124–130. For this reason, serological diagnosis has limited clinical and epidemiological value. For instance, only about 50–70% of women with persistent cervical HPV infection mount a detectable antibody response, and most women with transient infection do not have detectable antibodies or have them only for a short time131–135. Thus, the detection of HPV infection relies mainly on a molecular approach that amplifies the viral genome or mRNA, or detection of viral protein using immunoassays.
DNA-based assays
Because HPV cannot be cultured, all HPV assays currently in use rely on the detection of viral nucleic acids. They can be divided into: 1) target-amplification methods (PCR [polymerace chain reaction] with consensus or type-specific primers, and HPV mRNA amplification), and 2) signal amplification methods (liquid-phase or in situ hybridization). It is necessary to distinguish analytical sensitivity (minimum number of HPV genomes to be present in a sample to generate a positive test result) from clinical sensitivity (proportion of women with disease who test positive) (Table 1).
Table 1.
Parameters for assessing the clinical performance of HPV tests in cervical cancer screening.
| Terminology | Characteristics to measure |
|---|---|
| Sensitivity1 | The proportion (expressed as percent) of patients with a certain disease (e.g. CIN III or invasive cervical cancer) who have a POSITIVE HPV test. E.g., a sensitivity of 95% for CIN III means that 5 of 100 CIN III cases will be NEGATIVE (false-negative) and will be missed by this test. Sensitivity is affected mainly by the test itself and to some extent by the nature of samples submitted for testing. |
| Specificity2 | The proportion of subjects (expressed as percent) without a certain disease (e.g. CIN II or higher severity) who have a NEGATIVE HPV test. E.g., a specificity of 70% means that if 100 women without CIN II or higher severity participate in the screening, 30 will be POSITIVE (false-positive) by the test. Specificity is affected mainly by the test itself and to some extent by the nature of samples submitted for testing. |
| Positive predictive value | The chance (expressed as percent) that a POSITIVE HPV test result indicates the presence of a certain disease (e.g. invasive cervical cancer). In the context of cervical screening, this predictive value can be further divided into predictive value for the current situation or in the next 5 or 10 years. The positive predictive value of a test depends mainly on the prevalence of the disease in the target population. Cancer screening tests, including HPV, are bound to have low positive predictive value when applied to the general population. |
| Negative predictive value | The chance (expressed as percent) that a NEGATIVE HPV test result excludes the presence of a certain disease (e.g. CIN II or higher severity). Under the context of cervical screening, this predictive value can be further divided into predictive value for current situation or in the next 5 or 10 years. The negative predictive value of a test depends mainly on the prevalence of the disease in the target population. |
The definition presented here refers to “clinical” sensitivity. From the laboratory performance point of view, the concept of analytical sensitivity is often used, which means the lowest amount of analyte required in the specimen to generate a positive result, (also referred to as detection limit, e.g. 100 copies of viral DNA). For the HPV test, increasing the analytical sensitivity may not always increase the clinical sensitivity, but usually specificity is lost.
From the laboratory performance point of view, “specificity” means cross-reaction with similar targets. This is more relevant for assays intended for HPV typing. e.g. an assay with poor specificity may misidentify an HPV 58–positive sample as HPV 33.
The choice of the HPV test depends on the application (Table 1). Assays with high analytical sensitivity are crucial for molecular epidemiological studies and for evaluating vaccine efficacy. HPV typing assays with high analytical sensitivity and specificity are the key in virological surveillance, including the evaluation of vaccination impact on the prevalence of vaccine-covered types, identification of new types, discrimination of types in multiple infection, and monitoring of potential type replacement in the post-vaccine era.
On the other hand, when applied in the clinical situation for cervical cancer screening and post-treatment follow-up, assays with lower analytical sensitivity may produce a better positive predictive value136–138. It is important that HPV tests are clinically validated under context-specific conditions, especially in the target population.
Hybrid capture HPV DNA assay
Hybrid capture (HC2) is based on liquid phase hybridization using long synthetic RNA probes complementary to the genomic sequences of 13 high-risk types (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68) and five low-risk types (6, 11, 42, 43 and 44); these are used to prepare high-risk (B) and low-risk (A) probe cocktails that are used in two separate reactions139. In practice, often only the high-risk probes are used. DNA present in the biological specimen is hybridized in solution phase with each of the probe cocktails and forms specific HPV DNA-RNA hybrids. These hybrids are then captured by antibodies bound to the wells of microtiter plates that recognize specific RNA-DNA hybrids. After removal of excess antibodies and non-hybridized probes, the immobilized hybrids are detected by a series of reactions that give rise to a luminescent product that is detected by a luminometer. The intensity of emitted light, expressed as relative light units, is proportional to the amount of target DNA present in the specimen, and provides a semiquantitative measure of the viral load. The HC2 is currently available in a 96-well microplate format with automation that is easy to use in clinical settings. An automated robotic platform that allows handling of 96-well microplates has been developed for high-volume testing.
HC2 does not require special facilities to avoid cross-contamination because it does not rely on target amplification to achieve high sensitivity, as do PCR protocols. The recommended cut-of value for test-positive results is 1.0 relative light unit (equivalent to 1 pg HPV DNA per 1 mL of sampling buffer). Several studies have noted that the high-risk probe cocktail in HC2 cross-reacts with HPV types that are not represented in the probe mix. It has been reported that HC2 using the high-risk probe at a 1.0-pg/mL cut-off detected HPV types 53, 66, 67, 73, as well as other undefined types, and raising the cut-of to 10.0 pg/mL did not eliminate the cross reactivity to types 53 and 67140. Cross-reactivity of HC2 high-risk probe to HPV types that have a signifcant risk for cervical cancer may be considered as beneficial, but cross-reaction with low-risk types causing false-positive results may decrease the specificity and positive predictive value141.
Consensus, group- or type-specific PCR
Currently, the most important clinical application of HPV detection is to identify women with a higher risk of developing cervical intraepithelial neoplasia or invasive cervical cancer (Figure 2). HPV DNA detection systems that are designed to catch all the mucosal or anogenital HPV types are referred as “consensus”, those detecting high-risk or low-risk HPV types as a group are referred as “group-specific”, and those identify individual HPV types are referred as “type-specific”. A variety of commercial assays belonging to each category are available (Table 2).
Figure 2.
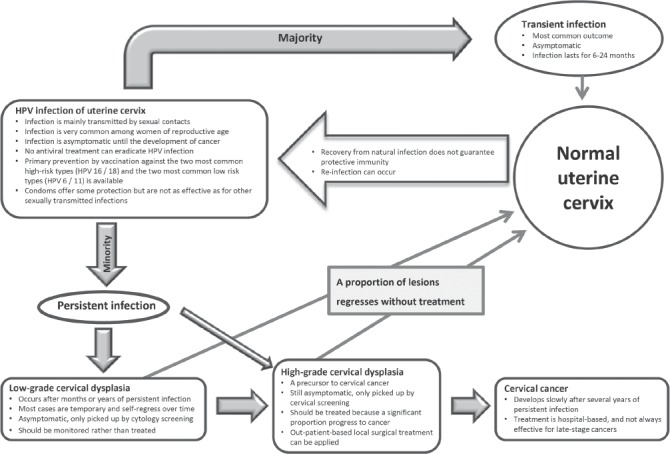
Natural history and management of cervical HPV infection.
Table 2.
Commercially a vailable HPV tests.
| Molecular target | Type differentiation | Test name | Principle | Manufacturer |
|---|---|---|---|---|
| HPV DNA (full genome) | High-risk types as a group, not type-differentiating | Hybrid Capture 2 HPV DNA Test | Hybridization | QIAGEN, Gaithersburg |
| CareHPV Test | hybridization | QIAGEN, Gaithersburg | ||
| HPV DNA (L1 ORF) | High-risk types as a group, not type-differentiating | Amplicor HPV Test | PCR | Roche, Branchburg |
| Cervista HPV HR Test1 | Hybridization (Invader)2 | Hologic, Madison | ||
| HPV DNA (L1 ORF) | Differentiate 13 or more high-risk types | CLART | Reverse line-blot Hybridization on PCR products | Genomica, Coslada |
| INNO-LiPa HPV Genotyping | Reverse line-blot Hybridization on PCR products | Innogenetics, Gent | ||
| Linear Array HPV Genotyping Test | Reverse line-blot Hybridization on PCR products | Roche, Branchburg | ||
| Digene HPV Genotyping RH Test (RUO) | Reverse line-blot Hybridization on PCR products | Digene, Hilden | ||
| HPV DNA (E1 ORF) | Differentiate 13 or more high-risk types | Infniti HPV-HR QUAD Assay | Microarray on PCR products | Autogenomics, Carlsbad |
| PapilloCheck | Microarray on PCR products | Greiner Bio-one, Frickenhausen | ||
| HPV DNA (L1 ORF) | Limited type differentiation - HPV 16/18 | Cervista HPV Test1 CORBAS 4800 HPV Test Real Time High Risk (HR) HPV Test |
Hybridization Real-time PCR Real-time PCR |
Hologic, Madison Roche, Pleasanton Abbott, Des Plaines |
| HPV E6/E7 mRNA | 14 high-risk types as a group, not type-differentiating | APTIMA HPV Assay | TMA | GenProbe, San Diego |
| HPV E6/E7 mRNA | Limited type differentiation – HPV 16/18/31/33/45 | NucliSENS EasyQ HPV3 | NASBA | BioMerieux, Marcy-l'Étoile |
| PreTect HPV-Proofer3 | NASBA | Norchip, Klokkarstua |
NASBA = nucleic acid sequence-based amplification; TMA = transcription-mediated amplification.
Also targets E6 and E7.
The “Invader” reaction involves two simultaneous isothermal reactions. A primary reaction is based on hybridization with two sequence-specific oligos to the same target, creating a single-nucleotide overlap. The overlap together with its 5' flap will be cleaved. In the secondary reaction, the cleaved flap combines with a fuorescence resonance energy transfer (FRET) probe that generates a fuorescent signal.
As a result, each released 5' flap from the primary reaction cycles on and of the FRET probes, enabling the secondary reaction to further amplify the target-specific signal to 1–10 million-fold.
Same technology marketed under different brand names in different countries.
Target regions
L1: The L1 region is the most frequently-used target for amplifying HPV genome from clinical samples (Figure 1). L1, on the one hand, is conserved enough for the design of consensus primers to amplify a broad spectrum of HPV types by using a single set of degenerated primers or a cocktail of primers142–149. The commonly-used primer sets are shown in Table 3. On the other hand, the L1 region also has sufficient sequence diversity to allow the identification of individual HPV types based on further analysis of the amplified products. Given the definition that L1 sequences of different HPV types should differ by more than 10%, type differentiation is achievable even based on a short fragment, less than 100-bp, of L1. HPV type can be identified by a few approaches. Type-specific restriction fragment length polymorphisms can be generated by using two restriction endonucleases Rsa I and Dde150,151. This method was more popular when sequencing was still expensive and labour intensive. The main disadvantage of restriction fragment length polymorphisms is the ambiguous restriction fragment patterns generated from co-infection with multiple types, especially when three or more types are present in a sample. This approach has gradually been replaced by sequencing because the cost for the latter has fallen substantially in recent years. However, sequencing of PCR product can reveal only the HPV type that predominates in a co-infection. Today, the best approach to identify multiple HPV types simultaneously from a sample is to perform hybridization with multiple HPV type-specific probes immobilized on strips, membranes or array slides (Table 2).
Table 3.
Commonly used primers targeting the L1 region of HPV genome.
| Primer name | Primer sequences | Remarks | References |
|---|---|---|---|
| MY09/11 | Forward (5'-GCMCAGGGWCATAAYAATGG-3') Reverse (5'-CGTCCMARRGGAWACTGATC-3') |
One of the commonly used first generation primers. Target a ∼450-bp fragment which is usually too long for formalin-fixed tissues. The amplified product allows type identification by restriction fragment length polymorphisms, hybridization with type-specific probes or by sequencing. |
143 |
| PGMY09/11 | PGMY11-A (5'-GCA CAG GGA CAT AAC AAT GG-3') PGMY11-B (5'-GCG CAG GGC CAC AAT AAT GG-3') PGMY11-C (5'-GCA CAG GGA CAT AAT AAT GG-3') PGMY11-D (5'-GCC CAG GGC CAC AAC AAT GG-3') PGMY11-E (5'-GCT CAG GGT TTA AAC AAT GG-3') PGMY09-F (5'-CGT CCC AAA GGA AAC TGA TC-3') PGMY09-G (5'-CGA CCT AAA GGA AAC TGA TC-3') PGMY09-H (5'-CGT CCA AAA GGA AAC TGA TC-3') PGMY09-I (5'-GCC AAG GGG AAA CTG ATC-3') PGMY09-J (5'-CGT CCC AAA GGA TAC TGA TC-3') PGMY09-K (5'-CGT CCA AGG GGA TAC TGA TC-3') PGMY09-L (5'-CGA CCT AAA GGG AAT TGA TC-3') PGMY09-M (5'-CGA CCT AGT GGA AAT TGA TC-3') PGMY09-N (5'-CGA CCA AGG GGA TAT TGA TC-3') PGMY09-P (5'-GCC CAA CGG AAA CTG ATC-3') PGMY09-Q (5'-CGA CCC AAG GGA AAC TGG TC-3') PGMY09-R (5'-CGT CCT AAA GGA AAC TGG TC-3') |
An improved version of MY09/MY11 targeting the 144 same 450-bp fragment. Use in the Linear Array HPV Genotyping Test (Roche, Branchburg), where the HPV types are identified by hybridization with type-specific probes. |
144 |
| GP5+/6+ | Forward (5'-TTT GTT ACT GTG GTA GAT ACT AC-3') Reverse (5'-GAA AAA TAA ACT GTA AAT CAT ATT C-3') |
Targets a ∼150-bp fragment suitable for formalin-fixed tissues. Commonly used in large-scale epidemiological studies. Reported to have sub-optimal sensitivity for a variant of HPV 52 commonly found in East Asia due to mutation at the primer binding site. |
145,153 |
| SPF | Forward primer (5'-GCI CAG GGI CAT AAC AAT GG-3') Two reverse primers: (5'-GTI GTA TCI ACA ACA GTA ACA AA-3') and (5'-GTI GTA TCI ACA TCA GTA ACA AA-3') |
Targets a short (∼65-bp) fragment. Good for samples with fragmented DNA such as formalin-fixed tissues. Modifed versions include SPF6 and SPF10 with the addition of more primers. SPF10 primers are used in the INNO-LiPa HPV Genotyping assay, but sequences not disclosed. |
146 |
| L1F/L1R | Forward (5'-CGT AAA CGT TTT CCC TAT TTT TTT-3') Reverse (5'-TAC CCT AAA GAC CCT ATA CTG-3') |
Targets a ∼255-bp fragment, a length that may not be retained in formalin-fixed tissues. | 147 |
While L1 is regarded as the best target for HPV genome detection, it is absent from a small proportion of invasive cervical cancer samples, probably due to disruption resulting from viral genome integration152. When such a situation is suspected, the constitutively retained genes, E6 or E7, can be amplifed to verify the presence of HPV infection. Furthermore, consensus primer sets may have different analytical sensitivities for different HPV types or variants147. When adopting assays developed elsewhere, evaluation based on samples collected from the local population is necessary before the assays are used clinically.
E6, E7: The E6 and E7 regions are good alternative targets for amplifying HPV genome from clinical specimens. Firstly, the nucleotide sequence diversity of E6 and E7 between HPV types allows the design of type-specific primers. Secondly, E6 and E7 gene expression is required to maintain the transformed phenotype of infected cells153–159. Therefore, both E6 and E7 genes are expected to be retained regardless of the status of viral integration. The constitutional presence of E6 and E7 genes makes them an appropriate target for viral load quantification160,161.
E2: Papillomavirus infection can exist in two forms. The vegetative phase, also known as the replicative or productive phase, is associated with a full episomal form of viral genome. This replicative phase is typically found in the upper layers of epithelium with differentiated keratinocytes, or in benign condyloma where a large amount of virus is released32,162–164. In the integrated form, the viral genome is disrupted and therefore the viral replication cycle cannot be completed. It is widely accepted that HPV-mediated cervical carcinogenesis proceeds via the integration of viral genome and disruption of the E2 ORF, thus releasing the suppressive control of E2 on the expression of viral oncogenes E6 and E7. Thus, E2 is often used as a surrogate marker to indicate the status of viral integration. By measuring the ratio between E2 and E6 (or E7) gene copy numbers, one can estimate the proportion of integrated viral genome present in a clinical specimen.
E6/E7 mRNA-based assays
E6 and E7 are oncoproteins involved in carcinogenesis. Persistent expression of E6 and E7 could serve as an indicator of progression from intraepithelial neoplasia to invasive cancer165–167. Detecting the mRNA encoded by E6 or E7 may, therefore, provide a better predictive value for malignant or high-grade lesions168–171. A few commercial assays based on this approach have been developed recently (Table 2)172,173. The PreTect HPV-Proofer (Norchip, Klokkarstua) and NucliSENS Easy Q HPV (BioMerieux, Marcy-l'Étoile) assays are based on the same technology, and are marketed under different brand names in different countries. PreTect Proofer and NucliSENS Easy Q detect E6/E7 mRNA from five high-risk types (HPV 16, 18, 31, 33 and 45) commonly found in high-grade lesions and cancers. The APTIMA HPV Assay (Gen-Probe) provides a broader coverage and targets mRNA of 14 high-risk HPV types. In a review on 11 studies examining these three currently-available commercial assays, the sensitivities for cervical intraepithelial neoplasia grade II and above (CIN [cervical intraepithelial neoplasia] II+) lesions ranged from 41% to 86% for the PreTect Proofer/EasyQ assay and from 90% to 95% for the APTIMA assay, whereas the specificities ranged from 63% to 97% for the PreTect Proofer/Easy Q assay and from 42% to 61% for the APTIMA assay174.
E6/E7 protein-based assays
Similarly, measuring the E6 or E7 proteins may provide a better predictive value than detecting viral DNA alone175,176. However, E6 and E7 proteins are known to be produced in small amounts in transformed cells. The sensitivity of the E6/E7 protein-based assay is a concern that needs to be addressed. Data on clinical evaluation of the E6/E7 protein-based assay is not yet available.
Building quality control capacity
The need to monitor vaccine effectiveness by effective surveillance programs and the increasing use of HPV assays in the field have stimuated the World Health Organization (WHO) to develop a structured Global Laboratory Network (WHO HPV LabNet). To date, this LabNet includes two Global Reference Laboratories (Sweden and USA) and eight Regional Reference Laboratories (Argentine, Australia, India, Japan, South Africa, Switzerland, Tailand and Tunisia). Work is currently being conducted in the areas of scientific and technical advice, quality assurance, training, and communication (http://www.who.int/biologicals/areas/human_papillomavirus/WHO_HPV_LabNet/en/index.html. Accessed on 9 July 2012). LabNet, with the collaboration of the National Institute of Biological Standards and Control (UK), is establishing international standards (IS) for HPV types, to harmonize HPV nucleic acid amplification technology-based assays. These efforts will be immensely helpful in monitoring the impact of HPV vaccination programs worldwide and in evaluating data on uniform platforms. One of the main aims is to harmonize various HPV detection assays and to minimize inter-laboratory variation by collectively establishing strong and effective quality control and quality assurance programs.
Clinical applications
HPV for primary screening
HPV infection progresses slowly to cervical cancer through stages of pathological changes that can be recognized from exfoliated cervical cells (Figure 2). Detection of lesions at the precancerous stage allows intervention by office-based local surgery. Until recently, cervical screening has been based solely on cytological examination of exfoliated cervical cells. The benefit of regular cervical cytology screening is undisputed177–181. Cervical cancer incidence decreases substantially after the introduction of cervical screening and there are consistent and marked differences in cervical cancer incidence rates between countries with and without organized screening programs61,182. While conventional Pap smear or liquid-based cytology is still the standard of care in many parts of the world, the intrinsic drawbacks of cytology-based screening call for replacement by HPV testing or the addition of adjunct markers183. Compared to molecular markers, cytology is more subjective and requires a stringent quality control and quality assurance program to maintain clinical performance. Cytology is relatively insensitive and is associated with an unavoidable portion of non-specific and self-limiting abnormal results. With the increased availability of high-through-put screening platforms (Table 2)184, a large number of large-scale studies have been conducted to investigate the value of using HPV DNA detection (mainly based on hybridization (hybrid capture) or PCR amplification targeting the L1 region) as a primary or supplementary tool for cervical screening.
Advantages
The overall advantages of HPV testing over cytology for screening of cervical cancer are: feasibility for high throughput, greater objectivity in result interpretation, high sensitivity, high negative predictive value, and ability to provide long-term risk stratification185–192. Furthermore, the performance of HPV assays is less subject to variation across centers. For instance, the reported sensitivity of cytology ranges from 33.8% to 94.0%, whereas that of the HPV assay (HC 2), used in the same series of studies across different continents, varies from 84.9% to 97.6%193–199. However, HPV infections are transient in most women and the prevalence of high-grade intraepithelial neoplasia or cancer among infected women is low. The low specificity and low positive predictive value are the major drawbacks of applying HPV DNA testing in clinical practice. A fine balance has to be established between the sensitivity and specificity of the HPV test to achieve a clinically-useful predictive value (Table 1).
Approaches to improve positive predictive value
Infection and disease prevalence
Several approaches can be considered to minimize the “background noise” and to improve the positive predictive value. The “background noise” depends on the prevalence of transient HPV infection among the target population for which the HPV test is being applied. Data on the age-specific prevalence of HPV infection and the age-specific incidence of CIN II/III and cervical cancer are essential to derive a cut-of for applying HPV testing (Figure 3)200. In general, HPV testing is not recommended for women below 30 years of age (or 35 in some countries) for whom transient infection is common187,201,202.
Figure 3.
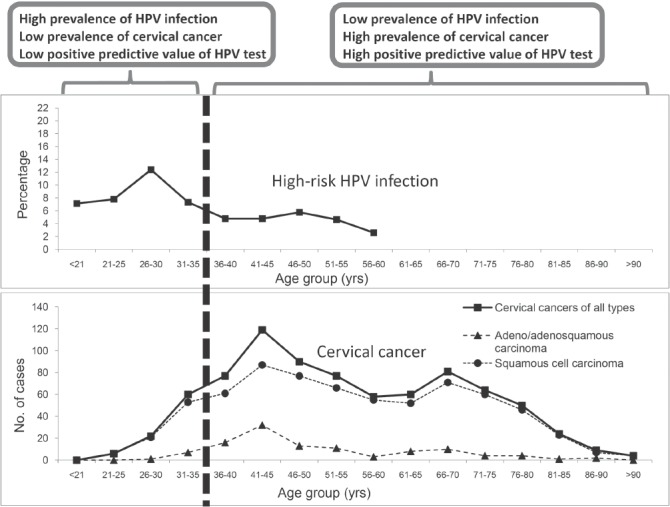
Improved positive predictive value of HPV test by refining the target population. Figure shows the age-specific prevalence of high-risk HPV infection and age-specific distribution of cervical cancer cases in Hong Kong196. Restricting HPV test to women aged ≥ 35 years avoids the age-related infection peak, and covers women with higher incidence of cervical cancer. By setting the testing population to women aged ≥ 35 years (in case of Hong Kong as used in this example) will therefore improve the positive predictive value of HPV testing for cervical cancer.
Super high-risk HPV types
At least 15 HPV types can be linked to cervical cancer with various degrees of risk association112. Initially, most HPV assays are designed to detect as many high-risk types as possible. In fact, the risk association among HPV types classified under this so-called “high-risk” group varies substantially. In recent years, it has been recognized that covering the HPV types with a small risk may jeopardize the overall positive predictive value of the assay. Thus type-specific assays, especially those targeting a group of HPV types with the highest risk association, have emerged. In this regard, HPV 16 and HPV 18 should be included as they are the two high-risk types most commonly found in cervical cancers across the world. HPV 31, 33 and 45 are the next group to be included, although their ranking showed some degrees of geographical variation203–207. HPV 52 and HPV 58 showed an even more skewed geographical distribution, and the clinical value of including these types into screening assays should be assessed based on the HPV type distribution data derived from the target population152,204,207–215.
An HPV16/18-specific test is expected to provide the highest positive predictive value. In the most recent guidelines from the United States, HPV16/18 testing is one of the options to triage women found to be HPV-positive but cytology-negative in primary screening using the co-test approach202.
Persistent infection
Since persistent infection is a pre-requisite for the development of cervical precancer and cancer, one can improve the positive predictive value by considering HPV testing results performed on specimens collected more than 12 months apart. If HPV is detected in both instances, the chance that it is a persistent rather than transient infection is higher. In this regard, an HPV type-specific test is necessary to differentiate repeated infections with different HPV types from genuine persistent infection with the same HPV type(s). It is only the latter that carries an increased risk of cancer development216. However, the limitation of this approach is that a proportion of women may not come back for the second test and may become lost to follow-up.
Reflex follow-up test
Another approach to improve the positive predictive value of HPV testing as a primary screening tool is to carry out a “reflex” follow-up test for HPV-positive samples. The “reflex” approach can prevent an extra visit and unnecessary anxiety while waiting for the follow-up test results. Biomarkers indicating the transforming activity of HPV can potentially serve this purpose102,217,218. The first approach is to detect direct indicators of HPV oncogene expression. At present, this mainly refers to the detection of mRNA encoded by the viral oncogenes, E6 and E7. Among women with normal cytology, atypical squamous cells of undetermined significance (ASCUS) and low-grade squamous intraepithelial lesions (LSIL), E6/E7 mRNA was detected in 30% of the HPV 16, 56% of the HPV 18 and 75% of the HPV 31 DNA-positive women, respectively. The mRNA test therefore potentially has a higher specificity compared to the HPV DNA test, and thus fewer patients would be referred for further testing or close follow-up219. Assays for direct measurement of E6 and E7 proteins from clinical specimens have been described and their clinical performance is being evaluated. The second approach is to detect biomarkers of increased cellular proliferation and chromosomal instability or those upregulated in response to HPV-encoded oncoproteins, including p16INK4a, Ki-67, topoisomerase IIA (TOP2), and minichromosome maintenance proteins (MCMP). Among these, p16INK4a is the most promising220,221. In many non-HPV-associated tumours, p16INK4a is inactivated by genetic deletion or hypermethylation, which leads to an increase in cyclin-dependent kinase activity and inactivation of Rb222. In contrast, in HPV-associated tumours, including cervical intraepithelial neoplasia and invasive cervical cancer, the inactivation of Rb by E7 leads to a marked overexpression of p16INK4a as a result of the lost of negative feedback regulation that depends on Rb activity223–225. It has been shown that, among 425 Pap-negative and HPV-positive women greater than 30 years of age, 25.4% were positive for p16/ki67 dual staining. The dual staining gave a sensitivity of 91.9% for CIN II and 96.4% for CIN III+ after a mean follow-up of 13.8 months (1–27 months)226. The result is encouraging but further studies are needed to support its clinical use.
Viral genome characterization
The patterns of viral integration at different stages of neoplastic progression have been investigated. Diverse, even conflicting, results have been reported. Some studies observed viral integration mainly from specimens of high-grade lesions227,228, whereas others found that viral integration takes place early during the course of infection and is detected in a substantial proportion of low-grade lesions229–232. It has been suggested that viral integration is a consequence rather than a cause of chromosomal instability233. Based on the available data, the main concern appears to be the lack of specificity. Basically, viral integration can be detected in normal and low-grade lesions, whereas cervical samples from cases of invasive cancer can harbour purely the episomal form of viral genome234–237.
DNA methylation is an epigenetic event that is linked to cancer development. A number of studies have been conducted to examine the association between the viral DNA methylation pattern and lesion severity. However, as with viral integration, most of the available data suggests that the patterns observed from low-grade and high-grade lesions are too diverse to achieve a clinically-useful predictive value238–242. Nevertheless, a recent study using a newer approach, pyrosequencing, has produced some promising results243. Viral integration and methylation have a strong biological basis, and further studies to explore their clinical application are worthwhile.
Co-test with cytology for primary screening
Co-test refers to the use of both HPV and cytology tests in parallel as first-line screening. The main advantage of co-test is the improvement in sensitivity for CIN II+ lesions; women who are double negative will have an extremely low-risk for CIN II+. The potential gain in cost-effectiveness from the expense of the extra test is the longer interval of safety supported by a double negative result. Data have suggested that the safety interval following an HPV-negative result could be as long as 5–7 years244–248. The most recent guidelines from the United Sates recommend HPV and cytology co-testing every 5 years for women aged 30–65 years201. However, the management of women with normal cytology but an HPV-positive result is an issue that is not yet completely resolved. The question is what proportion of the HPV-positive, cytology-normal women have transient HPV infection, and how many of them will turn out to be HPV-negative when the co-test is repeated in 1 year. This is fundamentally the same question when HPV testing is applied alone as a primary screening tool, and the answer varies substantially with the analytical sensitivity of the HPV assay and the prevalence of CIN II/III in the target population249. For patients with a normal Pap smear but an HPV-positive test, their risk of developing abnormal cytology and CIN II+ lesions are signifcantly higher than those with a double negative result. The increased risk was found to be HPV type-dependent, and the cumulative risk of those infected with HPV 16 reached 26% after a 13-year follow-up250. The United States Food and Drug Administration (US FDA) has approved the use of an HPV16/18 type-specific test in this particular clinical situation for women ≥ 30 years of age in order to identify individuals with a higher risk for developing disease in the future. According to the recent guidelines from the United States, women with HPV-positive and cytology-negative screening results can be either followed with the co-test 12 months later or triaged with the HPV16/18-specific test for referral to colposcopy201.
HPV as a triage for abnormal cytology
The first clinical application of HPV testing was on the triage of patients presented with ASCUS on Pap smear. Patients who were HPV-positive would be referred for colposcopy whilst those who were HPV-negative could be followed by repeating the Pap smear 12 months later251–254. This is still the most common application of HPV testing, and a large body of evidence is available to support returning women with negative HPV DNA results to a normal screening schedule.
The cost-effectiveness of using HPV testing to triage women with LSIL depends mainly on the context. For instance, it has been shown that about 80% of patients presenting with LSIL were HPV-positive; thus triage of LSIL by HPV testing was not recommended255. On the other hand, the US FDA has approved the use of HPV testing in post-menopausal women presenting with LSIL since the prevalence rate of HPV is low in this subset of patients. Therefore, data generated from context-specific studies are very important. The main variable is again the prevalence of HPV infection among the population tested. Such an approach may be effective for women well beyond the peak of infection, so that a substantial proportion of women will have an HPV-negative result and can be reassured256,257.
The underlying risk of having CIN II+ lesions among women with high-grade squamous intraepithelial lesions (HSIL) or atypical squamous cells is high, but HSIL (ASC-H) cannot be excluded, and colposcopy may be indicated for CIN II+ even if the HPV test result is negative258,259. HPV testing is also not very useful for women with atypical glandular cells because the underlying pathology may reside in the uterus. Whilst a positive HPV test suggests a cervical lesion, a negative test does not rule out such conditions as endometrial hyperplasia or cancer that are not HPV-related, and the risk of these is substantial in post-menopausal women260.
HPV for post-treatment surveillance
CIN II is often regarded as a threshold for treatment (Figure 2). The predominant mode is excision of the transformation zone using the loop electrosurgical excision procedure (LEEP). Since about 5 to 10% of patients have persistent or recurrent CIN II+ after LEEP, continual surveillance after treatment is needed. When compared to conventional cytology, HPV testing is more sensitive, carries a higher negative predictive value for recurrent or residual lesions, and is recommended by the American Congress of Obstetricians and Gynecologists for post-treatment follow-up261–263. In a cohort of 917 patients treated by LEEP, 81% were double-negative at 6 months after treatment and their risk of being diagnosed with CIN III+ was low. It has been suggested that patients with both negative cytology and negative HPV DNA can return to the 3-year screening program264.
Future challenges
The currently available prophylactic HPV vaccines are highly effective for the prevention of HPV16/18-related cervical neoplasia and offer some degree of cross-protection for lesions caused by related HPV types. A number of countries have implemented vaccination programs for adolescents and the coverage is expected to increase in the coming years265. These countries are also likely to be those running organized cervical screening programs265–268.
In the coming decades, several issues regarding cervical screening will need to be addressed269,270. The decrease in absolute incidence of cervical intraepithelial neoplasia and cervical cancer among those vaccinated will jeopardize the positive predictive value of any screening test. To achieve maximum cost-effectiveness, the screening strategy for those who have received the vaccine before their sexual debut should be different from those who have not271. These two populations will co-exist for a few decades and it will be a challenge to public health professionals to organize two systems in parallel for the same disease. Among those vaccinated, the chance of detecting a genuine abnormal signal (abnormal cytology results representing cervical precancer or cancer) will decrease, whereas the proportion of samples showing “noise” (abnormal cytology results due to inflammation and reactive changes that are self-limiting) will increase. The positive predictive value of minor cytological abnormalities will be even lower because of the reduced prevalence of CIN. As a result, because of its subjective nature, the reading and interpreting cytology results will be even more prone to human error. For these reasons, assays based on objective methods such as the detection of HPV and biomarkers will be advantageous.
Since HPV is a sexually-transmitted infection, using HPV testing for cervical screening may lead to anxiety and concerns about sexual relationships, and may have an emotional impact on the quality of life and a negative impact on mental health272.
Other practical challenges are the diversion of resources towards vaccination, and the resulting pressure for less frequent screening. At present, the recommended frequency in most countries is not less than once every 2–3 years. To lengthen the screening interval means getting closer to the interval required for advancing from low-grade to high-grade intraepithelial lesions or even to the development of invasive cancer. In other words, there may be just one chance to pick up at-risk women before the full development of invasive cancer. This will require a test with extreme sensitivity that is subject to a low positive predictive value, especially in the vaccinated population with a low incidence of disease. Lengthening of the interval for cytological examination is not advisable because the sensitivity of the test is low and repeated negative cytological tests are required to ensure no underlying precancerous or cancerous lesions. The high sensitivity of HPV testing, on the other hand, is more reassuring and allows lengthening of the screening interval. To overcome the low positive predictive value of highly-sensitive HPV testing in the vaccinated population with a low incidence of disease, a supplementary test for other biomarkers or co-testing with cytology is the way to move forward.
Conclusions
Technically speaking, HPV testing has several advantages over cytology-based screening, particularly in the situation where the incidence of cervical cancer and precancer decreases substantially following the widespread use of vaccination. However, although it is beyond the scope of this review, the psycho-social stigma associated with testing for a sexually-transmitted infection needs to be addressed. It has been shown that, even among well-educated women, most have not heard of HPV and do not know its association with cervical cancer273–276. Public education should go in parallel with the technical development in HPV testing. Moreover, whatever technology is used for screening, it is important to point out that the key for success is to reach a high coverage. Screening for cancer is like looking for a needle in a haystack, which is always challenging.
Glossary
Abbreviations
- ASCUS
atypical squamous cells of undetermined signifcance
- ASC-H
atypical squamous cells – cannot exclude high-grade squamous intraepithelial lesion
- CIN
cervical intraepithelial neoplasia
- EV
epidermodysplasia verruciformis
- HC2
hybrid capture assay version 2
- HPV
human papillomavirus
- HSIL
high-grade squamous intraepithelial lesion
- IS
international standard
- LEEP
loop electrosurgical excision procedure
- LSIL
low-grade squamous intraepithelial lesion
- LCR
long control region
- MCMP
minichromosome maintenance proteins
- ORF
open reading frame
- PCR
polymerase chain reaction
- Rb
retinoblastoma
- TOP2
topoisomerase II
- US FDA
United States Food and Drug Administration
- WHO
World Health Organization
Declaration of interest
None to declare
References
- 1.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 3.Syrjänen S. Current concepts on human papillomavirus infections in children. APMIS. 2010;118:494–509. doi: 10.1111/j.1600-0463.2010.02620.x. [DOI] [PubMed] [Google Scholar]
- 4.Partridge JM, Koutsky LA. Genital human papillomavirus infection in men. Lancet Infect Dis. 2006;6:21–31. doi: 10.1016/S1473-3099(05)70323-6. [DOI] [PubMed] [Google Scholar]
- 5.Feller L, Wood NH, Khammissa RA, Lemmer J. Human papillomavirus-mediated carcinogenesis and HPV-associated oral and oropharyngeal squamous cell carcinoma. Part 1: human papillomavirus-mediated carcinogenesis. Head Face Med. 2010;6:14. doi: 10.1186/1746-160X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feller L, Wood NH, Khammissa RA, Lemmer J. Human papillomavirus-mediated carcinogenesis and HPV-associated oral and oropharyngeal squamous cell carcinoma. Part 2: human papillomavirus associated oral and oropharyngeal squamous cell carcinoma. Head Face Med. 2010;6:15. doi: 10.1186/1746-160X-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher TQ, Derkay CS. Recurrent respiratory papillomatosis: update 2008. Curr Opin Otolaryngol Head Neck Surg. 2008;16:536–542. doi: 10.1097/MOO.0b013e328316930e. [DOI] [PubMed] [Google Scholar]
- 8.Dubina M, Goldenberg G. Viral-associated nonmelanoma skin cancers: a review. Am J Dermatopathol. 2009;31:561–573. doi: 10.1097/DAD.0b013e3181a58234. [DOI] [PubMed] [Google Scholar]
- 9.Feltkamp MC, de Koning MN, Bavinck JN, Ter Schegget J. Betapapillomaviruses: innocent bystanders or causes of skin cancer. J Clin Virol. 2008;43:353–360. doi: 10.1016/j.jcv.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Handisurya A, Schellenbacher C, Kirnbauer R. Diseases caused by human papillomaviruses (HPV) J Dtsch Dermatol Ges. 2009;7:453–466. doi: 10.1111/j.1610-0387.2009.06988.x. quiz 466, 467. [DOI] [PubMed] [Google Scholar]
- 11.Pfister H. Chapter 8: Human papillomavirus and skin cancer. J Natl Cancer Inst Monogr. 2003;31:52–56. doi: 10.1093/oxfordjournals.jncimonographs.a003483. [DOI] [PubMed] [Google Scholar]
- 12.zur Hausen H. Papillomaviruses in the causation of human cancers – a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 13.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy DR, Schiller JT. Prophylactic human papillomavirus vaccines. J Clin Invest. 2006;116:1167–1173. doi: 10.1172/JCI28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80:2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 16.Hazard K, Karlsson A, Andersson K, Ekberg H, Dillner J, Forslund O. Cutaneous human papillomaviruses persist on healthy skin. J Invest Dermatol. 2007;127:116–119. doi: 10.1038/sj.jid.5700570. [DOI] [PubMed] [Google Scholar]
- 17.Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol. 2000;74:11636–11641. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mammas IN, Sourvinos G, Spandidos DA. Human papilloma virus (HPV) infection in children and adolescents. Eur J Pediatr. 2009;168:267–273. doi: 10.1007/s00431-008-0882-z. [DOI] [PubMed] [Google Scholar]
- 19.Kirnbauer R, Lenz P, Okun MM. Human Papillomavirus. In: Bolognia J, Jorizzo J, Rapini R, editors. Dermatology, Vol. 1. London: Mosby; 2008. pp. 1183–1198. [Google Scholar]
- 20.McLaughlin JS, Shafritz AB. Cutaneous warts. J Hand Surg Am. 2011;36:343–344. doi: 10.1016/j.jhsa.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 21.Syrjänen KJ. Annual disease burden due to human papillomavirus (HPV) 6 and 11 infections in Finland. Scand J Infect Dis Suppl. 2009;107:3–32. doi: 10.1080/00365540902887730. [DOI] [PubMed] [Google Scholar]
- 22.Chan PK, Luk AC, Luk TN, Lee KF, Cheung JL, Ho KM, Lo KK. Distribution of human papillomavirus types in anogenital warts of men. J Clin Virol. 2009;44:111–114. doi: 10.1016/j.jcv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Oon SF, Winter DC. Perianal condylomas, anal squamous intraepithelial neoplasms and screening: a review of the literature. J Med Screen. 2010;17:44–49. doi: 10.1258/jms.2009.009058. [DOI] [PubMed] [Google Scholar]
- 24.Hsueh PR. Human papillomavirus, genital warts, and vaccines. J Microbiol Immunol Infect. 2009;42:101–106. [PubMed] [Google Scholar]
- 25.Ghaemmaghami F, Nazari Z, Mehrdad N. Female genital warts. Asian Pac J Cancer Prev. 2007;8:339–347. [PubMed] [Google Scholar]
- 26.Larson DA, Derkay CS. Epidemiology of recurrent respiratory papillomatosis. APMIS. 2010;118:450–454. doi: 10.1111/j.1600-0463.2010.02619.x. [DOI] [PubMed] [Google Scholar]
- 27.Bonagura VR, Hatam LJ, Rosenthal DW, de Voti JA, Lam F, Steinberg BM, Abramson AL. Recurrent respiratory papillomatosis: a complex defect in immune responsiveness to human papillomavirus-6 and -11. APMIS. 2010;118:455–470. doi: 10.1111/j.1600-0463.2010.02617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasca RA, Clarke RW. Recurrent respiratory papillomatosis. Arch Dis Child. 2006;91:689–691. doi: 10.1136/adc.2005.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feller L, Khammissa RA, Wood NH, Malema V, Meyerov R, Lemmer J. Focal epithelial hyperplasia (Heck disease) related to highly active antiretroviral therapy in an HIV-seropositive child. A report of a case, and a review of the literature. SADJ. 2010;65:172–175. [PubMed] [Google Scholar]
- 30.Feller L, Khammissa RA, Wood NH, Marnewick JC, Meyerov R, Lemmer J. HPV-associated oral warts. SADJ. 2011;66:82–85. [PubMed] [Google Scholar]
- 31.González J V, Gutiérrez RA, Keszler A, Colacino Mdel C, Alonio LV, Teyssie AR, Picconi MA. Human papillomavirus in oral lesions. Medicina (B Aires) 2007;67:363–368. [PubMed] [Google Scholar]
- 32.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 33.Grulich AE, Jin F, Conway EL, Stein AN, Hocking J. Cancers attributable to human papillomavirus infection. Sex Health. 2010;7:244–252. doi: 10.1071/SH10020. [DOI] [PubMed] [Google Scholar]
- 34.Ramoz N, Rueda LA, Bouadjar B, Montoya LS, Orth G, Favre M. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat Genet. 2002;32:579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- 35.Orth G. Genetics of epidermodysplasia verruciformis: Insights into host defense against papillomaviruses. Semin Immunol. 2006;18:362–374. doi: 10.1016/j.smim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Lazarczyk M, Cassonnet P, Pons C, Jacob Y, Favre M. The EVER proteins as a natural barrier against papillomaviruses: a new insight into the pathogenesis of human papillomavirus infections. Microbiol Mol Biol Rev. 2009;73:348–370. doi: 10.1128/MMBR.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterling JC. Human papillomaviruses and skin cancer. J Clin Virol. 2005;32:S67–71. doi: 10.1016/j.jcv.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 38.Patel T, Morrison LK, Rady P, Tyring S. Epidermodysplasia verruciformis and susceptibility to HPV. Dis Markers. 2010;29:199–206. doi: 10.3233/DMA-2010-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forslund O, Iftner T, Andersson K, Lindelof B, Hradil E, Nordin P, et al. Viraskin Study Group Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis. 2007;196:876–883. doi: 10.1086/521031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel AS, Karagas MR, Perry AE, Nelson HH. Exposure profles and human papillomavirus infection in skin cancer: an analysis of 25 genus beta-types in a population-based study. J Invest Dermatol. 2008;128:2888–2893. doi: 10.1038/jid.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asgari MM, Kiviat NB, Critchlow CW, Stern JE, Argenyi ZB, Raugi GJ, et al. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J Invest Dermatol. 2008;128:1409–1417. doi: 10.1038/sj.jid.5701227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akgül B, Cooke JC, Storey A. HPV-associated skin disease. J Pathol. 2006;208:165–175. doi: 10.1002/path.1893. [DOI] [PubMed] [Google Scholar]
- 43.Bleeker MC, Heideman DA, Snijders PJ, Horenblas S, Dillner J, Meijer CJ. Penile cancer: epidemiology, pathogenesis and prevention. World J Urol. 2009;27:141–150. doi: 10.1007/s00345-008-0302-z. [DOI] [PubMed] [Google Scholar]
- 44.Parkin DM, Bray F. Chapter 2: The burden of HPV-related cancers. Vaccine. 2006;24(Suppl 3):S3/11–25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 45.Muñoz N, Castellsagué X, de González AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24(Suppl 3):S3/1–10. doi: 10.1016/j.vaccine.2006.05.115. [DOI] [PubMed] [Google Scholar]
- 46.Chao A, Chen TC, Hsueh C, Huang CC, Yang JE, Hsueh S, et al. Human papillomavirus in vaginal intraepithelial neoplasia. Int J Cancer. 2012;131:E259–E268. doi: 10.1002/ijc.27354. [DOI] [PubMed] [Google Scholar]
- 47.Hildesheim A, Han CL, Brinton LA, Kurman RJ, Schiller JT. Human papillomavirus type 16 and risk of preinvasive and invasive vulvar cancer: results from a seroepidemiological case-control study. Obstet Gynecol. 1997;90:748–754. doi: 10.1016/S0029-7844(97)00467-5. [DOI] [PubMed] [Google Scholar]
- 48.Tavassoli FA, Devilee P, editors. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. Lyon: IARC Press; 2003. Tumours of the vulva. World Health Organization Classification of Tumours; pp. 313–333. [Google Scholar]
- 49.Junge J, Poulsen H, Horn T, Hørding U, Lundvall F. Human papillomavirus (HPV) in vulvar dysplasia and carcinoma in situ. APMIS. 1995;103:501–510. doi: 10.1111/j.1699-0463.1995.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 50.Alonso I, Fusté V, del Pino M, Castillo P, Torné A, Fusté P, et al. Does human papillomavirus infection imply a different prognosis in vulvar squamous cell carcinoma? Gynecol Oncol. 2011;122:509–514. doi: 10.1016/j.ygyno.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 51.Daling JR, Madeleine MM, Johnson LG, Schwartz SM, Shera KA, Wurscher MA, et al. Human papillomavirus, smoking, and sexual practices in the etiology of anal cancer. Cancer. 2004;101:270–280. doi: 10.1002/cncr.20365. [DOI] [PubMed] [Google Scholar]
- 52.Dietz CA, Nyberg CR. Genital, oral, and anal human papillomavirus infection in men who have sex with men. J Am Osteopath Assoc. 2011;111(3 Suppl 2):S19–S25. [PubMed] [Google Scholar]
- 53.Palefsky J. Can HPV vaccination help to prevent anal cancer? Lancet Infect Dis. 2010;10:815–816. doi: 10.1016/S1473-3099(10)70226-7. [DOI] [PubMed] [Google Scholar]
- 54.Smith PG, Kinlen LJ, White GC, Adelstein AM, Fox AJ. Mortality of wives of men dying with cancer of the penis. Br J Cancer. 1980;41:422–428. doi: 10.1038/bjc.1980.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picconi MA, Eiján AM, Distéfano AL, Pueyo S, Alonio LV, Gorostidi ST, et al. Human papillomavirus (HPV) DNA in penile carcinomas in Argentina: analysis of primary tumors and lymph nodes. J Med Virol. 2000;61:65–69. doi: 10.1002/(sici)1096-9071(200005)61:1<65::aid-jmv10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 56.Palefsky JM. Human papillomavirus-related disease in men: not just a women's issue. J Adolesc Health. 2010;46(4 Suppl):S12–S19. doi: 10.1016/j.jadohealth.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anic GM, Giuliano AR. Genital HPV infection and related lesions in men. Prev Med. 2011;53(Suppl 1):S36–S41. doi: 10.1016/j.ypmed.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 59.WHO. 2008–2013 Action Plan for the Global Strategy for the Prevention and Control of Noncommunicable Diseases. Geneva, Switzerland: World Health Organization; 2008. pp. 1–42. [Google Scholar]
- 60.Yang BH, Bray FI, Parkin DM, Sellors JW, Zhang ZF. Cervical cancer as a priority for prevention in different world regions: an evaluation using years of life lost. Int J Cancer. 2004;109:418–424. doi: 10.1002/ijc.11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 62.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 63.Chaturvedi AK, Engels EA, Pfeifer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 65.Allen CT, Lewis JS, Jr, El-Mofty SK, Haughey BH, Nussenbaum B. Human papillomavirus and oropharynx cancer: biology, detection and clinical implications. Laryngoscope. 2010;120:1756–1772. doi: 10.1002/lary.20936. [DOI] [PubMed] [Google Scholar]
- 66.Marur S, D'Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Syrjänen S, Lodi G, von Bültzingslöwen I, Aliko A, Arduino P, Campisi G, et al. Human papillomaviruses in oral carcinoma and oral potentially malignant disorders: a systematic review. Oral Dis. 2011;17(Suppl 1):58–72. doi: 10.1111/j.1601-0825.2011.01792.x. [DOI] [PubMed] [Google Scholar]
- 68.Toner M, O'Regan EM. Head and neck squamous cell carcinoma in the young: a spectrum or a distinct group? Part 1. Head Neck Pathol. 2009;3:246–248. doi: 10.1007/s12105-009-0135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leemans CR, Braakhuis BJ, Brakenhof RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 70.Howley PM, Lowy DR. Papillomavirus. In: Knipe DM, Howley PM, et al., editors. Fields Virology. 5th ed. Vol 2. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 2299–2354. [Google Scholar]
- 71.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doorbar J. The papillomavirus life cycle. J Clin Virol. 2005;32(Suppl 1):S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 73.Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into viruslike particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou J, Sun XY, Stenzel DJ, Frazer IH. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufcient for assembly of HPV virion-like particles. Virology. 1991;185:251–257. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]
- 75.Chen J, Ni G, Liu XS. Papillomavirus virus like particle-based therapeutic vaccine against human papillomavirus infection related diseases: immunological problems and future directions. Cell Immunol. 2011;269:5–9. doi: 10.1016/j.cellimm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 76.Villa LL. HPV prophylactic vaccination: The first years and what to expect from now. Cancer Lett. 2011;305:106–112. doi: 10.1016/j.canlet.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 77.Frazer IH. Cervical cancer vaccine development. Sex Health. 2010;7:230–234. doi: 10.1071/SH09132. [DOI] [PubMed] [Google Scholar]
- 78.Tumban E, Peabody J, Peabody DS, Chackerian B. A pan-HPV vaccine based on bacteriophage PP7 VLPs displaying broadly cross-neutralizing epitopes from the HPV minor capsid protein, L2. PLoS One. 2011;6:e23310. doi: 10.1371/journal.pone.0023310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Conway MJ, Cruz L, Alam S, Christensen ND, Meyers C. Cross-neutralization potential of native human papillomavirus N-terminal L2 epitopes. PLoS One. 2011;6:e16405. doi: 10.1371/journal.pone.0016405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caldeira Jdo C, Medford A, Kines RC, Lino CA, Schiller JT, Chackerian B, Peabody DS. Immunogenic display of diverse peptides, including a broadly cross-type neutralizing human papillomavirus L2 epitope, on virus-like particles of the RNA bacteriophage PP7. Vaccine. 2010;28:4384–4393. doi: 10.1016/j.vaccine.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stanley M. Prospects for new human papillomavirus vaccines. Curr Opin Infect Dis. 2010;23:70–75. doi: 10.1097/QCO.0b013e328334c0e1. [DOI] [PubMed] [Google Scholar]
- 82.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 83.Schefner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 84.White AE, Livanos EM, Tlsty TD. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 85.Havre PA, Yuan J, Hedrick L, Cho KR, Glazer PM. p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer Res. 1995;55:4420–4424. [PubMed] [Google Scholar]
- 86.Kessis TD, Connolly DC, Hedrick L, Cho KR. Expression of HPV16 E6 or E7 increases integration of foreign DNA. Oncogene. 1996;13:427–431. [PubMed] [Google Scholar]
- 87.Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tomas M, Banks L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- 89.Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 90.Reznikof CA, Yeager TR, Belair CD, Savelieva E, Puthenveettil JA, Stadler WM. Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res. 1996;56:2886–2890. [PubMed] [Google Scholar]
- 91.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 92.Dyson N, Guida P, Münger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arroyo M, Bagchi S, Raychaudhuri P. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex. Mol Cell Biol. 1993;13:6537–6546. doi: 10.1128/mcb.13.10.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smotkin D, Wettstein FO. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang JL, Tsao YP, Liu DW, Huang SJ, Lee WH, Chen SL. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J Biomed Sci. 2001;8:206–213. doi: 10.1007/BF02256414. [DOI] [PubMed] [Google Scholar]
- 97.Maufort JP, Shai A, Pitot HC, Lambert PF. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res. 2010;70:2924–2931. doi: 10.1158/0008-5472.CAN-09-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venuti A, Paolini F, Nasir L, Corteggio A, Roperto S, Campo MS, Borzacchiello G. Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer. 2011;10:140. doi: 10.1186/1476-4598-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 100.Romanczuk H, Tierry F, Howley PM. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- 103.de Villiers EM, Bernard HU, Broker T, Delius H, zur Hausen H. Family Papillomaviridae. In: Fauquet CM, Mayo MA, Manilof J, Desselberger U, Ball LA, editors. Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; 2005. pp. 239–255. [Google Scholar]
- 104.Van Regenmortel MH, Fauquet CM, Bishop DH, Carstens EB, Estes MK, Lemon SM, et al., editors. Virus Taxonomy. Seventh Report of the International Committee for the Taxonomy of Viruses. San Diego: Academic Press; 2000. Family Papillomaviridae; pp. 247–251. [Google Scholar]
- 105.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 106.Calleja-Macias IE, Villa LL, Prado JC, Kalantari M, Allan B, Williamson AL, et al. Worldwide genomic diversity of the high-risk human papillomavirus types 31, 35, 52, and 58, four close relatives of human papillomavirus type 16. J Virol. 2005;79:13630–13640. doi: 10.1128/JVI.79.21.13630-13640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chan PK, Luk AC, Park JS, Smith-McCune KK, Palefsky JM, Konno R, et al. Identification of human papillomavirus type 58 lineages and the distribution worldwide. J Infect Dis. 2011;203:1565–1573. doi: 10.1093/infdis/jir157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Z, Schifman M, Herrero R, Desalle R, Anastos K, Segondy M, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One. 2011;6:e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen Z, DeSalle R, Schifman M, Herrero R, Burk RD. Evolutionary dynamics of variant genomes of human papillomavirus types 18, 45, and 97. J Virol. 2009;83:1443–1455. doi: 10.1128/JVI.02068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prado JC, Calleja-Macias IE, Bernard HU, Kalantari M, Macay SA, Allan B, et al. Worldwide genomic diversity of the human papillomaviruses-53, 56, and 66, a group of high-risk HPVs unrelated to HPV-16 and HPV-18. Virology. 2005;340:95–104. doi: 10.1016/j.virol.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 111.Raiol T, Wyant PS, de Amorim RM, Cerqueira DM, Milanezi NG, Brígido Mde M, et al. Genetic variability and phylogeny of the high-risk HPV-31, -33, -35, -52, and -58 in central Brazil. J Med Virol. 2009;81:685–692. doi: 10.1002/jmv.21432. [DOI] [PubMed] [Google Scholar]
- 112.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah K V, et al. International Agency for Research on Cancer Multicenter Cervical Cancer Study Group Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 113.Schifman M, Herrero R, Desalle R, Hildesheim A, Wacholder S, Rodriguez AC, et al. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 114.Herrero R. Human papillomavirus (HPV) vaccines: limited cross-protection against additional HPV types. J Infect Dis. 2009;199:919–922. doi: 10.1086/597308. [DOI] [PubMed] [Google Scholar]
- 115.Wheeler CM, Castellsagué X, Garland SM, Szarewski A, Paavonen J, Naud P, et al. for the HPV PATRICIA Study Group. Cross-protective efcacy of HPV-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by non-vaccine oncogenic HPV types: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012;13:100–110. doi: 10.1016/S1470-2045(11)70287-X. [DOI] [PubMed] [Google Scholar]
- 116.Lehtinen M, Paavonen J, Wheeler CM, Jaisamrarn U, Garland SM, Castellsagué X, et al. for the HPV PATRICIA Study Group. Overall efcacy of HPV-16/18 AS04-adjuvanted vaccine against grade 3 or greater cervical intraepithelial neoplasia: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012;13:100–110. doi: 10.1016/S1470-2045(11)70286-8. [DOI] [PubMed] [Google Scholar]
- 117.Brown DR, Kjaer SK, Sigurdsson K, Iversen OE, Hernandez-Avila M, Wheeler CM, et al. The impact of quadrivalent human papillomavirus (HPV; types 6, 11, 16, and 18) L1 virus-like particle vaccine on infection and disease due to oncogenic nonvaccine HPV types in generally HPV-naive women aged 16–26 years. J Infect Dis. 2009;199:926–935. doi: 10.1086/597307. [DOI] [PubMed] [Google Scholar]
- 118.Wheeler CM, Kjaer SK, Sigurdsson K, Iversen OE, Hernandez-Avila M, Perez G, et al. The impact of quadrivalent human papillomavirus (HPV; types 6, 11, 16, and 18) L1 virus-like particle vaccine on infection and disease due to oncogenic nonvaccine HPV types in sexually active women aged 16–26 years. J Infect Dis. 2009;199:936–944. doi: 10.1086/597309. [DOI] [PubMed] [Google Scholar]
- 119.Schifman M, Cliford G, Buonaguro FM. Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer. 2009;4:8. doi: 10.1186/1750-9378-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B. El Ghissassi F; WHO International Agency for Research on Cancer. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 121.Chow LT, Broker TR. In vitro experimental systems for HPV: epithelial raft cultures for investigations of viral reproduction and pathogenesis and for genetic analyses of viral proteins and regulatory sequences. Clin Dermatol. 1997;15:217–227. doi: 10.1016/s0738-081x(97)00069-2. [DOI] [PubMed] [Google Scholar]
- 122.Dollard SC, Wilson JL, Demeter LM, Bonnez W, Reichman RC, Broker TR, Chow LT. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. OFF. Genes Dev. 1992;6:1131–1142. doi: 10.1101/gad.6.7.1131. [DOI] [PubMed] [Google Scholar]
- 123.Meyers C, Frattini MG, Hudson JB, Laimins LA. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 124.Stanley M. Immune responses to human papillomavirus. Vaccine. 2006;24(Suppl 1):S16–S22. doi: 10.1016/j.vaccine.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 125.Frazer IH. Interaction of human papillomaviruses with the host immune system: a well evolved relationship. Virology. 2009;384:410–414. doi: 10.1016/j.virol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 126.Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nat Rev Immunol. 2004;4:211–222. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 128.Theoflopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 129.Chang YE, Laimins LA. Microarray analysis identifes interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol. 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kobayashi A, Greenblatt RM, Anastos K, Minkof H, Massad LS, Young M, et al. Functional attributes of mucosal immunity in cervical intraepithelial neoplasia and effects of HIV infection. Cancer Res. 2004;64:6766–6774. doi: 10.1158/0008-5472.CAN-04-1091. [DOI] [PubMed] [Google Scholar]
- 132.Kirnbauer R, Hubbert NL, Wheeler CM, Becker TM, Lowy DR, Schiller JT. A virus-like particle enzyme-linked immunosorbent assay detects serum antibodies in a majority of women infected with human papillomavirus type 16. J Natl Cancer Inst. 1994;86:494–499. doi: 10.1093/jnci/86.7.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wikström A, van Doornum GJ, Quint WG, Schiller JT, Dillner J. Identification of human papillomavirus seroconversions. J Gen Virol. 1995;76:529–539. doi: 10.1099/0022-1317-76-3-529. [DOI] [PubMed] [Google Scholar]
- 134.Carter JJ, Wipf GC, Hagensee ME, McKnight B, Habel LA, Lee SK, et al. Use of human papillomavirus type 6 capsids to detect antibodies in people with genital warts. J Infect Dis. 1995;172:11–18. doi: 10.1093/infdis/172.1.11. [DOI] [PubMed] [Google Scholar]
- 135.Carter JJ, Koutsky LA, Hughes JP, Lee SK, Kuypers J, Kiviat N, Galloway DA. Comparison of human papillomavirus types 16, 18, and 6 capsid antibody responses following incident infection. J Infect Dis. 2000;181:1911–1919. doi: 10.1086/315498. [DOI] [PubMed] [Google Scholar]
- 136.Heideman DA, Hesselink AT, Berkhof J, van Kemenade F, Melchers WJ, Daalmeijer NF, et al. Clinical validation of the Cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol. 2011;49:3983–3985. doi: 10.1128/JCM.05552-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meijer CJ, Berkhof J, Castle PE, Hesselink AT, Franco EL, Ronco G, et al. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int J Cancer. 2009;124:516–520. doi: 10.1002/ijc.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meijer CJ, Berkhof H, Heideman DA, Hesselink AT, Snijders PJ. Validation of high-risk HPV tests for primary cervical screening. J Clin Virol. 2009;46(Suppl 3):S1–S4. doi: 10.1016/S1386-6532(09)00540-X. [DOI] [PubMed] [Google Scholar]
- 139.Lörincz A. Hybrid capture method for detection of human papillomavirus DNA in clinical specimens: a tool for clinical management of equivocal Pap smears and for population screening. J Obstet Gynaecol Res. 1996;22:629–36. doi: 10.1111/j.1447-0756.1996.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 140.Peyton CL, Schifman M, Lorincz AT, Hunt WC, Mielzynska I, Bratti C, et al. Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. Clin Microbiol. 1998;36:3248–3254. doi: 10.1128/jcm.36.11.3248-3254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Castle PE, Schifman M, Burk RD, Wacholder S, Hildesheim A, Herrero R, et al. Restricted cross-reactivity of hybrid capture 2 with nononcogenic human papillomavirus types. Cancer Epidemiol Biomarkers Prev. 2002;11:1394–1399. [PubMed] [Google Scholar]
- 142.Ting Y, Manos M. Detection and typing of genital human papillomaviruses. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 356–366. [Google Scholar]
- 143.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlée F, Hildesheim A, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38:357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Estrade C, Menoud PA, Nardelli-Haefiger D, Sahli R. Validation of a low-cost human papillomavirus genotyping assay based on PGMY PCR and reverse blotting hybridization with reusable membranes. J Clin Microbiol. 2011;49:3474–3481. doi: 10.1128/JCM.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 146.Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35:791–795. doi: 10.1128/jcm.35.3.791-795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chan PK, Cheung TH, Tam AO, Lo KW, Yim SF, Yu MM, et al. Biases in human papillomavirus genotype prevalence assessment associated with commonly used consensus primers. Int J Cancer. 2006;118:243–245. doi: 10.1002/ijc.21299. [DOI] [PubMed] [Google Scholar]
- 148.Kleter B, van Doorn LJ, ter Schegget J, Schrauwen L, van Krimpen K, Burger M, et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jeney C, Takács T, Sebe A, Schaf Z. Detection and typing of 46 genital human papillomaviruses by the L1F/L1R primer system based multiplex PCR and hybridization. J Virol Methods. 2007;140:32–42. doi: 10.1016/j.jviromet.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 150.Bernard HU, Chan SY, Manos MM, Ong CK, Villa LL, Delius H, et al. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170:1077–1085. doi: 10.1093/infdis/170.5.1077. [DOI] [PubMed] [Google Scholar]
- 151.Chan PK, Li WH, Chan MY, Ma WL, Cheung JL, Cheng AF. High prevalence of human papillomavirus type 58 in Chinese women with cervical cancer and precancerous lesions. J Med Virol. 1999;59:232–238. [PubMed] [Google Scholar]
- 152.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 153.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 154.McLaughlin-Drubin ME, Münger K. Oncogenic activities of human papillomaviruses. Virus Res. 2009;143:195–208. doi: 10.1016/j.virusres.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu Y, Chen JJ, Gao Q, Dalal S, Hong Y, Mansur CP, et al. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pim D, Banks L. Interaction of viral oncoproteins with cellular target molecules: infection with high-risk vs low-risk human papillomaviruses. APMIS. 2010;118:471–493. doi: 10.1111/j.1600-0463.2010.02618.x. [DOI] [PubMed] [Google Scholar]
- 157.Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, Tommasino M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes. 2010;40:1–13. doi: 10.1007/s11262-009-0412-8. [DOI] [PubMed] [Google Scholar]
- 158.Ganguly N, Parihar SP. Human papillomavirus E6 and E7 oncoproteins as risk factors for tumorigenesis. J Biosci. 2009;34:113–123. doi: 10.1007/s12038-009-0013-7. [DOI] [PubMed] [Google Scholar]
- 159.Howie HL, Katzenellenbogen RA, Galloway DA. Papillomavirus E6 proteins. Virology. 2009;384:324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gnanamony M, Peedicayil A, Subhashini J, Ram TS, Rajasekar A, Gravitt P, Abraham P. Detection and quantitation of HPV 16 and 18 in plasma of Indian women with cervical cancer. Gynecol Oncol. 2010;116:447–451. doi: 10.1016/j.ygyno.2009.10.081. [DOI] [PubMed] [Google Scholar]
- 161.Flores-Munguia R, Siegel E, Klimecki WT, Giuliano AR. Performance assessment of eight high-throughput PCR assays for viral load quantitation of oncogenic HPV types. J Mol Diagn. 2004;6:115–124. doi: 10.1016/S1525-1578(10)60499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chow LT, Broker TR, Steinberg BM. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010;118:422–449. doi: 10.1111/j.1600-0463.2010.02625.x. [DOI] [PubMed] [Google Scholar]
- 163.Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 164.Chow LT, Broker TR. Mechanisms and regulation of papillomavirus DNA replication. In: Campo MS, editor. Recent Advances in Papillomavirus Research. Norwich, UK: Caister Academic Press; 2006. pp. 53–71. [Google Scholar]
- 165.Sotlar K, Selinka HC, Menton M, Kandolf R, Bültmann B. Detection of human papillomavirus type 16 E6/E7 oncogene transcripts in dysplastic and nondysplastic cervical scrapes by nested RT-PCR. Gynecol Oncol. 1998;69:114–121. doi: 10.1006/gyno.1998.4994. [DOI] [PubMed] [Google Scholar]
- 166.Sotlar K, Stubner A, Diemer D, Menton S, Menton M, Dietz K, et al. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J Med Virol. 2004;74:107–116. doi: 10.1002/jmv.20153. [DOI] [PubMed] [Google Scholar]
- 167.Lie AK, Risberg B, Borge B, Sandstad B, Delabie J, Rimala R, et al. DNA- versus RNA-based methods for human papillomavirus detection in cervical neoplasia. Gynecol Oncol. 2005;97:908–915. doi: 10.1016/j.ygyno.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 168.Andersson S, Hansson B, Norman I, Gaberi V, Mints M, Hjerpe A, et al. Expression of E6/E7 mRNA from ‘high risk’ human papillomavirus in relation to CIN grade, viral load and p16INK4a. Int J Oncol. 2006;29:705–711. [PubMed] [Google Scholar]
- 169.Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, et al. Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of the Hybrid Capture 2 test for identification of women at risk of cervical cancer. J Clin Microbiol. 2010;48:2779–2785. doi: 10.1128/JCM.00382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Castle PE, Dockter J, Giachetti C, Garcia FA, McCormick MK, Mitchell AL, et al. A cross-sectional study of a prototype carcinogenic human papillomavirus E6/E7 messenger RNA assay for detection of cervical precancer and cancer. Clin Cancer Res. 2007;13:2599–2605. doi: 10.1158/1078-0432.CCR-06-2881. [DOI] [PubMed] [Google Scholar]
- 171.Cattani P, Zannoni GF, Ricci C, D'Onghia S, Trivellizzi IN, Di Franco A, et al. Clinical performance of human papillomavirus E6 and E7 mRNA testing for high-grade lesions of the cervix. J Clin Microbiol. 2009;47:3895–3901. doi: 10.1128/JCM.01275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dockter J, Schroder A, Hill C, Guzenski L, Monsonego J, Giachetti C. Clinical performance of the APTIMA HPV Assay for the detection of high-risk HPV and high-grade cervical lesions. J Clin Virol. 2009;45(Suppl 1):S55–S61. doi: 10.1016/S1386-6532(09)70009-5. [DOI] [PubMed] [Google Scholar]
- 173.Halfon P, Benmoura D, Agostini A, Khiri H, Martineau A, Penaranda G, Blanc B. Relevance of HPV mRNA detection in a population of ASCUS plus women using the NucliSENS EasyQ HPV assay. J Clin Virol. 2010;47:177–181. doi: 10.1016/j.jcv.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 174.Burger EA, Kornør H, Klemp M, Lauvrak V, Kristiansen IS. HPV mRNA tests for the detection of cervical intraepithelial neoplasia: a systematic review. Gynecol Oncol. 2011;120:430–438. doi: 10.1016/j.ygyno.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 175.Dreier K, Scheiden R, Lener B, Ehehalt D, Pircher H, Müller-Holzner E, et al. Subcellular localization of the human papillomavirus 16 E7 oncoprotein in CaSki cells and its detection in cervical adenocarcinoma and adenocarcinoma in situ. Virology. 2011;409:54–68. doi: 10.1016/j.virol.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ressler S, Scheiden R, Dreier K, Laich A, Müller-Holzner E, Pircher H, et al. High-risk human papillomavirus E7 oncoprotein detection in cervical squamous cell carcinoma. Clin Cancer Res. 2007;13:7067–7072. doi: 10.1158/1078-0432.CCR-07-1222. [DOI] [PubMed] [Google Scholar]
- 177.Franco EL, Duarte-Franco E, Rohan TE. Evidence-based policy recommendations on cancer screening and prevention. Cancer Detect Prev. 2002;26:350–361. doi: 10.1016/s0361-090x(02)00118-6. [DOI] [PubMed] [Google Scholar]
- 178.Sasieni P, Adams J. Efect of screening on cervical cancer mortality in England and Wales: analysis of trends with an age period cohort model. BMJ. 1999;318:1244–1245. doi: 10.1136/bmj.318.7193.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.van der Aa MA, Pukkala E, Coebergh JW, Anttila A, Siesling S. Mass screening programmes and trends in cervical cancer in Finland and the Netherlands. Int J Cancer. 2008;122:1854–1858. doi: 10.1002/ijc.23276. [DOI] [PubMed] [Google Scholar]
- 180.Bulkmans NW, Rozendaal L, Voorhorst FJ, Snijders PJ, Meijer CJ. Long-term protective efect of high-risk human papillomavirus testing in population-based cervical screening. Br J Cancer. 2005;92:1800–1802. doi: 10.1038/sj.bjc.6602541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sasieni PD, Cuzick J, Lynch-Farmery E. Estimating the efcacy of screening by auditing smear histories of women with and without cervical cancer. The National Co-ordinating Network for Cervical Screening Working Group. Br J Cancer. 1996;73:1001–1005. doi: 10.1038/bjc.1996.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.WHO/ICO Information Centre on HPV and Cervical Cancer. HPV and cervical cancer in the 2007 report. Vaccine. 2007;25:C1–C230. doi: 10.1016/S0264-410X(07)01183-8. [DOI] [PubMed] [Google Scholar]
- 183.Cuzick J, Clavel C, Petry KU, Meijer CJ, Hoyer H, Ratnam S, et al. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119:1095–1101. doi: 10.1002/ijc.21955. [DOI] [PubMed] [Google Scholar]
- 184.Poljak M, Kocjan BJ. Commercially available assays for multiplex detection of alpha human papillomaviruses. Expert Rev Anti Infect Ter. 2010;8:1139–1162. doi: 10.1586/eri.10.104. [DOI] [PubMed] [Google Scholar]
- 185.Naucler P, Ryd W, Törnberg S, Strand A, Wadell G, Elfgren K, et al. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N Engl J Med. 2007;357:1589–1597. doi: 10.1056/NEJMoa073204. [DOI] [PubMed] [Google Scholar]
- 186.Bulkmans NW, Berkhof J, Rozendaal L, van Kemenade FJ, Boeke AJ, Bulk S, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet. 2007;370:1764–1772. doi: 10.1016/S0140-6736(07)61450-0. [DOI] [PubMed] [Google Scholar]
- 187.Ronco G, Giorgi-Rossi P, Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, et al. New Technologies for Cervical Cancer screening (NTCC) Working Group. Efcacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 2010;11:249–257. doi: 10.1016/S1470-2045(09)70360-2. [DOI] [PubMed] [Google Scholar]
- 188.Cuzick J, Arbyn M, Sankaranarayanan R, Tsu V, Ronco G, Mayrand MH, et al. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008;26(Suppl 10):K29–K41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 189.Koliopoulos G, Arbyn M, Martin-Hirsch P, Kyrgiou M, Prendiville W, Paraskevaidis E. Diagnostic accuracy of human papillomavirus testing in primary cervical screening: a systematic review and meta-analysis of non-randomized studies. Gynecol Oncol. 2007;104:232–246. doi: 10.1016/j.ygyno.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 190.Cuschieri KS, Cubie HA. The role of human papillomavirus testing in cervical screening. J Clin Virol. 2005;32(Suppl 1):S34–S42. doi: 10.1016/j.jcv.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 191.Wright TC., Jr. HPV DNA testing for cervical cancer screening. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95(Suppl 1):S239–S246. doi: 10.1016/S0020-7292(06)60039-8. [DOI] [PubMed] [Google Scholar]
- 192.Dehn D, Torkko KC, Shroyer KR. Human papillomavirus testing and molecular markers of cervical dysplasia and carcinoma. Cancer. 2007;111:1–14. doi: 10.1002/cncr.22425. [DOI] [PubMed] [Google Scholar]
- 193.Bigras G, de Marval F. The probability for a Pap test to be abnormal is directly proportional to HPV viral load: results from a Swiss study comparing HPV testing and liquid-based cytology to detect cervical cancer precursors in 13,842 women. Br J Cancer. 2005;93:575–581. doi: 10.1038/sj.bjc.6602728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ronco G, Segnan N, Giorgi-Rossi P, Zappa M, Casadei GP, Carozzi F, et al. New Technologies for Cervical Cancer Working Group. Human papillomavirus testing and liquid-based cytology: results at recruitment from the new technologies for cervical cancer randomized controlled trial. J Natl Cancer Inst. 2006;98:765–774. doi: 10.1093/jnci/djj209. [DOI] [PubMed] [Google Scholar]
- 195.Belinson J, Qiao YL, Pretorius R, Zhang WH, Elson P, Li L, et al. Shanxi Province Cervical Cancer Screening Study: a cross-sectional comparative trial of multiple techniques to detect cervical neoplasia. Gynecol Oncol. 2001;83:439–444. doi: 10.1006/gyno.2001.6370. [DOI] [PubMed] [Google Scholar]
- 196.Herrero R, Hildesheim A, Bratti C, Sherman ME, Hutchinson M, Morales J, et al. Population-based study of human papillomavirus infection and cervical neoplasia in rural Costa Rica. J Natl Cancer Inst. 2000;92:464–474. doi: 10.1093/jnci/92.6.464. [DOI] [PubMed] [Google Scholar]
- 197.Cuzick J, Szarewski A, Cubie H, Hulman G, Kitchener H, Luesley D, et al. Management of women who test positive for high-risk types of human papillomavirus: the HART study. Lancet. 2003;362:1871–1876. doi: 10.1016/S0140-6736(03)14955-0. [DOI] [PubMed] [Google Scholar]
- 198.Petry KU, Menton S, Menton M, van Loenen-Frosch F, de Carvalho Gomes H, Holz B, et al. Inclusion of HPV testing in routine cervical cancer screening for women above 29 years in Germany: results for 8466 patients. Br J Cancer. 2003;88:1570–1577. doi: 10.1038/sj.bjc.6600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wright TC, Jr, Denny L, Kuhn L, Pollack A, Lorincz A. HPV DNA testing of self-collected vaginal samples compared with cytologic screening to detect cervical cancer. JAMA. 2000;283:81–86. doi: 10.1001/jama.283.1.81. [DOI] [PubMed] [Google Scholar]
- 200.Chan PK, Chang AR, Yu MY, Li WH, Chan MY, Yeung AC, Cheung TH, et al. Age distribution of human papillomavirus infection and cervical neoplasia reflects caveats of cervical screening policies. Int J Cancer. 2010;126:297–301. doi: 10.1002/ijc.24731. [DOI] [PubMed] [Google Scholar]
- 201.Goldie SJ, Gafkin L, Goldhaber-Fiebert JD, Gordillo-Tobar A, Levin C. Mahé C, Wright TC; Alliance for Cervical Cancer Prevention Cost Working Group. Cost-effectiveness of cervical-cancer screening in five developing countries. N Engl J Med. 2005;353:2158–2168. doi: 10.1056/NEJMsa044278. [DOI] [PubMed] [Google Scholar]
- 202.Saslow D, Solomon D, Lawson HW, Killackey M, Kulasingam SL, Cain J, et al. ACS-ASCCP-ASCP Cervical Cancer Guideline Committee. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. CA Cancer J Clin. 2012;62:147–172. doi: 10.3322/caac.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, et al. Retrospective International Survey and HPV Time Trends Study Group. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 204.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Cliford GM. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128:927–935. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 205.Insinga RP, Liaw KL, Johnson LG, Madeleine MM. A systematic review of the prevalence and attribution of human papillomavirus types among cervical, vaginal, and vulvar precancers and cancers in the United States. Cancer Epidemiol Biomarkers Prev. 2008;17:1611–1622. doi: 10.1158/1055-9965.EPI-07-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cliford GM, Smith JS, Plummer M, Muñoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chan PK, Cheung TH, Li WH, Yu MY, Chan MY, Yim SF, et al. Attribution of human papillomavirus types to cervical intraepithelial neoplasia and invasive cancers in Southern China. Int J Cancer. 2012;131:692–705. doi: 10.1002/ijc.26404. [DOI] [PubMed] [Google Scholar]
- 208.Yip YC, Ngai KL, Vong HT, Tzang LC, Ji S, Yang M, Chan PK. Prevalence and genotype distribution of cervical human papillomavirus infection in Macao. J Med Virol. 2010;82:1724–1729. doi: 10.1002/jmv.21826. [DOI] [PubMed] [Google Scholar]
- 209.Hlaing T, Yip YC, Ngai KL, Vong HT, Wong SI, Ho WC, et al. Distribution of human papillomavirus genotypes among cervical intraepithelial neoplasia and invasive cancers in Macao. J Med Virol. 2010;82:1600–1605. doi: 10.1002/jmv.21847. [DOI] [PubMed] [Google Scholar]
- 210.Chan PK, Ho WC, Yu MY, Pong WM, Chan AC, Chan AK, Cheung, et al. Distribution of human papillomavirus types in cervical cancers in Hong Kong: current situation and changes over the last decades. Int J Cancer. 2009;125:1671–1677. doi: 10.1002/ijc.24495. [DOI] [PubMed] [Google Scholar]
- 211.Chan PK, Lam CW, Cheung TH, Li WW, Lo KW, Chan MY, et al. Association of human papillomavirus type 58 variant with the risk of cervical cancer. J Natl Cancer Inst. 2002;94:1249–1253. doi: 10.1093/jnci/94.16.1249. [DOI] [PubMed] [Google Scholar]
- 212.Huang S, Afonina I, Miller BA, Beckmann AM. Human papillomavirus types 52 and 58 are prevalent in cervical cancers from Chinese women. Int J Cancer. 1997;70:408–411. doi: 10.1002/(sici)1097-0215(19970207)70:4<408::aid-ijc6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 213.Wu EQ, Zhang GN, Yu XH, Ren Y, Fan Y, Wu YG, et al. Evaluation of high-risk human papillomaviruses type distribution in cervical cancer in Sichuan province of China. BMC Cancer. 2008;8:202. doi: 10.1186/1471-2407-8-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ding DC, Hsu HC, Huang RL, Lai HC, Lin CY, Yu MH, Chu TY. Type-specific distribution of HPV along the full spectrum of cervical carcinogenesis in Taiwan: an indication of viral oncogenic potential. Eur J Obstet Gynecol Reprod Biol. 2008;140:245–251. doi: 10.1016/j.ejogrb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 215.Asato T, Maehama T, Nagai Y, Kanazawa K, Uezato H, Kariya K. A large case-control study of cervical cancer risk associated with human papillomavirus infection in Japan, by nucleotide sequencing-based genotyping. J Infect Dis. 2004;189:1829–1832. doi: 10.1086/382896. [DOI] [PubMed] [Google Scholar]
- 216.Castle PE, Rodríguez AC, Burk RD, Herrero R, Wacholder S, Alfaro M, et al. Proyecto Epidemiológico Guanacaste (PEG) Group. Short term persistence of human papillomavirus and risk of cervical precancer and cancer: population based cohort study. BMJ. 2009;339:b2569. doi: 10.1136/bmj.b2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007;23:315–330. doi: 10.1155/2007/678793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cuschieri K, Wentzensen N. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2008;17:2536–2545. doi: 10.1158/1055-9965.EPI-08-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Molden T, Kraus I, Karlsen F, Skomedal H, Nygård JF, Hagmar B. Comparison of human papillomavirus messenger RNA and DNA detection: a cross-sectional study of 4,136 women >30 years of age with a 2-year follow-up of high-grade squamous intraepithelial lesion. Cancer Epidemiol Biomarkers Prev. 2005;14:367–372. doi: 10.1158/1055-9965.EPI-04-0410. [DOI] [PubMed] [Google Scholar]
- 220.Gupta N, Srinivasan R, Rajwanshi A. Functional biomarkers in cervical precancer: an overview. Diagn Cytopathol. 2010;38:618–623. doi: 10.1002/dc.21270. [DOI] [PubMed] [Google Scholar]
- 221.Carozzi F, Confortini M, Dalla Palma P, Del Mistro A, Gillio-Tos A, De Marco L, et al. New Technologies for Cervival Cancer Screening (NTCC) Working Group. Use of p16-INK4A overexpression to increase the specificity of human papillomavirus testing: a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2008;9:937–945. doi: 10.1016/S1470-2045(08)70208-0. [DOI] [PubMed] [Google Scholar]
- 222.Klaes R, Friedrich T, Spitkovsky D, Ridder R, Rudy W, Petry U, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001;92:276–284. doi: 10.1002/ijc.1174. [DOI] [PubMed] [Google Scholar]
- 223.Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153:1741–1748. doi: 10.1016/S0002-9440(10)65689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Keating JT, Cviko A, Riethdorf S, Riethdorf L, Quade BJ, Sun D, et al. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol. 2001;25:884–891. doi: 10.1097/00000478-200107000-00006. [DOI] [PubMed] [Google Scholar]
- 225.Nakao Y, Yang X, Yokoyama M, Ferenczy A, Tang SC, Pater MM, Pater A. Induction of p16 during immortalization by HPV 16 and 18 and not during malignant transformation. Br J Cancer. 1997;75:1410–1416. doi: 10.1038/bjc.1997.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Petry KU, Schmidt D, Scherbring S, Luyten A, Reinecke-Lüthge A, Bergeron C, et al. Triaging Pap cytology negative, HPV positive cervical cancer screening results with p16/Ki-67 dual-stained cytology. Gynecol Oncol. 2011;121:505–509. doi: 10.1016/j.ygyno.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 227.Hudelist G, Manavi M, Pischinger KI, Watkins-Riedel T, Singer CF, Kubista E, Czerwenka KF. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol Oncol. 2004;92:873–880. doi: 10.1016/j.ygyno.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 228.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–3884. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 229.Kulmala SM, Syrjänen SM, Gyllensten UB, Shabalova IP, Petrovichev N, Tosi P, et al. Early integration of high copy HPV16 detectable in women with normal and low grade cervical cytology and histology. J Clin Pathol. 2006;59:513–517. doi: 10.1136/jcp.2004.024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gallo G, Bibbo M, Bagella L, Zamparelli A, Sanseverino F, Giovagnoli MR, et al. Study of viral integration of HPV-16 in young patients with LSIL. J Clin Pathol. 2003;56:532–536. doi: 10.1136/jcp.56.7.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cheung JL, Cheung TH, Tang JW, Chan PK. Increase of integration events and infection loads of human papillomavirus type 52 with lesion severity from low-grade cervical lesion to invasive cancer. J Clin Microbiol. 2008;46:1356–1362. doi: 10.1128/JCM.01785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cheung JL, Cheung TH, Ng CW, Yu MY, Wong MC, Siu SS, et al. Analysis of human papillomavirus type 18 load and integration status from low-grade cervical lesion to invasive cervical cancer. J Clin Microbiol. 2009;47:287–293. doi: 10.1128/JCM.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307–313. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- 234.Arias-Pulido H, Peyton CL, Joste NE, Vargas H, Wheeler CM. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J Clin Microbiol. 2006;44:1755–1762. doi: 10.1128/JCM.44.5.1755-1762.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ho CM, Chien TY, Huang SH, Lee BH, Chang SF. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102:54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 236.Cheung JL, Lo KW, Cheung TH, Tang JW, Chan PK. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194:1706–1712. doi: 10.1086/509622. [DOI] [PubMed] [Google Scholar]
- 237.Chan PK, Cheung JL, Cheung TH, Lo KW, Yim SF, Siu SS, Tang JW. Profle of viral load, integration, and E2 gene disruption of HPV58 in normal cervix and cervical neoplasia. J Infect Dis. 2007;196:868–875. doi: 10.1086/520884. [DOI] [PubMed] [Google Scholar]
- 238.Bhattacharjee B, Sengupta S. CpG methylation of HPV 16 LCR at E2 binding site proximal to P97 is associated with cervical cancer in presence of intact E2. Virology. 2006;354:280–285. doi: 10.1016/j.virol.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 239.Badal V, Chuang LS, Tan EH, Badal S, Villa LL, Wheeler CM, et al. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J Virol. 2003;77:6227–6234. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ding DC, Chiang MH, Lai HC, Hsiung CA, Hsieh CY, Chu TY. Methylation of the long control region of HPV16 is related to the severity of cervical neoplasia. Eur J Obstet Gynecol Reprod Biol. 2009;147:215–220. doi: 10.1016/j.ejogrb.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 241.Kalantari M, Calleja-Macias IE, Tewari D, Hagmar B, Lie K, Barrera-Saldana HA, et al. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78:12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. Methylation patterns of papillomavirus DNA, its infuence on E2 function, and implications in viral infection. J Virol. 2003;77:12450–12459. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schifman M, Wentzensen N, et al. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a costarican population. J Natl Cancer Inst. 2012;104:556–565. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sherman ME, Lorincz AT, Scott DR, Wacholder S, Castle PE, Glass AG, et al. Baseline cytology, human papillomavirus testing, and risk for cervical neoplasia: a 10-year cohort analysis. J Natl Cancer Inst. 2003;95:46–52. doi: 10.1093/jnci/95.1.46. [DOI] [PubMed] [Google Scholar]
- 245.Kjaer S, Høgdall E, Frederiksen K, Munk C, van den Brule A, Svare E, et al. The absolute risk of cervical abnormalities in high-risk human papillomavirus-positive, cytologically normal women over a 10-year period. Cancer Res. 2006;66:10630–10636. doi: 10.1158/0008-5472.CAN-06-1057. [DOI] [PubMed] [Google Scholar]
- 246.Dillner J, Rebolj M, Birembaut P, Petry KU, Szarewski A, Munk C, et al. Joint European Cohort Study. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ. 2008;337:a1754. doi: 10.1136/bmj.a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wright TC, Jr, Schifman M, Solomon D, Cox JT, Garcia F, Goldie S, et al. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet Gynecol. 2004;103:304–309. doi: 10.1097/01.AOG.0000109426.82624.f8. [DOI] [PubMed] [Google Scholar]
- 248.Cervical cytology screening. ACOG Practice Bulletin no. 109. Obstet Gynecol. 2009;114:1409–20. doi: 10.1097/AOG.0b013e3181c6f8a4. [DOI] [PubMed] [Google Scholar]
- 249.Kinney W, Stoler MH, Castle PE. Special commentary: patient safety and the next generation of HPV DNA tests. Am J Clin Pathol. 2010;134:193–199. doi: 10.1309/AJCPRI8XPQUEAA3K. [DOI] [PubMed] [Google Scholar]
- 250.Kjær SK, Frederiksen K, Munk C, Iftner T. Long-term absolute risk of cervical intraepithelial neoplasia grade 3 or worse following human papillomavirus infection: role of persistence. J Natl Cancer Inst. 2010;102:1478–1488. doi: 10.1093/jnci/djq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wright TC, Jr, Massad LS, Dunton CJ, Spitzer M, Wilkinson EJ, Solomon D. 2006 ASCCP-Sponsored Consensus Conference. 2006 consensus guidelines for the management of women with abnormal cervical screening tests. J Low Genit Tract Dis. 2007;11:201–222. doi: 10.1097/LGT.0b013e3181585870. [DOI] [PubMed] [Google Scholar]
- 252.Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. ASCUS-LSIL Triage Study (ALTS) Group. Am J Obstet Gynecol. 2003;188:1383–1392. doi: 10.1067/mob.2003.457. [DOI] [PubMed] [Google Scholar]
- 253.Arbyn M, Buntinx F, Van Ranst M, Paraskevaidis E, Martin-Hirsch P, Dillner J. Virologic versus cytologic triage of women with equivocal Pap smears: a meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia. J Natl Cancer Inst. 2004;96:280–293. doi: 10.1093/jnci/djh037. [DOI] [PubMed] [Google Scholar]
- 254.Castle PE, Sideri M, Jeronimo J, Solomon D, Schifman M. Risk assessment to guide the prevention of cervical cancer. Am J Obstet Gynecol. 2007;197(356):e1–6. doi: 10.1016/j.ajog.2007.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Arbyn M, Sasieni P, Meijer CJ, Clavel C, Koliopoulos G, Dillner J. Chapter 9: Clinical applications of HPV testing: a summary of meta-analyses. Vaccine. 2006;24(S3/):78–89. doi: 10.1016/j.vaccine.2006.05.117. [DOI] [PubMed] [Google Scholar]
- 256.Ronco G, Cuzick J, Segnan N, Brezzi S, Carozzi F, Folicaldi S, et al. NTCC working group. HPV triage for low grade (L-SIL) cytology is appropriate for women over 35 in mass cervical cancer screening using liquid based cytology. Eur J Cancer. 2007;43:476–480. doi: 10.1016/j.ejca.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 257.Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. J Natl Cancer Inst. 2000;92:397–402. doi: 10.1093/jnci/92.5.397. [DOI] [PubMed] [Google Scholar]
- 258.Castle PE, Fetterman B, Tomas Cox J, Shaber R, Poitras N, Lorey T, Kinney W. The age-specific relationships of abnormal cytology and human papillomavirus DNA results to the risk of cervical precancer and cancer. Obstet Gynecol. 2010;116:76–84. doi: 10.1097/AOG.0b013e3181e3e719. [DOI] [PubMed] [Google Scholar]
- 259.Sherman ME, Castle PE, Solomon D. Cervical cytology of atypical squamous cells-cannot exclude high-grade squamous intraepithelial lesion (ASC-H): characteristics and histologic outcomes. Cancer. 2006;108:298–305. doi: 10.1002/cncr.21844. [DOI] [PubMed] [Google Scholar]
- 260.Castle PE, Fetterman B, Poitras N, Lorey T, Shaber R, Kinney W. Relationship of atypical glandular cell cytology, age, and human papillomavirus detection to cervical and endometrial cancer risks. Obstet Gynecol. 2010;115:243–248. doi: 10.1097/AOG.0b013e3181c799a3. [DOI] [PubMed] [Google Scholar]
- 261.Paraskevaidis E, Arbyn M, Sotiriadis A, Diakomanolis E, Martin Hirsch P, Koliopoulos G, et al. The role of HPV DNA testing in the follow-up period after treatment for CIN: a systematic review of the literature. Cancer Treat Rev. 2004;30:205–211. doi: 10.1016/j.ctrv.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 262.Arbyn M, Paraskevaidis E, Martin-Hirsch P, Prendiville W, Dillner J. Clinical utility of HPV-DNA detection: triage of minor cervical lesions, follow-up of women treated for high-grade CIN: an update of pooled evidence. Gynecol Oncol. 2005;99(3 Suppl 1):S7–S11. doi: 10.1016/j.ygyno.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 263.Jones J, Saleem A, Rai N, Shylasree TS, Ashman S, Gregory K, et al. Human papillomavirus genotype testing combined with cytology as a ‘test of cure’ post treatment: The importance of a persistent viral infection. J Clin Virol. 2011;52:88–92. doi: 10.1016/j.jcv.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 264.Kitchener HC, Walker PG, Nelson L, Hadwin R, Patnick J, Anthony GB, et al. HPV testing as an adjunct to cytology in the follow up of women treated for cervical intraepithelial neoplasia. BJOG. 2008;115:1001–1007. doi: 10.1111/j.1471-0528.2008.01748.x. [DOI] [PubMed] [Google Scholar]
- 265.Dorleans F, Giambi C, Dematte L, Cotter S, Stefanof P, Mereckiene J, et al. VENICE 2 project gatekeepers group. The current state of introduction of human papillomavirus vaccination into national immunisation schedules in Europe: first results of the VENICE2 2010 survey. Euro Surveill. 2010;15:19730. doi: 10.2807/ese.15.47.19730-en. [DOI] [PubMed] [Google Scholar]
- 266.European Medicines Agency. Cervarix human papillomavirus vaccine [types 16, 18] (recombinant, adjuvanted, adsorbed). Available at: http://www.emea.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/000721/human_med_000694.jsp&mid=WC0b01ac058001d124. Accessed on 9 July 2012.
- 267.European Medicines Agency. Gardasil human papillomavirus vaccine [types 6, 11, 16, 18] (recombinant, adsorbed). Available at: http://www.emea.europa.eu/ema/index.jsp?curl=pages/medicines/human/medicines/000703/human_med_000805.jsp&mid=WC0b01ac058001d124. Accessed on 9 July 2012.
- 268.Brotherton JM, Deeks SL, Campbell-Lloyd S, Misrachi A, Passaris I, Peterson K, et al. Interim estimates of human papillomavirus vaccination coverage in the school-based program in Australia. Commun Dis Intell. 2008;32:457–61. [PubMed] [Google Scholar]
- 269.Franco EL, Cuzick J. Cervical cancer screening following prophylactic human papillomavirus vaccination. Vaccine. 2008;26:A16–23. doi: 10.1016/j.vaccine.2007.11.069. [DOI] [PubMed] [Google Scholar]
- 270.Franceschi S, Denny L, Irwin KL, Jeronimo J, Lopalco PL, Monsonego J, et al. Eurogin 2010 roadmap on cervical cancer prevention. Int J Cancer. 2011;128:2765–2774. doi: 10.1002/ijc.25915. [DOI] [PubMed] [Google Scholar]
- 271.Mayrand MH, Franco EL. Integrating novel primary- and secondary-prevention strategies: the next challenge for cervical cancer control. Future Oncol. 2010;6:1725–1733. doi: 10.2217/fon.10.141. [DOI] [PubMed] [Google Scholar]
- 272.Drolet M, Brisson M, Maunsell E, Franco EL, Coutlée F, Ferenczy A, et al. The psychosocial impact of an abnormal cervical smear result. Psychooncology. 2011 doi: 10.1002/pon.2003. Jun 21. doi: 10.1002/pon.2003. [DOI] [PubMed] [Google Scholar]
- 273.Wardle J, Waller J, Brunswick N, Jarvis MJ. Awareness of risk factors for cancer among British adults. Public Health. 2001;115:173–174. doi: 10.1038/sj/ph/1900752. [DOI] [PubMed] [Google Scholar]
- 274.Waller J, McCafery K, Forrest S, Szarewski A, Cadman L, Wardle J. Awareness of human papillomavirus among women attending a well woman clinic. Sex Transm Infect. 2003;79:320–322. doi: 10.1136/sti.79.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Dell DL, Chen H, Ahmad F, Stewart DE. Knowledge about human papillomavirus among adolescents. Obstet Gynecol. 2000;96:653–656. doi: 10.1016/s0029-7844(00)01009-7. [DOI] [PubMed] [Google Scholar]
- 276.Anhang R, Goodman A, Goldie SJ. HPV communication: review of existing research and recommendations for patient education. CA Cancer J Clin. 2004;54:248–259. doi: 10.3322/canjclin.54.5.248. [DOI] [PubMed] [Google Scholar]


