Abstract
During the Late phase of the Human Immunodeficiency Virus Type-1 (HIV-1) replication cycle, viral Gag proteins and the intact RNA genome are trafficked to specific sub-cellular membranes where virus assembly and budding occurs. Targeting to the plasma membranes of T cells and macrophages is mediated by interactions between the N-terminal matrix (MA) domain of Gag and cellular phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] molecules. However, in macrophages and dendritic cells, a subset of Gag proteins appears to be targeted to tetraspanin enriched viral compartments, a process that appears to be mediated by MA interactions with the Delta subunit of the cellular Adaptor Protein AP-3 (AP-3δ). We cloned, overexpressed and purified the protein interactive domain of AP-3δ and probed for MA binding by NMR. Unexpectedly, no evidence of binding was observed in these in vitro experiments, even at relatively high protein concentrations (200 μM), suggesting that AP-3δ plays an alternative role in HIV-1 assembly.
Introduction
HIV-1 infects, replicates within, and eventually kills CD4+-bearing T-cells, macrophages, and dendritic cells. Virus assembly in T-cells occurs predominantly at the plasma membrane (PM) (18), a process initiated by the binding of a ribonucleoprotein complex comprising the viral genome and a small number of Gag proteins (24–26) to raft-like assembly sites on the PM (8, 11, 15, 28, 32, 36, 39, 40, 51, 52, 55). The targeting of Gag to the PM is mediated by phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] (37), a cellular factor that serves as a major landmark for the PM. NMR studies indicate that PI(4,5)P2 can function both as a direct membrane anchor and as a trigger for myristate exposure, and the exposure of the saturated 1′-acyl chain of PI(4,5)P2 and the saturated myristyl group to the membrane were proposed to serve as a mechanism for targeting the Gag:Genome complex to raft-like regions of the PM (47). PI(4,5)P2 and raft-associated lipids are enriched in HIV-1 envelopes (6), consistent with this assembly mechanism, and the Gag proteins of other retroviruses appear to be targeted to assembly sites in a similar PI(4,5)P2-dependent manner (17, 46).
In macrophages and dendritic cells, the mechanisms of Gag trafficking and assembly are less well understood (1, 2, 5, 9, 13, 20, 21, 33, 34, 38, 41–43, 49, 54, 55). In these cell types, a large proportion of Gag proteins appears to be targeted predominantly to the PM or to PM-derived invaginations and internal compartments, some of which do not appear to be accessible to the extracellular milieu (23). Gag co-localizes at internal assembly sites with tetraspanins (19, 53), which appear to play roles in virus release at sites of cell-cell contact (7, 14, 16, 27, 48, 56). Recent studies indicate that Gag molecules are targeted to tetraspanin-containing assembly sites by the cellular Adapter Protein AP-3 (10, 12). APs are heterotetrameric complexes that facilitate intracellular vesicle transport by selecting vesicle cargo through interactions with signal motifs in cytoplasmic domains of transmembrane proteins, recruiting clathrin for the formation of clathrin coated vesicles (CCV) and recruiting accessory proteins for vesicle formation (31, 44, 50). AP-2 regulates receptor-mediated endocytosis at the plasma membrane and has been reported to interact with signal motifs in the cytoplasmic tail of HIV-1 gp41 thereby targeting the glycoprotein to endosomes (3, 4, 35) where it can presumably be incorporated into virions destined for the exosome pathway. AP-3 is more specifically localized to the trans-Golgi network and to peripheral endosomes, and has been shown to mediate the intracellular trafficking of CD63 to late endosomes (45) where it co-localizes strongly with Gag (33). Significantly, downregulation of the AP-3 δ subunit (AP-3δ) results in attenuation of virus particle release from HIV-1 infected dendritic cells (12), and a combination of biochemical and mutagenesis studies suggested that targeting is mediated by interactions between the amino-terminal residues of the MA domain of Gag and the unstructured protein interactive domain of AP-3δ (10). To further characterize proposed interactions between MA and AP-3δ, we cloned, expressed and purified a peptide fragment corresponding to the protein interactive domain (PID, residues 641–742) of AP-3δ (AP-3δPID) for NMR-based structural studies.
Materials and Methods
DNA encoding the 107 residue protein-interactive domain of AP-3δ was subcloned into a pGEX-6P-1 (Amersham Pharmacia) vector as a C-terminal Glutathione S-transferase (GST) fusion protein. Cells were grown in LB or M9 minimal media supplemented with 99.9 % enriched 15N-ammonium chloride as the sole nitrogen (Isotec). Protein expression was induced in shake flasks with 1 mM IPTG. The cells were harvested and lysed with a microfluidizer (Micorfluidics), clarified by centrifugation, and the target protein was applied to the Glutathione Sepharose 4B matrix (Amersham). The resin was washed extensively with PBS, and an on-column cleavage step was performed with PreScission protease (Amersham) in cleavage buffer releasing the target protein from the resin. The cloning vector and subsequent cleavage resulted in the addition of 5 nonnative residues (GPLGS) to the N-terminus of the protein. Unlabeled and 15N-isotopically labeled myr(−)MA and MA were prepared as previously described (30). AP-3δPID and the Matrix proteins were exchanged into NMR buffer (50 mM sodium phosphate, pH 5.5, 5 mM DTT, and 10% D2O). NMR data were obtained with a Bruker Avance 600 MHz NMR spectrometer equipped with a cryogenic probe (protein concentrations of 50–200 μM; 35 °C).
Results
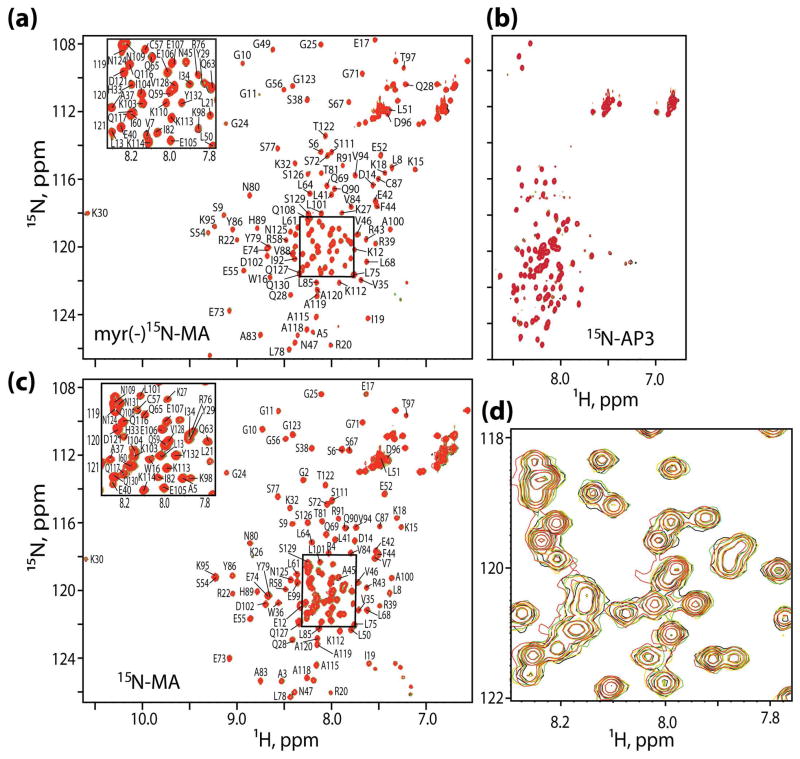
The AP-3δPID construct expressed well, was water-soluble, and exhibited the expected molecular mass (Electrospray mass spectrometry: MWobs = 13,204.1 Da; MWcalc = 13,203.7 Da). We initially titrated AP-3δPID (natural isotopic abundance) into a solution containing the 15N-labeled myr(−)MA and probed for binding by 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectroscopy. As shown in Figure 1a, the AP-3δPID titrations did not lead to significant perturbations of the myr(−)MA 1H-15N HSQC spectrum. We also titrated myr(−)MA (natural isotopic abundance) into 15N-labeled AP-3δPID, Figure 1b. The 1H-15N HSQC spectrum of AP-3δPID exhibited limited chemical shift dispersion consistent with an unstructured, random coil conformation, as expected. Addition of myr(−)MA did not lead to significant changes in the AP-3δPID HSQC spectrum. These data collectively indicate that AP-3δPID and HIV-1 myr(−)MA do not interact with each other. To determine if the lack of binding was due to the missing N-terminal myristyl group, we also titrated AP-3δPID (natural isotopic abundance) into a solution containing the 15N-labeled myristylated MA protein. As shown in Figure 1c, AP-3δPID did not perturb the 1H-15N HSQC spectrum of 15N-labeled myristylated MA, indicating that these proteins also do not interact.
Figure 1.
1H-15N HSQC NMR titration experiments showing that myristylated and unmyristylated forms of HIV-1 MA do not interact with the protein-interactive domain of AP-3δ. (a)15N labeled myr(−)MA titrated with unlabeled AP-3δPID ; AP-3δPID: MA ratios: 0:1 (black), 0.5:1 (yellow), 1:1 (green), and 2:1 (red). (b) 15N labeled AP-3δPID titrated with unlabeled HIV-1 myr(−)MA; MA: AP-3δPID ratios: 0:1 (black), 0.5:1 (yellow), 1:1 (green), and 2:1 (red). (c,d) 15N-labeled myristylated MA titrated with AP-3δPID (natural isotopic abundance); AP-3δPID:MA ratios: 0:1 (black), 0.5:1 (yellow), 1:1 (green), and 2:1 (red). The expansion in (d) is provided to more clearly distinguish between the highly similar spectra.
Discussion
Considerable evidence had previously been obtained supporting a role for HIV-1 MA:AP-3δ interactions in intracellular Gag trafficking and virus release. First, a yeast two-hybrid screen of a HeLa cDNA library using full-length Gag as bait identified the δ subunit of AP-3 as a Gag-interacting protein (10). Additional screens and truncation experiments identified residues of the protein interaction domain of AP-3 as the Gag binding site (10). In addition, directed yeast two-hybrid experiments with Gag-deletion constructs indicated that the N-terminal α-helix of the MA domain is involved in binding, and the protein interactive domain of AP-3δ was found to interact with GST-Gag fragments that contained MA (including a GST-MA fragment) (10). Finally, downregulation of AP-3δ resulted in significant reduction in virus release from HIV-1 infected dendritic cells (12) (although bfefeldin A induced dissociation of AP-3 from membranes does not appear to inhibit virus production in HeLa cells or primary monocyte-derived macrophages (22)). We were therefore surprised by the current NMR results, which show definitively that neither the myristylated or unmyristylated forms of HIV-1 MA interact with the unstructured protein-interactive domain of AP-3δ. The current experiments were conducted at the relatively high protein concentrations where relatively weak interactions (Kd values of ~ 500 mM) can be readily detected. It therefore appears that the intracellular association of Gag and AP-3, and the dependence of AP-3 on virus release from dendritic cells, either do not involve interactions between MA and the protein interactive domain of AP-3δ or require other factors that are not present in our in vitro NMR assay (discussed further below).
AP-3 is one of many cellular proteins that have been implicated in aspects of intracellular Gag trafficking and virus assembly and release (for example, see (1, 5, 8, 21)). For example, the cellular “tail interacting protein of 47 kDaltons” (TIP47) was also implicated as a MA-binding partner, based in part on a yeast two-hybrid screen (29). We recently obtained NMR evidence that the intact TIP47 molecule, which contains a membrane-binding domain, is capable of interacting with MA, but the cellular receptor-binding domain of TIP47 and a TIP47 construct that lacks the membrane-spanning domain do not bind MA (Eric Freed, M.F. Summers, and co-workers, manuscript in preparation). It appears that MA interactions with TIP47 are promoted in aqueous solution by non-native interactions with residues that would normally be sequestered within the membrane, which may explain the yeast 2-hybrid results obtained for this system. Non-native interactions in the absence of cellular constituents might also explain why MA:AP-3δ interactions were observed in the yeast 2-hybrid screens, whereas MA:AP-3δPID binding could not be detected in solution by NMR techniques.
In summary, although there is now good evidence that HIV-1 Gag is targeted primarily to the PM or PM-derived internal membranes in macrophages and dendritic cells, the roles of AP-3 in membrane targeting and virus assembly remain unclear. Quantitative binding studies involving cellular factors, and perhaps with larger and more biologically relevant fragments of Gag, are warranted.
Highlights.
Previously proposed AP-3:MA binding model tested by NMR.
Findings unexpectedly show that the Protein Interactive Domain of AP-3 does not bind HIV-1 MA.
Findings suggest AP-3 may play other roles in Gag trafficking and HIV-1 assembly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benaroch P, Billard E, Gaudin R, Schindler M, Jouve M. HIV-1 assembly in macrophages. Retrovirology. 2010;7:29. doi: 10.1186/1742-4690-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 2009;5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boge M, Wyss S, Bonifacino JS, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 4.Byland R, Vance PJ, Hoxie JA, Marsh M. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell. 2007;18:414–425. doi: 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annual review of microbiology. 2008;62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 6.Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR. Retroviruses Human Immunodeficiency Virus and Murine Leukemia Virus are enriched in phosphoinositides. J Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Dziuba N, Friedrich B, von Lindern J, Murray JL, Rojo DR, Hodge TW, O’Brien WA, Ferguson MR. A critical role for CD63 in HIV replication and infection of macrophages and cell lines. Virology. 2008;379:191–196. doi: 10.1016/j.virol.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chukkapalli V, Ono A. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J Mol Biol. 2011;410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, Peters TR, Dermody TS, Woodruff E, Wang JJ, Spearman P. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120:663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 11.Finzi A, Orthwein A, Mercier J, Cohen EA. Productive Human Immunodeficiency Virus Type 1 assembly takes place at the plasma membrane. J Virol. 2007;81:7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia E, Nikolic DS, Piguet V. HIV-1 replication in dendritic cells occurs through a tetraspanin-containing compartment enriched in AP-3. Traffic. 2008;9:200–214. doi: 10.1111/j.1600-0854.2007.00678.x. [DOI] [PubMed] [Google Scholar]
- 13.Gould SJ, Booth AM, Hildreth JE. The Trojan exosome hypothesis. Proc Natl Acad Sci USA. 2003;100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, Ott DE, Freed EO. Real-time visualization of HIV-1 Gag trafficking in infected macrophages. PLoS Pathogens. 2008;4:e1000015. doi: 10.1371/journal.ppat.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J Mol Biol. 2006;359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Grigorov B, Attuil-Audenis V, Perugi F, Nedelec M, Watson S, Pique C, Darlix JL, Conjeaud H, Muriaux D. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamard-Peron E, Julliard F, Saad JS, Roy C, Roingeard P, Summers MF, Darlix JL, Picart C, Muriax D. Targeting of Murine Leukemia Virus Gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the Matrix. J Virol. 2010;84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermida-Matsumoto L, Resh MD. Localization of Human Immunodeficiency virus Type 1 Gag and env at the Plasma Membrane by Confocal Imagine. J Virol. 2000;74:8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J Virol. 2009;83:5375–5387. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi A, Freed EO. HIV-1 Gag trafficking. Future HIV Therapy. 2007;1:427–438. [Google Scholar]
- 22.Joshi A, Garg H, Nagashima K, Bonifacino JS, Freed EO. GGA and Arf proteins modulate retrovirus assembly and release. Mol Cell. 2008;30:227–238. doi: 10.1016/j.molcel.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe. 2007;2:85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jouvenet N, Neil SJD, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biology. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krementsov DN, Weng J, Lambele M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology. 2009;6:64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung K, Kim JO, Ganesh L, Kabat J, Schwartz O, Nabel GJ. HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe. 2008;3:285–292. doi: 10.1016/j.chom.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Verges S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci USA. 2006;103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 31.Nakatsu F, Ohno H. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct Funct. 2003;28:419–429. doi: 10.1247/csf.28.419. [DOI] [PubMed] [Google Scholar]
- 32.Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 34.Nydegger S, Foti M, Derdowski A, Spearman P, Thali M. HIV-1 Egress is Gated Through Late Endosomal Membranes. Traffic. 2003;4:902–910. doi: 10.1046/j.1600-0854.2003.00145.x. [DOI] [PubMed] [Google Scholar]
- 35.Ohno H, Aguilar RC, Fournier MC, Hennecke S, Cosson P, Bonifacino JS. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 36.Ono A. HIV-1 assembly at the plasma membrane. Vaccine. 2010;28S:B55–B59. doi: 10.1016/j.vaccine.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono A, Freed EO. Cell-Type-Dependent Tageting of Human Immunodeficiency Virus Type 1 Assembly to the Plasma Membrane and the Multivesicular body. J Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 41.Pelchen-Matthews A, Krameer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol. 2003;162:443–445. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 Gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- 43.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 44.Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 45.Rous BA, Reaves JJ, Ihrke G, Briggs JA, Gray SR, Stephens DJ, Banting G, Luzio JP. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13:1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol. 2008;82:1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of Retroviral Replication in Living Cells Reveals Budding into Multivesicular Bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 50.Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suyama M, Daikoku E, Goto T, Sano K, Morikawa Y. Reactivation from latency displays HIV particle budding at plasma membrane, accompanying CD44 upregulation and recruitment. Retrovirology. 2009;6:63. doi: 10.1186/1742-4690-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waki K, Freed EO. Macrophages and Cell-Cell Spread of HIV-1. Viruses. 2010;2:1603–1620. doi: 10.3390/v2081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welsch S, Groot F, Krausslich HG, Keppler OT, Sattentau QJ. Architecture and regulation of the HIV-1 assembly and holding compartment in macrophages. J Virol. 2011;85:7922–7927. doi: 10.1128/JVI.00834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich HG. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathogens. 2007;3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol. 2009;83:7467–7474. doi: 10.1128/JVI.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]