SUMMARY
Patterning of the dorsal-ventral axis in the early Drosophila embryo depends on the nuclear distribution of the Dorsal transcription factor. Using live two-photon light-sheet microscopy, we quantified the nuclear Dorsal gradient in space and time and found that its amplitude and basal levels display oscillations throughout early embryonic development. These dynamics raise questions regarding how cells can reproducibly establish patterns of gene expression from a rapidly varying signal. We therefore quantified domains of Dorsal target genes, discovering their expression patterns are also dynamic. Computational modeling of this system reveals a correlation between Dorsal gradient dynamics and changes in target gene expression and suggests that these dynamics, together with time averaging of noise, results in the formation of graded gene expression borders in regions where the gradient is nearly flat. We propose that mRNA levels remain plastic during transient signaling events, allowing tissues to refine patterns in the face of genetic or environmental variation.
INTRODUCTION
In a developing organism, tissues have long been proposed to be patterned by spatially graded signals that specify cell fate in a concentration-dependent manner. Classically, these “morphogens” have been defined as originating from a defined source and forming a graded distribution by diffusion and degradation; however, in recent years it has become clear that morphogens can become spatially organized by a variety of mechanisms. Two of the best-characterized morphogen gradients pattern the anterior-posterior (AP) and dorsal-ventral (DV) axes of the Drosophila early embryo: the Bicoid and Dorsal transcription factor gradients (reviewed in Porcher and Dostatni, 2010 and Reeves and Stathopoulos, 2009). Their graded distributions are established using very different mechanisms. Bicoid is locally translated because its mRNA contains a localization sequence; whereas, Dorsal is localized to the nucleus more strongly in the ventral regions of the embryo because of localized Toll-receptor associated signaling. Live imaging has revealed significant dynamics in the exact levels of Bicoid (e.g., Gregor et al., 2007b; Little et al., 2011); however, the dynamics of target gene expression examined in fixed embryos suggest that the levels of Bicoid are important, but not the only defining factor in the expression of target genes (e.g., Jaeger et al., 2004; Ochoa-Espinosa et al., 2009). In contrast, no study to date has investigated systematically temporal features of the Dorsal gradient and its relationship to the expression of its target genes.
The role of Dorsal in the expression of its target genes has been conceptualized as the concentration-dependent activation of genes, divided into three broad categories (Types I, II, and III) based on both their domains of expression and their presumed threshold-dependent responsiveness to Dorsal levels (see Figures 1A and 1B; reviewed in Reeves and Stathopoulos, 2009 and Stathopoulos and Levine, 2005). Type I genes, such as twist (twi) and snail (sna), are expressed in ventral regions of the embryo in a domain where the levels of nuclear Dorsal are high (up to ~20% DV position; where 0% is the ventral-most position and 100% is the dorsal-most position). Type II genes like ventral nervous system defective (vnd), are thought to be expressed in ventrolateral domains (dorsal boundaries at ~33% DV position) through the combined actions of enhancers that are of intermediate affinity to Dorsal and that are repressed by the Snail transcription factor in the ventral-most regions. Type III genes are expressed in domains with boundaries past 45% DV position, and can be further subdivided into two categories: those that are activated by Dorsal (Type III+, such as short-gastrulation [sog]) and those that are repressed by Dorsal (Type III−, such as zerknüllt [zen] and decapentaplegic [dpp]). Presumably, the lowest levels of Dorsal are sufficient to determine the spatial extent of Type III target genes, but the roles played by other factors remain unclear (e.g., Jiang and Levine, 1993; Liberman and Stathopoulos, 2009).
Figure 1. Quantitative Analysis of Nuclear Dorsal Levels and Expression Domains of Dorsal-Ventrally Expressed Genes.
(A) Cross-section of late nc 14 embryo hybridized with sna (red), vnd (blue), ind (cyan), sog (green), and dpp (yellow) antisense RNA probes.
(B) Plot of domains of gene expression for Dorsal target genes. Data comes from averages of >10 embryos for each gene. The solid black curve is average Dorsal gradient as measured from cross-sections (see Figure 6A). The dotted Dorsal curve is from Bothma et al. (2010). The region where the Dorsal input is questionable appears in gray.
(C) Antibody staining in a fixed Dorsal-Venus embryo cross-section (clockwise from upper left): anti-Dorsal, anti-Venus, anti-histone H3, and merge between anti-Dorsal (magenta) and anti-Venus (green).
(D) Quantification of fluorescent antibody staining from part C. Each dot corresponds to the intensity in a nucleus for anti-Dorsal (red) and anti-Venus (dark green). Error bars denote the standard error of the intensity of the pixels in each nucleus (also in E). The two solid curves represent best-fit curves for anti-Venus intensity (green) and anti-Dorsal (magenta). This demonstrates that anti-Venus is slightly wider (see inset).
(E) The normalized intensity of anti-Venus plotted against anti-Dorsal for each nucleus. Note that, in intermediate intensities, the curve falls below the 45° line (orange), indicating that anti-Venus is brighter on average than anti-Dorsal, and thus the gradient is wider.
(F) Box plot of gradient widths (σ, see Equation 1 in Experimental Procedures) for various antibody stainings, live imaging analysis, and maternal genetic backgrounds. Numbers indicate sample size. Plus signs indicate outliers. Asterisks indicate statistical significance (p < 10−6). Whole mount data from (Liberman et al., 2009). Embryo in (A) reprinted with permission from Reeves and Stathopoulos (2009). Graded Dorsal and differential gene regulation in the Drosophila embryo. In Perspectives on Generation and Interpretation of Morphogen Gradients, J. Briscoe, P. Lawrence, and J.-P. Vincent (Plainview, NY: Cold Spring Harbor Press).
To study the role of nuclear Dorsal in controlling gene expression, a number of studies have attempted to measure nuclear Dorsal, using either antibody stainings in fixed tissues (Chung et al., 2011; Liberman et al., 2009; Zinzen et al., 2006) or imaging of Dorsal-GFP in live embryos (DeLotto et al., 2007; Kanodia et al., 2009). Our previous study in fixed embryos showed that the Dorsal gradient was more narrow than often described, resulting in a relatively flat distribution more than 110 μm from the ventral midline (40% DV position); this raised the question of how Dorsal could specify gene expression in this domain (Liberman et al., 2009). In contrast, others reported broader gradients in live embryos, arguing that the Dorsal concentration could carry positional information up to at least 60% DV position; furthermore, they argued that the Dorsal nuclear concentration continued to decline all the way to the dorsal midline (compare dotted versus solid black curves in Figure 1B; Chung et al., 2011; Kanodia et al., 2009). Both sets of studies suggested that the Dorsal nuclear gradient is dynamic, varying in time both within nuclear cycles and from one nuclear cycle to the next (DeLotto et al., 2007; Kanodia et al., 2009; Liberman et al., 2009).
Previous live studies of Dorsal nuclear concentration and dynamics (DeLotto et al., 2007; Kanodia et al., 2009) employed a Dorsal-GFP fusion that results in a measurably wider gradient than wild-type and also fails to fully complement dorsal null mutants (Liberman et al., 2009). Furthermore, these studies employed conventional confocal microscopy, in which nuclear motion and limited light penetration both complicated an accurate measurement of the Dorsal-GFP nuclear gradient in both time and space (DeLotto et al., 2007; Kanodia et al., 2009). Thus, the dynamics of the Dorsal gradient have not yet been satisfactorily measured, nor has it been investigated how these dynamics might impact domains of gene expression.
In this study, we employed a dorsal-venus fusion transgene and improved microscopy to address two outstanding questions regarding the Dorsal gradient: first, how does a highly dynamic morphogen signal specify gene expression domains, and second, how does a narrow gradient deliver precise positional information to the entire DV axis. We find that the Dorsal nuclear gradient varies in both time and space during nuclear cycles (nc) 11–14, and that the expression of Dorsal target genes is often as dynamic as the gradient. Furthermore, we suggest that the graded boundaries in the expression patterns of Type III genes result from a time-integration of the nearly-flat gradient tail. We used a threshold-based model to show that much of the dynamics and sharpness of Dorsal target gene expression patterns can be accounted for by the dynamics and shape of the Dorsal nuclear gradient.
RESULTS
Use of a Dorsal-Venus Fusion to Monitor Dorsal in Living Embryos with Light-Sheet Microscopy
To create a transgene encoding a fully functional Dorsal-Venus fusion protein, we BAC-recombineered 25 kb of genomic DNA surrounding the dorsal locus with sequences encoding the venus yellow fluorescent protein optimized for Drosophila, inserted in-frame at the C terminus of the Dorsal protein (see Experimental Procedures). This dorsal-venus transgene fully complements null mutations in dorsal when present at one copy, similar to an unmodified dorsal rescue transgene. In contrast, neither the 25 kb dorsal-gfp construct we constructed nor previous dorsal-gfp cDNA based constructs complement the dorsal mutant at one copy (Liberman et al., 2009; see Experimental Procedures). Live imaging and immunostaining demonstrate that Dorsal-Venus exhibits a distribution more similar to wild-type Dorsal than Dorsal-GFP. We defined a quantitative measure of the width of the gradient by fitting the data to Equation 1 (see Experimental Procedures), resulting in a metric of σ (Liberman et al., 2009). This analysis shows that embryos carrying Dorsal-Venus 25 kb rescue construct have a width of σ = 0.16 ± 0.01 (standard deviation), which is much more similar to the width of the wild-type Dorsal gradient (σ = 0.14 ± 0.01) than that from embryos carrying a 25 kb Dorsal-GFP construct that we made (σ = 0.20 ± 0.02) (see Figures 1C–1F). The similarity of the Dorsal-Venus distribution to that of Dorsal offers us the opportunity to accurately assess the spatiotemporal behavior of functional Dorsal nuclear gradients using live imaging.
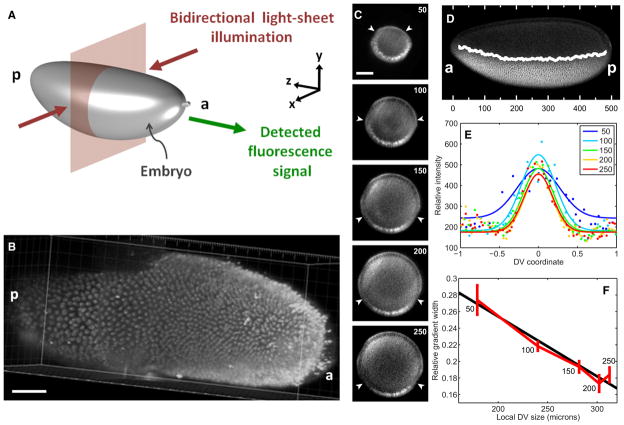
To quantitatively measure the levels of Dorsal-Venus in early embryos, we imaged embryos from mothers containing one copy of a dorsal-venus transgene, one copy of the endogenous dorsal gene, and one copy of an H2A-RFP transgene to label all of the nuclei so they can be unambiguously segmented (Materials and Methods). Using two-photon scanned light-sheet microscopy (2P-SPIM) (Figures 2A and 2B; Truong et al., 2011), which provides superior resolution at high sample depth compared to conventional confocal microscopy, we imaged end-on cross-sections of the nc 14 embryo to determine the Dorsal-Venus nuclear gradient between 50 and 250 μm from the anterior pole. We found the gradient changes with AP location, becoming increasingly wider at 100 μm or closer to the pole (Figures 2C–2F). Therefore, our quantitative analysis of the Dorsal gradient in the rest of this study will be based on measurements made from images of H2A-RFP and Dorsal-Venus during nc 11–14 in optical cross-sections of embryos 150 μm from the anterior pole (i.e., just posterior to the presumptive cephalic furrow; see Figure 3A and Movie S1 available online).
Figure 2. Two-Photon Light-Sheet Microscopy Reveals Anterior-Posterior Modulation of the Dorsal Gradient Width.
(A) Schematic of the illumination and detection geometry used in live imaging of Dorsal-Venus embryos.
(B) Three-dimensionally-rendered volume image stack of the nuclei-labeled H2A-RFP signal from a live embryo at nc 14 demonstrates that the nuclei-resolving resolution is achieved up to at least 250 μm from the anterior end (or ~50%) of the embryo. The optical distortions seen beyond 250 μm are due to the coverglass that supports the embryo.
(C) Images of an embryo at 50–250 μm from the anterior pole. The white arrowheads denote the location where nuclear and cytoplasmic Dorsal-Venus become approximately equal in intensity.
(D) Visual illustration of the gradient width as a function of AP position in a whole mount embryo fluorescently stained against anti-Dorsal. Hash marks indicate distance (in μm) from the anterior pole. The white curve represents the approximate location where the nuclear and cytoplasmic intensity become equal.
(E) Quantification of the Dorsal-Venus nuclear gradients from (C).
(F) The width of the gradients (σ, see Equation 1 in Experimental Procedures) from (C) in units relative to the local DV size, plotted against the local DV size. Numbers next to the points denote distance from anterior pole. Error bars represent 68% confidence interval in computing σ. a: anterior, p: posterior. Scale bar represents 50 μm.
Figure 3. The Dorsal-Venus Nuclear Gradient Is Dynamic, with Increasing Amplitude, Decreasing Basal Levels, and Constant Width.
(A) Snapshots of the Dorsal-Venus nuclear gradient for nuclear cycles 11–14 imaged at 150 μm using two-photon light-sheet microscopy. These snapshots were taken at peak values of the gradient amplitude for each nuclear cycle. Scale bar represents 50 μm.
(B) Quantification of the Dorsal-Venus nuclear gradient from snapshots shown in (A).
(C) Evolution of gradient amplitude (blue), basal levels (orange), and gradient width (red) from nuclear cycle 11 through gastrulation for the embryo shown in (A).
(D) Normalized gradient amplitude and basal levels from a single embryo zoomed in on time points between 53 and 41 min before gastrulation. The mitosis between nc 13 and 14 interphases takes place between ~50–46 min before gastrulation. The vertical dashed lines represent the time points depicted in the following panels.
(E–I) Snapshots of the Dorsal-Venus gradient at the time points shown in (D). The time points progress from the end of nc 13 interphase (E), the beginning of the following mitosis (F), the middle of mitosis (G), the end of mitosis (H), and the beginning of nc 14 interphase (I). Even in (H), a detectable ventral-to-dorsal gradient is present. Blue curves represent raw data. Red curves represent the Gaussian-like fit (Equation 1). See also Figure S1 and Movie S1.
Dynamic Properties of the Dorsal-Venus Nuclear Gradient
We analyzed overall spatial properties of the Dorsal nuclear gradient throughout the course of nc 11–14 by collecting 2P-SPIM time lapse images. The image processing tools extracted the nuclear Dorsal-Venus signal by segmenting the nuclear regions based on the H2A-RFP images. The parameters of the gradient amplitude (A), basal levels (B), and width (σ) were determined using the Gaussian-fitting described in the Experimental Procedures, and each can vary over time (see Figure 3B and Equation 1 in the Experimental Procedures). The gradient amplitude [A(t)] increases from nuclear cycle to nuclear cycle (blue curve in Figure 3C), agreeing with previous data using fixed samples (Liberman et al., 2009) and predictions from modeling studies (Kanodia et al., 2009). Moreover, the Dorsal-Venus gradient amplitude exhibits a “saw-tooth” pattern over time, never reaching steady state, consistent with single-nucleus traces from other live studies (DeLotto et al., 2007). This pattern can be explained by the nuclei filling with Dorsal-Venus relatively slowly throughout each nuclear cycle interphase, then rapidly equilibrating with the cytoplasm when the nuclear envelopes break down at the beginning of mitosis.
In contrast to the filling of the ventral-most nuclei with Dorsal-Venus, the dorsal-most nuclei appear to begin each interphase with “too much” Dorsal-Venus. As interphase proceeds, these dorsal-most nuclei slowly evacuate nuclear Dorsal, causing the Dorsal levels in these nuclei (i.e., the “basal levels” of the gradient, B(t); see orange trace in Figure 3C) to decrease during interphase. When mitosis begins, basal levels rapidly increase. This counter-action between the Dorsal levels building in the ventral-most nuclei and declining in the dorsal-most, fits with the notion that the nuclei begin each interphase with Dorsal levels equilibrated with the cytoplasm, and only after an intact nuclear envelope forms can selective nuclear import/export processes develop the nuclear concentration gradient. With the successive import of Dorsal in ventral nuclei during each syncytial cycle, a cytoplasmic Dorsal gradient also develops that can be seen by the end of nc 13 mitosis when the nuclear envelope breaks down (Figures 3D–3I). It was previously unappreciated that the overall levels of Dorsal protein are nonuniform along the DV axis.
Our analysis of the Dorsal-Venus gradient over time shows a remarkably constant gradient width [σ(t)] across all time (inter-phase only, red trace in Figure 3C), implying the Dorsal nuclear gradient always becomes nearly flat at the same location (~40% DV position). This result was also suggested by our previous fixed tissue analysis, in which the gradient width appeared to be unchanging from nuclear cycle to nuclear cycle (Liberman et al., 2009). However, the previous results obtained with fixed tissues left open the possibility that gradient widths could change either subtly across nuclear cycles or significantly within a nuclear cycle. The live imaging done here dismisses that possibility.
Measurements of the Dorsal-Venus nuclear gradient in three live embryos revealed similar results (see Figure S1 available online). Together, these results underscore two questions regarding the action of the Dorsal gradient. First, how does a constantly-changing morphogen gradient specify domains of gene expression? Gene expression patterns might be established early then depend on cis-regulatory action, as has been proposed for the Bicoid network (e.g., Bergmann et al., 2007; Jaeger et al., 2004). Alternatively, mRNA transcription of the target genes might constantly change, driven by the dynamic changes in nuclear Dorsal. Second, how does a signal as narrow as the Dorsal nuclear gradient control the expression pattern of genes past 40% DV position (e.g., the Type III genes)? In other words, how could Dorsal provide reliable positional information given that its gradient is nearly flat?
Dorsal Target Gene Expression Patterns in Space and Time
To address the first question, we examined gene expression patterns in manually cross-sectioned, wild-type embryos using multiplex in situ hybridization during nc 11–14 (see Figure 4 and Supplemental Experimental Procedures). As nuclear cycles get progressively longer, we divided nc 13 and 14 into early/late and early/mid/late time points, respectively, based on nuclear morphology and density (Figure 4A). The brightness and contrast of images in Figures 4A–4E were intentionally adjusted to visually highlight distinctions; whereas, the profiles in Figure 4F come from analysis of the raw images (see Supplemental Experimental Procedures). In order to obtain a representative sampling of target gene dynamics, we examined four classical Dorsal-target genes of Type I (sna), II (vnd), III+ (sog), and III− (zen) patterns. Genetic and cis-regulatory analyses support the view that these genes are Dorsal targets (reviewed in Reeves and Stathopoulos, 2009).
Figure 4. Changes in mRNA Patterns Are Identifiable Both between and within Nuclear Cycles.
Wild-type embryos from double in situ and antibody fluorescent stainings were manually cross-sectioned and imaged.
(A) Nuclei were labeled with α-histone H3 to determine embryo stage.
(B–E) Expression of sna (B, Type I), vnd (C, Type II) sog (D, Type III+) and zen (E, Type III−) throughout nuclear cycles 11–14. Embryos are oriented with ventral side down.
(F) Profiles of each gene (color-coded) reflect the expression averaged from 4–13 embryos at each nuclear cycle. Embryos were costained with sna and vnd or with sog and zen. D, dorsal; V, ventral. Brightness and contrast of embryo cross-sections have been adjusted for visual clarity. See Experimental Procedures for analysis of raw data. See also Figure S2.
Each of these classical Dorsal target genes have dynamic patterns of expression. zen is expressed in the dorsal half of the embryo as early as nc 11, and builds in time until mid-to-late nc 14, when its expression pattern refines into a narrow stripe (Figure 4E), presumably from Dpp signaling (Rushlow et al., 2001). On the other hand, the expression patterns of sna, vnd, and sog exhibit more complex dynamics (Figures 4B, 4C, and 4D, respectively). sog transcripts, nuclear-localized and likely nascent, are observed as early as nc 12. The mature (nonnuclear) mRNA for all three of these genes is first seen in nc 13 (Figures 4B–4D; see also Figure S2A). During this nuclear cycle, both sog and vnd are initially expressed in ventral regions where sna normally would repress them. As nc 13 progresses, the levels of sog and vnd increase only outside the sna domain, presumably because increasing activity of Sna repression limits expression in ventral regions. At the onset of nc 14, sog and vnd patterns are present with more uniform expression in ventral and lateral domains; little evidence of Sna-mediated repression is apparent. However, as nc 14 continues, and the levels of both sog and vnd increase, ventral repression becomes apparent again.
Our data demonstrate that domains of gene expression change during the transition between nc 13 and nc 14, in that repression in ventral regions is no longer apparent at the start of nc 14 (Figure 4F and Figure S2). One possible explanation is that expression observed at the onset of nc 14 corresponds to early transcripts that avoid Sna repression, because Sna levels in early nc 14 are not high enough to repress sog and vnd.
To investigate further, we examined sog transcript localization more closely. At the beginning of nc 13, sog is present only as nuclear dots, likely sites of nascent transcription in the nucleus, whereas sog in late nc 13 is localized outside the nucleus near the apical membrane. We interpret nonnuclear sog transcripts as being complete mRNAs, as they appear to have been exported from the nucleus. At the onset of nc 14, sog is once again colocalized with nuclear staining and nonnuclear transcripts are absent (Figures 5A–5C); nonnuclear transcripts appear again at mid nc 14 (Figure 4D). Moreover, the early nc 14 pattern shows little sign of Sna-mediated ventral repression. These results are consistent with the idea that the sog transcript is degraded in between nc 13 and nc 14.
Figure 5. Dynamics of Nascent sog Correlate with Sna Protein.
(A) Z stack projections (19.5 μm) of sog mRNA.
(B) Z stack projection merge of sog mRNA (red) and nuclear Histone H3 staining (green).
(C) Zoomed in image of (B) to the area of sog expression.
(D–F) Same as (A–C), respectively, except with sna.
(G) Profiles (light green) of Sna-GFP from all embryos analyzed (n) and the average curve (dark green).
(H) Same as (G) except for intronic sog. Additional embryos stained with intronic sog but not GFP are included.
(I) Box plot of peak Sna-GFP levels shows upward progression during nc 11–14. Red spots indicate outliers.
(J) The intensity of intronic sog at the ventral midline is plotted against the peak intensity of Sna-GFP. As the nuclear cycles progress, a decrease in intronic sog expression correlates with an increase in Sna-GFP. Only embryos costained with both intronic sog and Sna-GFP are included. See also Figure S3.
We also inspected sna mRNA localization, thinking that if all transcripts are degraded at the nc 13/nc 14 transition, then perhaps this could explain the loss of Sna-mediated repression in ventral regions. Instead, sna transcripts exhibit a different trend: they switch back and forth between being delocalized (early nc 13, early nc 14) and apically localized (late nc 13, mid/ late nc 14 Figures 4B and 5D–5F). Little is known regarding the relationship between sna transcript localization/stability, but it has been observed that for a number of other genes, mRNA localization is thought to affect function (Lécuyer et al., 2007). We did not find evidence during our time-course that sna transcripts are completely degraded, as our results suggest for sog, nevertheless we continued to test our working hypothesis, which was that decreased Sna levels result in derepression of genes at the start of nc 14.
We hypothesized that insufficient levels of Sna protein (rather than transcript) at the start of nc 14 might account for lack of ventral repression, therefore we examined levels of Sna protein within embryos relative to sog transcript. Embryos carrying a Sna-GFP rescue construct (Dunipace et al., 2011) were immunostained with anti-GFP, which is more robust than any anti-Sna antibody we have tried, and coprocessed with a riboprobe to the 5′ intron of sog (Figure S3A), which provides a near-real-time assay of active sog transcription. In analyzing these embryos, we found that Sna-GFP levels increase starting from nc 13 and reach peak intensity by the end of nc 14 (Figures 5G and 5I). Although a slight dip in average Sna levels is observed between nc 13 and 14, the levels are highly variable, perhaps because the staging of our fixed embryos is not fine enough to capture the most rapid dynamics (e.g., Figure S2B). Nevertheless, in each embryo, we found a strong and consistent negative correlation between Sna-GFP and intronic sog in the ventral-most nuclei (Figures 5J and S3B–S3F), suggesting that a threshold amount of Sna activity is required to extinguish de novo sog transcription.
Lack of Precision of the Dorsal Gradient May Explain Graded Expression Profiles of Type III Genes
Our previous analysis of the Dorsal gradient led us to believe that its tails were flat, suggesting it could not provide the positional information necessary to specify the domains of expression of target genes such as sog and zen (Liberman et al., 2009). The live imaging of Dorsal-Venus performed here provided clear insights into Dorsal gradient dynamics, but did not have enough signal-to-noise ratio to yield quantitative information on the gradient tail within the domain of low nuclear Dorsal levels. To circumvent this technical challenge and provide further insight into levels of nuclear Dorsal present in this domain, we quantified the Dorsal gradient in 153 fixed, manually cross-sectioned wild-type embryos at mid to late nc 14 (see Experimental Procedures).
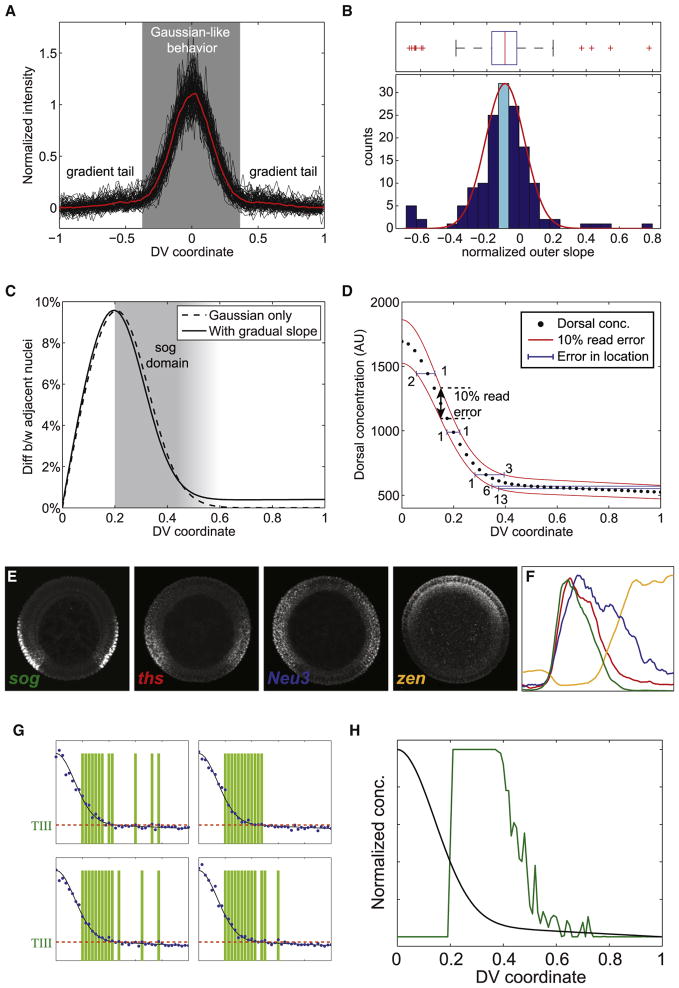
These measurements revealed that the Dorsal gradient displays two regimes (see Figure 6A). From the ventral midline up to 40% DV position, the gradient adopts a narrow Gaussian-like shape (σ ~0.14; see Figure 1F). More dorsally, however, the gradient becomes more linear and can be empirically characterized by a constant slope (M) multiplied by x (the position along the DV axis; see Equation 2 in Experimental Procedures). The average slope of the gradient tail (normalized by the gradient amplitude) is −0.1 ± 0.03 (95% confidence interval; see histogram in Figure 6B). Thus, on average, nuclear Dorsal levels slowly decrease with increasing x. However, there is a considerable amount of variance in the distribution as 25% of embryos were measured to have a positive slope to the tail. Although a portion of this variance reflects measurement error, error cannot account for the mean slope being definitively negative (see Figure S4).
Figure 6. A Shallow Gradient Can Deliver Positional Information with Limited Precision, Supporting Graded Expression Patterns.
(A) Normalized Dorsal nuclear gradients (black) for 153 fixed, manually cross-sectioned embryos. Average gradient in red.
(B) Histogram of the normalized tail slope for all embryos from (A). The cyan histogram bar denotes the 95% confidence interval of the mean. The box plot above the histogram depicts the bulk of the data falling within the box-and-whiskers, with a handful of extreme outliers both above and below the bulk of the data (red “+” signs). The red curve overlying the histogram depicts a normal distribution with the same mean and variance as the bulk of the data.
(C) Plot of difference in Dorsal levels seen by adjacent nuclei versus DV coordinate.
(D) Potential errors in gene expression boundary placement due to 10% stochastic fluctuations in Dorsal readout (red curves). From left to right, error bars in x denote the error for a gene presumptively placed at 10%, 20%, 33%, 50%, and 70% DV position. Numbers indicate rough numbers of nuclei. Each black dot represents a nucleus. Forty nuclei are plotted, in keeping with a typical nc 14 nuclear density.
(E) Mature, Dorsal-dependent expression of Type III genes sog, zen, ths, and Neu3. zen pattern is shown before Dpp-dependent refinement occurs.
(F) Profiles of genes shown in (E). Note differing locations of the dorsal boundaries and graded borders.
(G) A noisy gradient tail may result in graded boundaries of Type III genes. Simulations of four instances of Type III+ gene activation as a result of reading out a noisy gradient (blue dots indicate the readout of each nucleus). In each case, gene expression is either active in a nucleus (green bar) or not, depending on whether the read Dorsal signal is above the threshold (red dotted line). Nuclei closer to the ventral side will be activated more often.
(H) Final output of a Type III gene relative to Dorsal gradient (simulated data). This pattern is the result of time-averaging of the activation states of the nuclei (examples in G) and the basal levels decreasing within a nuclear cycle (example in Figure 7E). The graded boundary of the Type III gene is determined by how long and how often nuclei read a signal above the threshold. See also Figure S4.
With a gradually sloping tail and ventral, narrow Gaussian, it seems questionable that Dorsal could deliver precise positional information to lateral and dorso-lateral positions. To address this quantitatively, we evaluated the relative difference in Dorsal concentration (Δc/c) that would be seen by neighboring nuclei (see Equation 4 in Experimental Procedures). In the gradient tail, at the dorsal border of sog (x = 50% DV position), the difference in Dorsal levels that adjacent nuclei see is less than 1% (Figure 6C). This is problematic as previous work has suggested that it is unlikely nuclei can reliably interpret concentration changes of less than 10% (e.g., Gregor et al., 2007a).
Another approach to examine the expected imprecision of the gene expression boundaries was to determine the effects of stochastic fluctuations in reading a shallow Dorsal gradient. In Figure 6D, we plot the Dorsal concentration for 40 nuclei along the semicircumference of DV axis (using Equation 2 in Experimental Procedures) and increase and decrease this amount by 10%, the level of read error suggested by previous studies (red curves in Figure 6D; Gregor et al., 2007a). With these fluctuations, the error in x for placing a gene expression boundary outside the steep Gaussian-like regime is six or more nuclei, even with a gradient tail that reliably slopes downward. The implication is that Type III genes are located in a region where it is difficult for Dorsal to specify sharp, precise boundaries.
To investigate this issue further, we performed in situ hybridization of manually cross-sectioned nc 14 embryos with the anti-sense riboprobes of Type III transcripts. This analysis revealed that, in general, Type III genes (e.g., sog, ths, Neu3, and zen) possess graded borders at different DV positions (see Figures 6E and 6F). Perhaps the lack of precision in positional information results in the graded borders of these genes in dorsal regions (see Figures 6G and 6H). In other words, the noisy Dorsal gradient will activate gene expression in all nuclei in the graded border, but some more frequently than others depending on whether Dorsal levels are above a threshold. This would lead to a time-averaging mechanism in which mistakes are smoothed out as mRNA accumulates (see Figures 6G and 6H and S5) (Tostevin et al., 2007).
Simulations of Dorsal-Dependent Patterning
Live imaging of the Dorsal-Venus nuclear gradient has revealed complex dynamical behavior, with the gradient amplitude increasing and the basal levels decreasing over time. In addition, carefully examining mRNA expression in cross-sectioned embryos revealed graded boundaries of the Type III genes as well as gene expression dynamics across and within nuclear cycles. Together, these observations seem related, yet it is not intuitively obvious what quantitative effect the Dorsal gradient dynamics may have on the gene expression patterns. To test the plausibility of a causal relationship between the spatiotemporal dynamics of the gradient and its targets, we constructed a model of the wild-type Dorsal gradient based on our live and fixed tissue data (see Equation 2 and Experimental Procedures). Moreover, we formulated a threshold-based model of mRNA dynamics to simulate expression patterns of sna, vnd, sog, and zen over nc 11–14, according to the network depicted in Figure 7A (see Equation 3 in Experimental Procedures). In our model, the mRNA lifetimes and the thresholds that dictate gene transcription were fit such that the simulations would optimally match the experimental data shown in Figure 4F (see Figures 7C and 7D and Experimental Procedures).
Figure 7. Expression of DV Genes in Space and Time Correlates with Dynamic Nuclear Dorsal Levels.
(A) Signaling network used in the model simulations. Arrows indicate activation; blunt arrows indicate repression.
(B) Heat map of the simulated Dorsal gradient. Black curves denote constant Dorsal concentration contours, corresponding to the thresholds chosen for sna, vnd, and sog/zen.
(C) The observed (top row) and simulated (bottom) profiles of sna, vnd, sog, and zen.
(D) The mid nc 14 Dorsal target genes sna (red), vnd (blue), sog (green), and zen (yellow). Circles denote averages of fluorescent in situ hybridization patterns from >10 embryos and solid curves denote simulation results.
(E) Simulation of dynamic Dorsal morphogen gradient. TI, II, and III were placed as in (B), using the final gradient.
(F) Simulations of threshold responses to Dorsal gradients from the nc 11–14 near the end of each nuclear cycle when the gradient amplitude is peaking. The horizontal lines correspond to the Type I (red), Type II (blue), and Type III (green) thresholds (contours of B). The location these thresholds are crossed by the nc 11–14 Dorsal gradients are given by the vertical lines.
(G) Simulated sna expression pattern for early, mid, and late nc 14.
(H) The amount of simulated Dorsal (black), sog (green), and sna (red) a ventral nucleus sees. The dashed horizontal lines correspond to the sna and sog thresholds.
(I) Boxplot of location of sna boundary in wild-type embryos, staged within nuclear cycle 14. Numbers indicate sample size.
(J) Simulated vnd expression pattern for early, mid, and late nc 14.
(K) The amount of simulated Dorsal (black, gray) and vnd (blue, cyan) seen over time by nuclei at 30% and 35% DV position, respectively. The dashed horizontal line corresponds to the vnd threshold.
(L) Boxplot of location of vnd boundary in wild-type embryos, staged within nuclear cycle 14.
(M) Simulated sog expression pattern for early, mid, and late nc 14.
(N) The amount of simulated Dorsal (black, gray) and sog (dark and light green) seen over time by nuclei at 35% and 55% DV position, respectively. The dashed horizontal line corresponds to the sog threshold.
(O) Boxplot of location of intronic sog boundary in wild-type embryos, staged within nuclear cycle 14. See also Figure S5.
The Dorsal levels simulated in space and time are shown in Figure 7B, in which the black curves are contours of constant Dorsal levels, corresponding to the fitted thresholds for gene expression (Type I, II, and III from left to right). These threshold contours suggest that gene expression boundaries will move in time as a result of the dynamics of the Dorsal gradient. This is demonstrated more clearly in Figure 7F, which depicts simulated Dorsal gradients near the end of nc 11–14 (horizontal arrows correspond to the signaling thresholds). The signaling threshold for Type II genes is located near 33% DV position throughout all four nuclear cycles, even in the face of the dynamics of the Dorsal gradient (Figures 7F and 7J–7L), whereas the DV positions of Type I and Type III genes change significantly over nuclear cycles (red and green arrows in Figure 7F). In particular, during nc 14, the movement of the thresholds predicts Type I genes to expand, Type II genes to remain fixed, and Type III+ genes to begin the cycle expressed even in the dorsal-most nuclei (Figure 7E).
Simulations of gene expression patterns agree with these general predictions of threshold-dependent patterning. During nc 14, the simulated sna boundary moves dorsally (Figure 7G), the vnd boundary does not move (Figures 7J and 7K), and the sog domain begins broad, then the dorsal portion retracts (Figures 7M and 7N). These predictions prompted us to investigate the nc 14 dynamics of these three genes in more detail using fixed embryos. The vnd dorsal boundary remains static in early and mid nc 14, and expands only slightly in late nc 14 (Figure 7L). However, sna and sog expression is dynamic. As predicted based from our simulations, we found the sna domain expands during nc 14 (Figure 7I); and this specific result is supported by another study published recently (McHale et al., 2011). Furthermore, although sog mature transcripts are expressed in a constant domain (data not shown), importantly the boundary identified using an intronic sog probe, which serves as a “real-time” proxy for responsiveness of transcription, shows the pattern retracts (Figure 7O).
Additionally, the threshold-based simulations offer a plausible explanation for the observed on/off cycling of Sna activity and its consequences on the ventral repression of sog and vnd. Simulated sna is expressed strongly in late nc 13, but decreases subsequently during nc 13 mitosis simply because transcription ceases whereas degradation continues (Figure 7H). Afterward, sna levels must build again in early nc 14 before it can repress sog and vnd. In the intervening time, sog (and vnd; not shown) is transcribed in the ventral-most nuclei (Figure 7H).
We identified several differences between the simulations and the observed patterns of gene expression, most notably that sog expression is difficult to accurately simulate. Although the simulations correctly predict some aspects of the dorsal border of sog, in that it is graded and placed past 40% DV position (as explained by Figures 6G, 6H and S5), the simulated border is more graded than seen experimentally; the entire dorsal portion of the embryo expresses sog strongly. If the threshold parameter for sog were raised slightly higher to attempt to restrict sog expression more ventrally, only a very narrow final domain of sog is present (data not shown). The differences we see between the simulations and analysis of fluorescent in situ hybridization experiments may stem from a variety of reasons, including roles for other activators and repressors in supporting expression. For example, if a dorsally-acting factor (such as zen or another gene expressed in a similar domain) were to repress sog, then the lower threshold combined with the action of such a dorsally-acting repression could support expression of sog in a domain more comparable to the endogenous pattern (see Figure S5B).
Although Dorsal is an important player in patterning of these genes, there are indeed other inputs required for full DV patterning; for example, as stated above, some additional input is required to explain the sog dorsal boundary. Nevertheless this simple model incorporating only the interactions in Figure 7A does remarkably well, in that the model was able to demonstrate the plausibility that the observed gene expression dynamics is driven by the Dorsal gradient dynamics. In particular, the model successfully predicted that Type I patterns expand during inter-phase, Type II patterns remain static, and Type III+ patterns begin nuclear cycles broadly expressed then retract.
DISCUSSION
The observations that morphogen gradients are dynamic have raised questions about the influence of time on pattern formation (reviewed in Kutejova et al., 2009). In this study, we investigate quantitatively how the nuclear distribution of Dorsal, which rapidly changes throughout the nuclear divisions in the Drosophila blastoderm, gives rise to precise gene expression patterns. Our findings reveal that, in contrast to Bicoid, whose nuclear distribution stabilizes relatively quickly both between and within nuclear cycles (Little et al., 2011; Lucchetta et al., 2008), the Dorsal gradient is highly dynamic, exhibiting a temporally oscillating pattern of nuclear Dorsal concentrations that never reaches a steady state. The dynamics within a nuclear cycle result from the slow net nuclear import of Dorsal throughout each nuclear cycle interphase, followed by an abrupt export of Dorsal when the nuclear envelopes break down at the beginning of mitosis. We suggest these slow dynamics associated with nuclear localization of Dorsal relate to Toll-mediated signaling being required for its ability to gain competence to enter the nuclei. In addition, early nuclear cycles may concentrate an initially uniform distribution of Dorsal onto the ventral side of the embryo thereby redistributing the overall concentration of Dorsal protein over time.
The dynamics of the nuclear Dorsal gradient appear to determine the temporal evolution of gene expression. Our observations show that Dorsal target gene expression follows a dynamic pattern similar to the Dorsal gradient, both within and across nuclear cycles. An implication of these observations is that gene expression patterns are able to switch their on/off state in response to changes in the concentration of Dorsal. This is similar to the manner that target genes respond to Activin in the Xenopus embryo (Gurdon et al., 1995). However, in contrast to Activin-dependent patterning, Dorsal target genes do not appear to exhibit a “ratchet effect,” as it is possible to turn on/off gene expression by changing the levels of Dorsal above/below an activation threshold. Thus, the response of target gene expression to Dorsal levels appears to be a real-time response. Our data further suggest that the activity of Sna protein is also transient and/or that repression is delayed compared to onset of transcription, as both sog and vnd are ventrally-repressed by the end of nc 13 and derepressed in some early nc 14 embryos.
The highly dynamic patterning of genes along the dorsal-ventral axis documented in this study could possibly allow for fine-tuning of gene expression patterns to respond to feedback and/or buffer against genetic and environmental perturbation. In support of this hypothesis, many genes expressed at this stage along the DV axis support relatively short transcripts of less than 5 kb (such as sna, twi, vnd, rho, brk, wntD, and zen to name a few), and thus are able to respond quickly to changes in the Dorsal gradient and/or in cis-regulation of other DV genes. In contrast, genes with larger transcripts, such as sog or Neu3 of 20+ kb in length, will take 15+ min to transcribe at a rate of 1.1–1.4 kb/min (Shermoen and O’Farrell, 1991; Thummel et al., 1990). Because any incomplete nascent transcripts are most likely aborted and degraded upon cell division (Rothe et al., 1992; Shermoen and O’Farrell, 1991), these long transcripts are particularly constrained by the rapid (~10 min) mitotic cycles of the early embryo. In addition, the action of Sna repression through transcriptional inhibition could be delayed in genes with long transcripts (McHale et al., 2011). Thus, transcription length can impact a gene’s response to other factors (e.g., Sna) and, together with Dorsal, is proposed to play an important role in regulating the timing of gene expression.
The nuclear distribution of Dorsal does not reach a steady state and yet the expression patterns of most Dorsal target genes appear to stop changing at mid-to-late nc 14. This may result from the fact that the Dorsal gradient changes more slowly during this period and/or that at this point patterning may have been stabilized by the logic of the cis-regulatory network. At the onset of gastrulation, when Dorsal levels plummet, some patterns are extinguished whereas others are retained. We suggest those that are retained must rely on a Dorsal-independent mechanism to support expression along the DV axis. For example, several genes (e.g., sim and vnd) switch to autoregulatory feedback mechanisms to retain expression within the same domain and thereby no longer depend on Dorsal to support activation (Nambu et al., 1991; Von Ohlen et al., 2007).
The low slope of the Dorsal gradient in the lateral and dorso-lateral regions of the embryos makes it unlikely that the Dorsal morphogen gradient can specify precise domains of gene expression (i.e., sharp boundaries) here. Each Type III gene analyzed in this study exhibited a graded border, and our results lead us to propose two mechanisms that may contribute to this pattern. First, although a recent study showed that stochastic gene expression along the dorsal-ventral axis relates to polymerase pausing (Boettiger and Levine, 2009), we highlight that stochastic expression is a common phenomenon associated with most genes of Type III pattern. In addition, a time-averaging mechanism (Tostevin et al., 2007), we propose, will give rise to a graded expression response at the gradient tails. Second, if the basal levels of the Dorsal gradient decrease within a nuclear cycle, then the location where the Dorsal gradient crosses the putative Type III threshold will retreat from the dorsal midline to ~50% DV position as demonstrated here with intronic sog. In this case, assuming transcripts are stable within a nuclear cycle, nuclei that transiently saw enough Dorsal to express the gene for only a given time window will be part of the graded domain. In all, our data suggest that both noise and dynamics may be factors contributing to proper patterning of genes beyond the spatial range of a morphogen. The graded nature of Type III patterns may influence their functions; for instance in the case of sog to support an inverse gradient of TGF-β signaling (e.g., Dorfman and Shilo, 2001).
As both the Dorsal gradient as well as its target genes change in time, this suggests a correlation between Dorsal levels and gene expression dynamics. Our model demonstrates that Dorsal gradient dynamics can plausibly account for the observed expression patterns in nc 13 and 14 for sna, vnd, and zen, capturing the general oscillatory nature of DV gene expression and, in particular, provides insight that ventral patterns expand and more dorsal patterns retract. However, the failure of the simulations to reproduce the dorsal border of sog could be explained by a missing component to the modeling caused by our limited understanding of the process, such as a dorsally-acting repressor. This would be consistent with other patterning systems in which cross-repressive interactions between target genes are important factors (Jaeger et al., 2004). Alternatively, the behavior of the dorsal border of sog could be explained by the additional input of activators such as Zelda (Liberman and Stathopoulos, 2009) or by a gradient tail that is steeper than our measurements suggest. Therefore, although genes exhibit dynamics in their expression that generally correlate with changing Dorsal levels, there is clearly more to understand.
It is becoming increasingly clear that steady state models of morphogen gradients ignore crucial developmental events and that modeling of systems that takes into consideration the dynamics is informative (e.g., Bergmann et al., 2007; Bolouri and Davidson, 2003; Jaeger and Reinitz, 2006; Kutejova et al., 2009; Lek et al., 2010). In some cases, the dynamics of morphogen gradients are instrumental in the establishment of “memory-like” patterns (Dessaud et al., 2010; Nahmad and Stathopoulos, 2009). In contrast, the on/off cycling of gene expression associated with the Dorsal system demonstrates plasticity rather than memory. We surmise this plasticity may be a critical design feature for the subtle fine-tuning of gene expression domains, or early initiation of genetic regulatory pathways that must operate in the short developmental time period of the Drosophila blastoderm.
EXPERIMENTAL PROCEDURES
Construction of the Dorsal-Venus Construct
The 25 kb dorsal-venus and dorsal-GFP transgenes were generated using recombineering mediated gap repair performed using SW105 cells as previously described (Venken et al., 2006). The BAC encompassing the dorsal gene (BACR07M13) was obtained from the BacPac Resource Center and the attB-P[acman]-ApR was modified to contain ~600 bp homology arms to the region of interest. Seamless insertion of venus or gfp just before the stop codon of dl was performed using the galK system (Warming et al., 2005). A 6XGly sequence was added before the start of both the Venus and GFP sequences using PCR. The final constructs were isolated and electroporated into EPI300 cells (Epicenter) and the copy number was induced using Fosmid Autoinduction Solution (Epicenter) according to the manufacturer’s instructions. The constructs were isolated using Nucleobond EF plasmid midi prep kits (ClonTech) and injected into line 23648 (BDSC) at a concentration of 0.5–1 μg/μl in water using standard techniques. All primers used for gap repair and recombineering are described in Supplemental Experimental Procedures.
These dorsal constructs fused to a fluorescent protein were inserted into the 86Fb landing site on the third chromosome (Bischof et al., 2007; Groth et al., 2004) and crossed into dl1 and dl4 mutant backgrounds (Bloomington stock center) to assay for ability to complement the dl mutant.
Live Imaging
Live imaging of the Dorsal-Venus embryos was carried out using two-photon scanned light-sheet microscopy (Truong et al., 2011). Using a custom-built microscopy setup, 3-micron-thick light sheets were used to illuminate bidirectionally from two opposing sides of the embryo, creating an optical section that was perpendicular to the embryo’s AP-axis (Figure 2A). The illumination light, derived from a femtosecond-pulsed laser (Chameleon UltraII, Coherent) was set at 960 nm to simultaneously induce fluorescence from Venus-labeled Dorsal proteins and RFP-labeled nuclei (H2A-RFP; Bloomington Stock Center) via two-photon excitation. To image the dynamic Dorsal-Venus nuclear gradient as shown in Figure 3, pre-nc10 embryos were mounted horizontally with heptane-glue on a coverglass with about two-thirds of the embryo’s anterior body extending beyond the edge of the coverglass. The coverglass was mounted in a 25°C water-filled chamber and oriented so that the embryo’s anterior end faced the detection optics. Imaging was conducted at a single focal plane, 150 μm from the embryo’s anterior end; in 15 s time intervals; with 6 s of illumination/exposure time. The fluorescence from Dl-Venus and H2A-RFP were spectrally separated and imaged simultaneously onto neighboring regions of the recording camera (iXon-DU885, Andor Technology) with a spectral splitter (DV2, Photometrics).
Image Analysis
In our analysis of Dorsal gradients, we took a similar approach as described in Liberman et al. (2009), which is outlined here. First, the background was subtracted to set regions outside the embryo to black. Next, nuclei were detected (Figure 1C, lower right), and the Dorsal fluorescence (Figure 1C, top) was normalized to the nuclear data. For analysis of antisense RNA hybridized embryos, intensity as a function of arc distance was measured in an annular region around the periphery of the embryo. See also Supplemental Experimental Procedures for more details.
Empirical Fits of the Dorsal Gradient
The Dorsal nuclear gradient empirically conformed to Gaussian-like curves as previously explained (Liberman et al., 2009):
| (Equation 1) |
Each gradient is thus represented spatially by three parameters (A, B, and σ). The first two parameters, A and B, describe the “amplitude” and “basal levels” of the gradient, respectively. The basal levels (B) can be thought of as the amount of nonzero Dorsal that is present in the dorsal-most nuclei. The amplitude (A) can be thought of as the amount of nuclear Dorsal present in the ventral-most nuclei greater than that found in the dorsal-most. σ is a measure of the spatial range of the gradient (gradient width).
To account for the gradual slope after 40% DV length, a correction to the Gaussian-like behavior (Equation 1) was made:
| (Equation 2) |
This final parameter, M, multiplies the absolute distance from the ventral midline, and denotes the value of the gradual slope found after the Gaussian term decays to zero. A linear function for the gradient tail was chosen over other one-parameter realizations because it is the simplest representation of a function with a nonzero slope, and can be interpreted as a one-term Taylor expansion of the real functional form of the gradient tail. Although this leads to a nonzero derivative at the dorsal midline, and thus is nonphysical, the goal of this parameterization of the gradient tail is simply to distinguish between a flat and a nonflat tail, and is sufficient for that test (see Figure S4).
Simulation of the Wild-Type Dorsal Gradient
We used Equation 2 to simulate the wild-type Dorsal gradient in space and time (see Figure 7). We extracted A(t) and B(t) averaging the data from three live embryo measurements and σ = 0.14 from wild-type fixed embryos. We chose M(t) = −0.1A(t) to reflect the mean value of the normalized outer slope.
Simulation of mRNA Dynamics
mRNA is described by the following equation:
| (Equation 3) |
where i = sna, vnd, sog, or zen. fi is the mRNA production rate, modeled as a hard threshold function. For sna, this function is equal to 1 if c(x,t) > θsna, and zero otherwise. In the case of zen, fzen is equal to 1 if c(x,t) < θzen, and zero otherwise. For sog and vnd, fi is equal to 1 if both c(x,t) > θi and [sna] < 0.5, and zero otherwise. The input c(x,t) is the simulated Dorsal nuclear gradient with 10% standard deviation Gaussian noise added. The production of mRNA only occurs during interphase. The parameter τi is the lifetime of mRNA species i. The values of θi and τi were fitted to the data from Figure 4F. For further details on analysis of mRNA dynamics, see Supplemental Experimental Procedures.
Calculations of Precision
To be able to read distinct Dorsal levels, nuclei that are spaced by a distance of Δx must be able to measure Dorsal levels to within a relative error (Δc/c) given by the following equation:
| (Equation 4) |
During nc 14, the internuclear distance is ~7 μm, and the equation for c(x) is given by Equation 2, with A = 1,080, B = 520, σ = 0.14, and M = −89. Here, A, B, and M are in arbitrary units, and σ is in units relative to the length of the DV axis. These parameters are the average values for the fixed, nc 14 data set.
Manual Cross-Sections of Embryos
For cross-section imaging, stained embryos in glycerol were manually cut with a 0.10 mm blade under a dissecting microscope to remove the anterior and posterior ends, leaving a section 100–200 μm in width that corresponds to 150–200 μm from the embryo poles. These cross-sections were then aligned on a glass slide and mounted in glycerol with a coverslip. Two pieces of double-sided tape were used as a spacer between the microscope slide and coverslip. Z stacks of 15–20 μm were imaged using a Zeiss LSM 5 Pascal.
Supplementary Material
Acknowledgments
We are grateful to Scott Fraser (Caltech) for advice on experimental approach and comments on the manuscript. We also thank Leslie Dunipace (Caltech) for reagents and advice on BAC recombineering, Anil Ozdemir (Caltech) for reagents, and Greg Bietel (Northwestern University) for providing the Venus DNA sequence. The anti-Dorsal antibody developed by Ruth Steward was obtained from the DSHB, developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology. This work was supported by the Jane Coffin Childs Memorial Fellowship for Medical Research to G.T.R, by the Caltech Beckman Institute and NIH Center for Excellence in Genomic Science grant P50HG004071 to T.V.T. to provide funds to build the two-photon light-sheet microscope, and by grant R01 GM077668 from the NIGMS to A.S.
Footnotes
Supplemental Information includes five figures, Supplemental Experimental Procedures, and one movie and can be found with this article online at doi:10.1016/j.devcel.2011.12.007.
References
- Bergmann S, Sandler O, Sberro H, Shnider S, Schejter E, Shilo BZ, Barkai N. Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol. 2007;5:e46. doi: 10.1371/journal.pbio.0050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 in-tegrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Levine M, Boettiger A. Morphogen gradients: limits to signaling or limits to measurement? Curr Biol. 2010;20:R232–R234. doi: 10.1016/j.cub.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, Lu H. A microfluidic array for large-scale ordering and orientation of embryos. Nat Methods. 2011;8:171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLotto R, DeLotto Y, Steward R, Lippincott-Schwartz J. Nucleocytoplasmic shuttling mediates the dynamic maintenance of nuclear Dorsal levels during Drosophila embryogenesis. Development. 2007;134:4233–4241. doi: 10.1242/dev.010934. [DOI] [PubMed] [Google Scholar]
- Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R, Shilo BZ. Biphasic activation of the BMP pathway patterns the Drosophila embryonic dorsal region. Development. 2001;128:965–972. doi: 10.1242/dev.128.6.965. [DOI] [PubMed] [Google Scholar]
- Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4075–4084. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007a;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007b;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Mitchell A, Mahony D. Direct and continuous assessment by cells of their position in a morphogen gradient. Nature. 1995;376:520–521. doi: 10.1038/376520a0. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Reinitz J. On the dynamic nature of positional information. Bioessays. 2006;28:1102–1111. doi: 10.1002/bies.20494. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, Manu, Myasnikova E, Vanario-Alonso CE, Samsonova M, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- Kanodia JS, Rikhy R, Kim Y, Lund VK, DeLotto R, Lippincott-Schwartz J, Shvartsman SY. Dynamics of the Dorsal morphogen gradient. Proc Natl Acad Sci USA. 2009;106:21707–21712. doi: 10.1073/pnas.0912395106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutejova E, Briscoe J, Kicheva A. Temporal dynamics of patterning by morphogen gradients. Curr Opin Genet Dev. 2009;19:315–322. doi: 10.1016/j.gde.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lek M, Dias JM, Marklund U, Uhde CW, Kurdija S, Lei Q, Sussel L, Rubenstein JL, Matise MP, Arnold HH, et al. A homeodomain feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development. 2010;137:4051–4060. doi: 10.1242/dev.054288. [DOI] [PubMed] [Google Scholar]
- Liberman LM, Stathopoulos A. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev Biol. 2009;327:578–589. doi: 10.1016/j.ydbio.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Reeves GT, Stathopoulos A. Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proc Natl Acad Sci USA. 2009;106:22317–22322. doi: 10.1073/pnas.0906227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Vincent ME, Ismagilov RF. A precise Bicoid gradient is nonessential during cycles 11–13 for precise patterning in the Drosophila blastoderm. PLoS ONE. 2008;3:e3651. doi: 10.1371/journal.pone.0003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale P, Mizutani CM, Kosman D, Mackay DL, Belu M, Hermann A, McGinnis W, Bier E, Hwa T. Gene length may contribute to graded transcriptional responses in the Drosophila embryo. Dev Biol. 2011;360:230–240. doi: 10.1016/j.ydbio.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahmad M, Stathopoulos A. Dynamic interpretation of hedgehog signaling in the Drosophila wing disc. PLoS Biol. 2009;7:e1000202. doi: 10.1371/journal.pbio.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci USA. 2009;106:3823–3828. doi: 10.1073/pnas.0807878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher A, Dostatni N. The bicoid morphogen system. Curr Biol. 2010;20:R249–R254. doi: 10.1016/j.cub.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Reeves GT, Stathopoulos A. Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol. 2009;1:a000836. doi: 10.1101/cshperspect.a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pehl M, Taubert H, Jäckle H. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992;359:156–159. doi: 10.1038/359156a0. [DOI] [PubMed] [Google Scholar]
- Rushlow C, Colosimo PF, Lin MC, Xu M, Kirov N. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 2001;15:340–351. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Burtis KC, Hogness DS. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell. 1990;61:101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]
- Tostevin F, ten Wolde PR, Howard M. Fundamental limits to position determination by concentration gradients. PLoS Comput Biol. 2007;3:e78. doi: 10.1371/journal.pcbi.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Von Ohlen TL, Harvey C, Panda M. Identification of an upstream regulatory element reveals a novel requirement for Ind activity in maintaining ind expression. Mech Dev. 2007;124:230–236. doi: 10.1016/j.mod.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.