Abstract
Heparin and related heparan sulfate interact with a number of cytokines and growth factors thereby playing an essential role for many physiological and pathophysiological processes by involving both signal transduction and the regulation of the tissue distribution of cytokines/growth factors. Follistatin (FS) is an autocrine protein with a heparin-binding motif that serves to regulate the cell proliferative activity of the paracrine hormone, and member of the TGF-β family, activin A (ActA). Follistatin is currently under investigation as an antagonist of another TGF-β family member myostatin (Mstn) to promote muscle growth in diseases associated with muscle atrophy. In the present study, we employ surface plasmon resonance (SPR) spectroscopy to dissect the binding interactions between the heparin polysaccharide and both free follistatin (FS288), and its complexes (FS288-ActA, FS288-Mstn). FS288 complexes show much higher heparin binding affinity than FS288 alone. SPR solution competition studies using heparin oligosaccharides showed that the binding of FS288 and its complex to heparin is chain length dependent. Full chain heparin or large oligosaccharides, having 18 to 20 sugar residues show the highest binding activity for FS288 and FS288-ActA, whereas smaller heparin molecules could interact with the FS288-Mstn complex. These interactions were also analyzed in normal physiological buffers and at different salt concentrations and pH values. Unbound follistatin was much more sensitive to all salt concentrations above 150 mM. Heparin binding of the FS288-ActA complex was disrupted at 500 mM salt, whereas it was actually increased for the FS288-Mstn complex. At acidic pH values, heparin binding to FS288 and FS288-ActA binding was enhanced. While slightly acidic pH values (pH 6.2 and 5.2) enhanced FS288-Mstn binding to heparin, at pH 4 heparin-binding was inhibited. Overall these studies demonstrate that specific ligand binding to FS288 differentially regulates its affinity and behavior for heparin molecules.
Keywords: Follistatin, myostatin, activin A, heparin, surface plasmon resonance
Heparin and heparan sulfate (HS) are structurally related members to a family of polyanionic, polydisperse, linear polysaccharides called glycosaminoglycans (GAGs), which perform a variety of critical biological functions. They consist of repeating disaccharide subunits of 1→4 linked hexuronic acid, β-D-glucuronic acid (GlcA) or α-L-iduronic acid (IdoA) and glucosamine, α-D-N–acetylglucosamine (GlcNAc) or α-D-N-sulfoglucosamine (GlcNS).1, 2 Heparin and HS are biosynthesized as proteoglycans (PGs) using the same biosynthetic pathway.3 While heparin-PGs are stored in mast cell granules, HS-PGs are ubiquitously expressed on the cell surface of most cell types and in the extracellular matrix (ECM). Interactions between heparin/HS with proteins mediate diverse biological processes including development, angiogenesis, anticoagulation, inflammation, cancer and microbial/viral pathogenesis.4–9 Intensive biochemical and genetic studies have shown that HSPGs play crucial roles in regulating key developmental signaling pathways, such as the pathways for fibroblast growth factors (FGFs), Hedgehog (Hh), and transforming growth factor-β (TGF-β).10 In the FGF signaling pathway, FGF receptor (FGFR) dimerization with FGF requires the association of heparin or highly sulfated HS polysaccharide chains of HSPGs, e.g. FGF1 signaling is transmitted across the cell membrane through the formation of a ternary complex of FGF1, FGFR1, and HS.11–13 In the Hh signaling pathway, HSPGs are essential for proper Hh distribution, stabilization, and signaling activity.14 HSPGs are also believed to facilitate Hh ligand presentation to responding cells, and participate as part of a larger receptor complex.10,15 Thus, an understanding of heparin/HS-protein interactions at the molecular level is of fundamental importance to biology and will aid in the development of highly specific therapeutic agents to these pathways.4,9
Transforming growth factor- β (TGF-β) is the prototypical molecule of a superfamily of ligands including TGF-β isoforms, activins, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs).16,17 Ligands of TGF-β superfamily of growth factors initiate signal transduction through a bewildering complexity of ligand–receptor interactions, which regulate a diverse set of cellular and physiological functions, including early embryonic development, cellular growth and proliferation, differentiation, migration and death.18–19 Myostatin (Mstn), also known as growth and differentiation factor-8 (GDF-8), is a TGF-β family member that has been identified as a strong inhibitor of muscle growth.20 Mstn knock-out mice exhibit muscles that are 2–3 times larger than those of wild-type (WT) mice.21 A similar phenotype was observed by transgenic overexpression of Mstn inhibitors (antagonists), such as follistatin (FS).22 FS is a secreted polypeptide that regulates several signaling pathways through its ability to inactivate TGF-β-like growth factor molecules such as activin or bone morphogenetic proteins by sequestering ligands in a tight, nearly irreversible inactive complex.23 Previous biochemical studies indicated that the different follistatin isoforms (FS288, FS303 and FS315) have different binding affinities for heparin, which potentially influence the physiological functions and locations of these isoforms.23 It was proposed that FS288 represents a predominantly cell-bound bioneutralizing antagonist since FS288 has the highest affinity to heparin. Structural studies have determined that FS288 completely surrounds the ligand and blocks both the type I and type II receptor binding sites. Furthermore, our structural studies on myostatin in complex with the antagonist FS288 revealed a unique continuous electropositive surface created when Mstn binds FS288.20 This feature significantly increases the affinity of the complex for heparin over FS288 alone or FS288-ActA. As such, this is likely the reason that FS288-Mstn complexes are degraded more readily when applied to the cell surface than the corresponding FS288-ActA complexes. 20
The goal of current study is to analyze the molecular interactions of heparin with FS288 and its complexes with ActA and Mstn (FS288-ActA, FS288-Mstn), under normal physiological and non physiological conditions (high salt concentrations and low pH conditions). The Biacore system (BIAcore 3000) was employed for the study. The system utilizes the Surface Plasmon Resonance (SPR) phenomena and allows a direct quantitative analysis of the label-free molecular interactions in real-time.
EXPERIMENTAL PROCEDURES
Materials
Sensor SA Chip was from GE Healthcare (Uppsala, Sweden). Heparin sodium salt and low molecular weight heparin (LMWH) were obtained from porcine intestinal mucosa (Celsus Laboratories, Cincinnati, OH). Heparin oligosaccharides from (dp2 to dp 20) were prepared from controlled partial heparin lyase 1 treatment of bovine lung heparin (Sigma) followed by size fractionation. 24 SPR measurements were performed on a BIAcore 3000 (GE Healthcare, Uppsala, Sweden) operated using the version software. Buffers were filtered and degassed for SPR assay.
Protein expression and purification
FS288, ActA and Mstn, proteins were expressed utilizing stable CHO cells and purified as described previously. 20, 25 Briefly, FS288 was purified through binding a heparin affinity column followed by binding a cation exchange column. ActA was purified through affinity chromatography using an NHS-Sepharose column coupled with FS288. ActA was eluted with 50mM glycine, 0.03% Tween 80, pH 2.5. Mstn was purified through sequential anion and cation exchange column followed by a C4 reverse phase step. Purified proteins were concentrated and buffer exchanged into 10mM HEPES, 150mM NaCl, pH 7.5.
Complexes of FS288-ActA and FS288-Mstn were prepared by mixing FS288 at a 2.5:1 molar ratio with each ligand and separating the complexes of either FS288-ActA or FS288-Mstn from unbound FS through application of samples on an S200 Hiload Sephacryl column.
Preparation of heparin biochip
Biotinylated heparin was prepared by reacting sulfo-N-hydroxysuccinimide long-chain biotin (Piece, Rockford, IL) with free amino groups of unsubstituted glucosamine residues in the polysaccharide chain following a published procedure. 26 The biotinylated heparin was immobilized to streptavidin (SA) chip based on the manufacturer’s protocol. The successful immobilization of heparin was confirmed by the observation of a ~250 resonance unit (RU) increase in the sensor chip. The control flow cell (FC1) was prepared by 1 min injection with saturated biotin.
Kinetic measurement of interaction between heparin and protein using BIAcore
The protein samples were diluted in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4,). Different dilutions of protein samples were injected at a flow rate of 40 µl/min for 3 mins. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 2 min dissociation time, the sensor surface was regenerated by injecting with 40 µl of 2 M NaCl to get fully regenerated surface. The response was monitored as a function of time (sensorgram) at 25 °C.
SPR solution competition study of heparin oligosaccharides
To measure the chain length dependence of the heparin-protein interactions, SPR solution competition study was performed by pre-equilibrating different heparin oligosaccharides with protein followed by subsequent injection of this solution over the heparin immobilized sensor chip. Different chain lengths of heparin (full size, LMWH and heparin oligosaccharides degree of polymerization (dp2 to dp20) at 50 nM was pre-mixed with 5 nM of FS288 or FS288: ActA complex in HBS-EP buffer and injected over heparin chip at a flow rate of 40 µl/min. After each run, the dissociation period and regeneration protocol were performed as described above. The SPR experiments were performed in triplicate.
Effect of buffer conditions on heparin-protein interactions
To measure the effect of buffer conditions on heparin-protein interactions, samples of the standard SPR HBS-EP buffer (0.01 M HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4,) were modified to contain 300 mM, 500 mM and 1000 mM NaCl, or adjusted to pH 6.2, pH 5.2 or pH 4.0. Three protein samples, FS288, FS288-ActA complex, and FS288-Mstn complex, were diluted (1:9) in the five buffer conditions and injected at a flow rate of 40 µl/min for 3 min. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 2 min dissociation time, the sensor surface was regenerated by injecting with 40 µl of 10 mM of glycine, 2 M NaCl to fully regenerate the surface.
RESULTS AND DISCUSSION
Kinetics measurement of heparin-FS288, heparin-FS288 ligand complex interactions
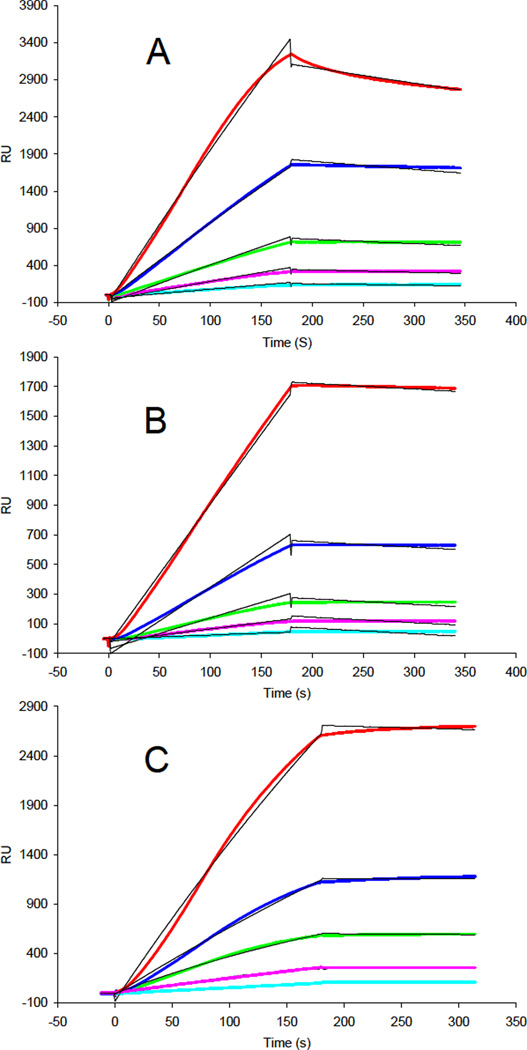
The profiles of binding kinetics were measured for FS288 and its complexes to immobilized heparin using SPR to provide a quantitative picture of these interactions. Sensorgrams of heparin FS288, FS288-ActA and FS288-Mstn complexes interactions are shown in Figures 1. The binding kinetics (Table 1) parameters were determined by globally fitting the sensorgram curves to a 1:1 Langmuir model (black fitting lines in Figure 1) from BIA evaluation software. FS288 and FS288-complexes showed very different heparin binding kinetics and affinity (Figure 1 and Table 1). The FS288-complexes showed 100-fold greater heparin-binding affinity than FS288 alone (KD = 4.7 ×10−10 M, and 4.0 ×10−10 M vs. KD = 5.6 ×10−8 M). The enhanced heparin affinity of the complexes can be ascribed to a much slower koff than observed for the FS288. The high binding affinity of the FS288-Mstn complex to heparin is consistent with previous findings demonstrating the electrostatic surface potential of the FS288-Mstn complex was remarkably electropositive.20 The results are also consistent with the data from our earlier study showing that FS288 released prior to FS288-Mstn complex from a heparin column.20
Figure 1.
A. SPR sensorgrams of FS288 heparin interaction. Concentrations of the protein (from top to bottom): 20, 10, 5, 2.5 and 1.25 nM, respectively. B. SPR sensorgrams of FS288-ActA complex heparin interaction. Concentrations of the protein (from top to bottom): 20, 10, 5, 2.5 and 1.25 nM, respectively. C. SPR sensorgrams of Mstn-FS288 complex heparin interaction. Concentrations of the protein (from top to bottom): 20, 10, 5, 2.5 and 1.25 nM, respectively.
Table 1.
Summary of kinetic data of protein- heparin interactions*
| Proteins | kon (1/MS) | koff (1/S) | KD (M) |
|---|---|---|---|
| FS288 | 1.1 ×104 (±669) | 6.0 ×10−4 (±1.4×10−5) | 5.6 ×10−8 |
| FS288-ActA complex | 3.3 ×103 (±118) | 1.6 ×10−6(±5.2 ×10−7) | 4.7 ×10−10 |
| FS288-Mstn complex | 4.85 ×104(±3.68×104) | 1.96 ×10−5(±2.5×10−5) | 4.0 ×10−10 |
The data with (±) in parentheses are the standard errors (SE) from the global fitting of sensorgrams from different concentrations protein binding.
SPR solution competition study of heparin oligosaccharides
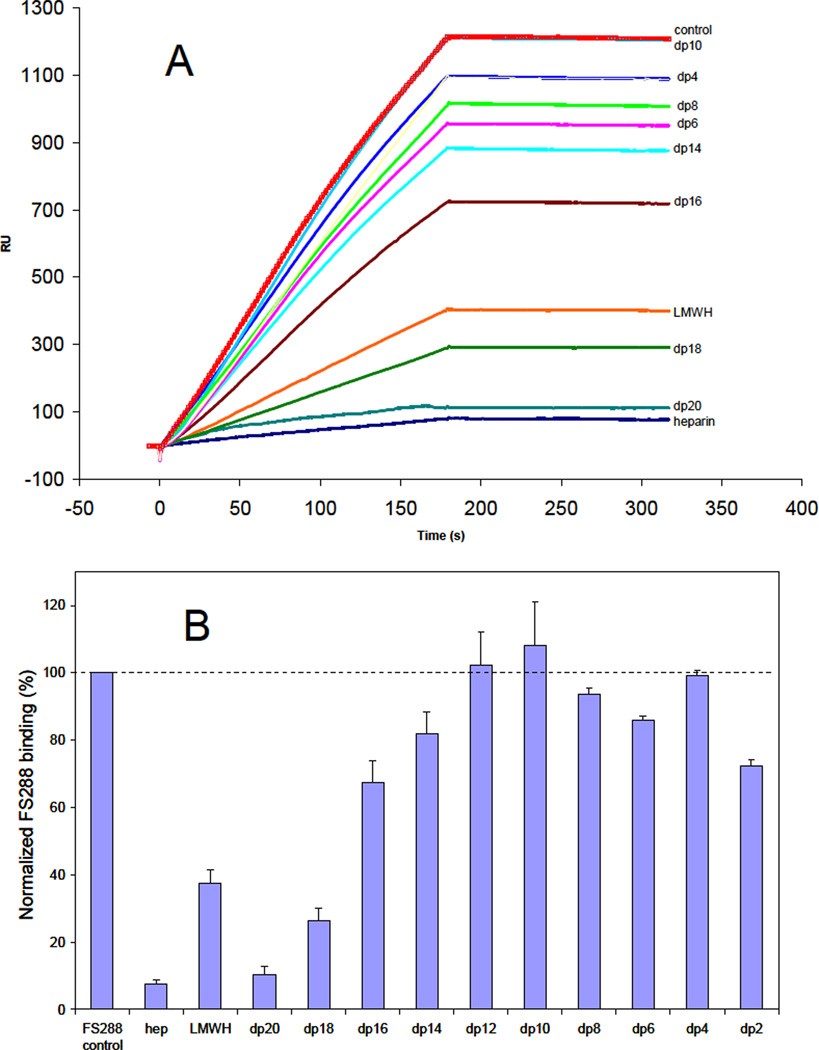
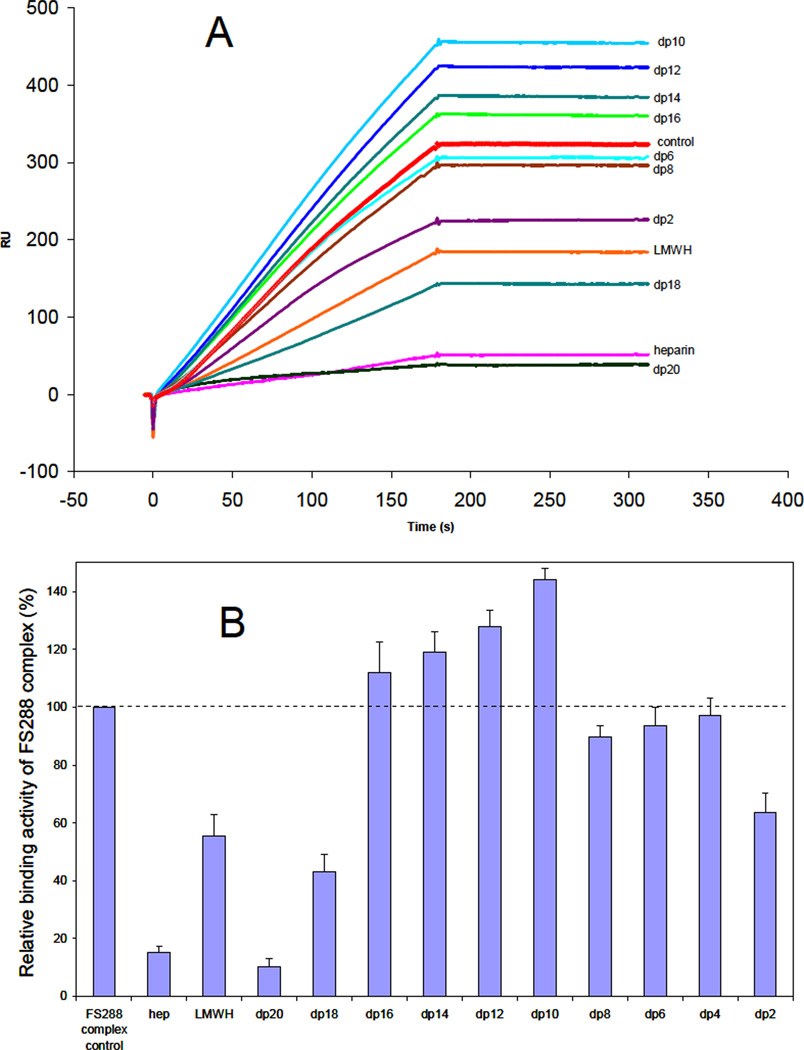
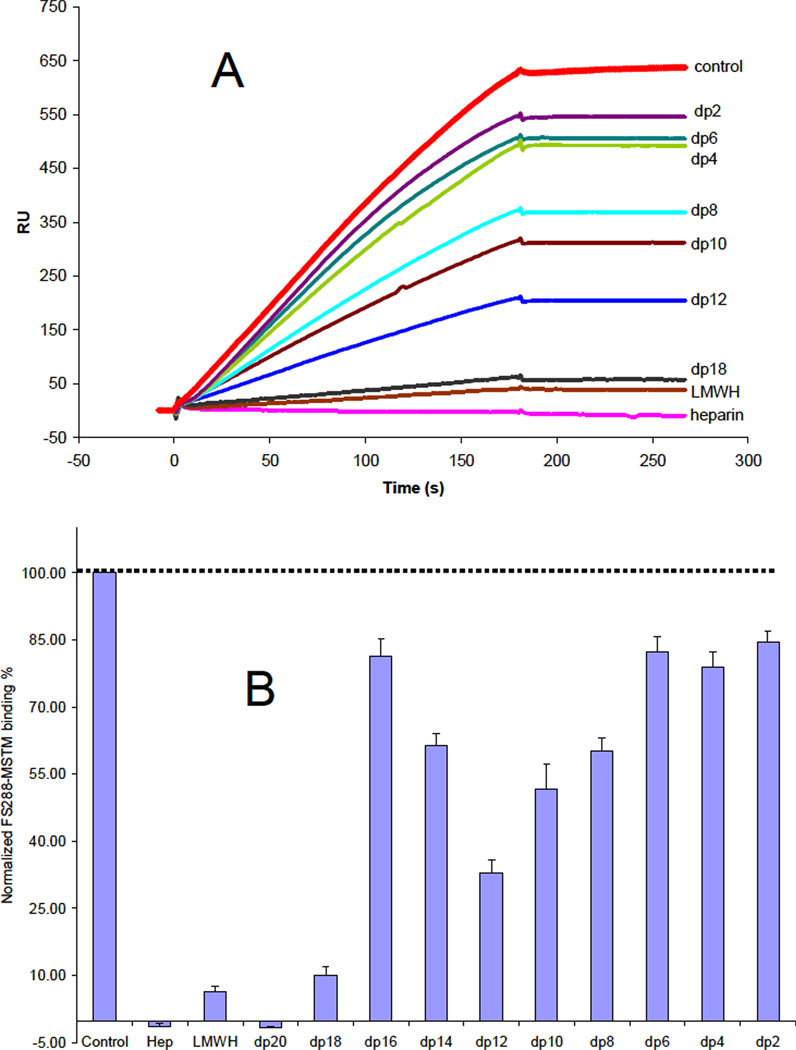
While the amino acid residues of FS involved in heparin binding have been identified, 27 little is known about the structural requirements of heparin molecules that bind to FS and the structure of the ligand-bound state is unclear.20 An SPR competition assay was used to examine the chain length dependence of heparin critical for binding. FS288 and its complexes were each mixed with heparin-derived oligosaccharides of various sizes in HBS-EP buffer and injected over a heparin chip. Once the active binding site on FS288 (or FS288 complexes) is occupied by heparin-derived oligosaccharide in the solution, its binding to the surface-immobilized heparin should decrease, resulting in a reduction in SPR signal. Heparin-derived oligosaccharides of different sizes, identified for the number of sugar residues in each (dp2 to dp20) were used in this competition experiment. The results of this SPR solution competition study are shown in Figures 2, 3 and 4. The results show that the binding of FS288 and its FS288-ActA, and FS288-Mstn complexes to heparin is chain length dependent. In all cases, heparin, LMWH and large oligosaccharides dp18 and dp20 strongly inhibited the binding of FS288 and FS288-ActA, and FS288-Mstn complexes to heparin immobilized on the chip. These findings on the heparin size requirement for binding are in good agreement with our previous structural measurements.20 In the FS288-Mstn complex the structure suggests that a composite heparin-binding site, corresponding to a continuous electropositive crevice that spans the complex, is generated in which a single heparin molecule could bind. The crevice measures ~ 60A wide and would fit a heparin molecule ~14–16 sugars in length, similar to that reported in the FGF–FGF receptor heparin complex.28 Of particular interest was the disparity observed for the ability of dodecasaccharide (dp12) to block binding to the heparin chip. For the FS288-Mstn complex over half of the binding interaction was lost in the presence of the dp12, where as Fs288 and FS288-ActA were unaffected. This suggests that the overall size requirement of heparin binding might be unique for different FS-ligand complexes.
Figure 2.
A. Binding preferences of FS288 to various chain lengths of heparin using a SPR solution competition assay. B. Bar graphs (based on triplicate experiments) displaying the inhibition activity of different sized heparin (in solution) on FS288 binding to immobilized heparin (on chip surface).
Figure 3.
A. Binding preferences of FS288-ActA complex to various chain lengths of heparin using a SPR solution competition assay. B. Bar graphs (based on triplicate experiments) displaying the inhibition activity of different sized heparin (in solution) on the complex binding to immobilized heparin (on chip surface).
Figure 4.
A. Binding preferences of FS288-Mstn complex to various chain lengths of heparin using a SPR solution competition assay. B. Bar graphs (based on triplicate experiments) displaying the inhibition activity of different sized heparin (in solution) on the complex binding to immobilized heparin (on chip surface).
Effect of buffer conditions on the interactions
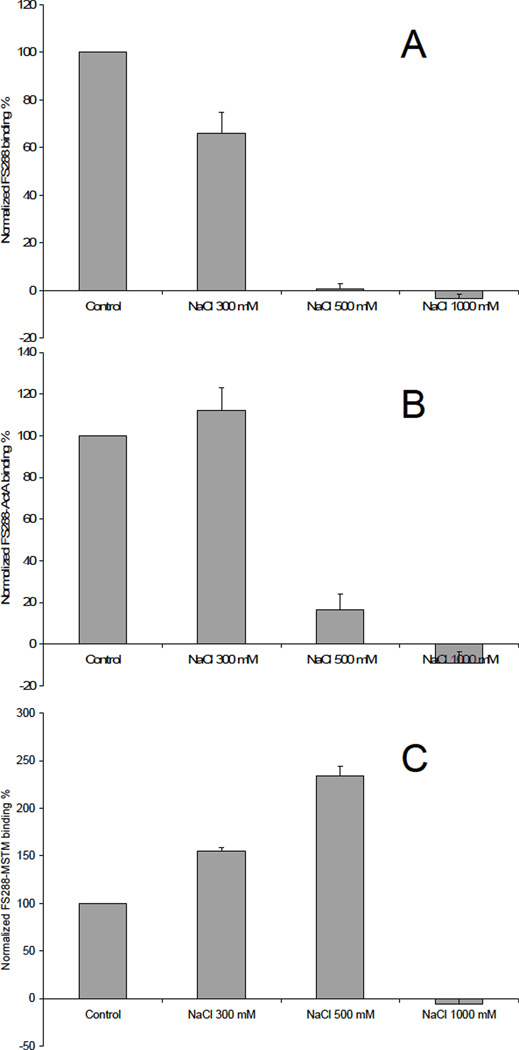
Binding buffers with different salt (NaCl) concentrations (300 mM, 500 mM and 1000 mM NaCl) and at different pH values (pH 6.2, pH 5.2 and pH4.0) were used for the SPR analysis to assess the effect of buffer on heparin-protein interactions. The results of heparin-protein interactions in various buffers are shown in Figures 5 and 6. High salt concentration (1 M) inhibited all the protein/complex binding to heparin, suggesting that this is primarily an electrostatically driven interaction. At 300 mM salt, FS288 binding to heparin was inhibited, but FS288-ActA and FS288-Mstn complexes binding to heparin were promoted. Surprisingly, the strongest interaction of FS288-Mstn complex binding to heparin was observed at 500 mM salt, whereas this concentration of salt abolished binding of both FS288 and FS288-ActA to heparin. This observation of FS288 and FS288-ActA heparin binding in the presence of NaCl is consistent with previous fluorescence polarization measurement.29
Figure 5.
A. Binding preference of FS288 in various NaCl concentrations. Bar graphs (based on triplicate experiments) displaying the FS288 binding to immobilized heparin (on chip surface). B. Binding preference of FS288-ActA complex in various NaCl concentrations. Bar graphs (based on triplicate experiments) displaying the FS288-ActA complex binding to immobilized heparin (on chip surface). C. Binding preference of FS288-Mstn complex in various NaCl concentrations. Bar graphs (based on triplicate experiments) displaying the FS288-Mstn complex binding to immobilized heparin (on chip surface).
Figure 6.
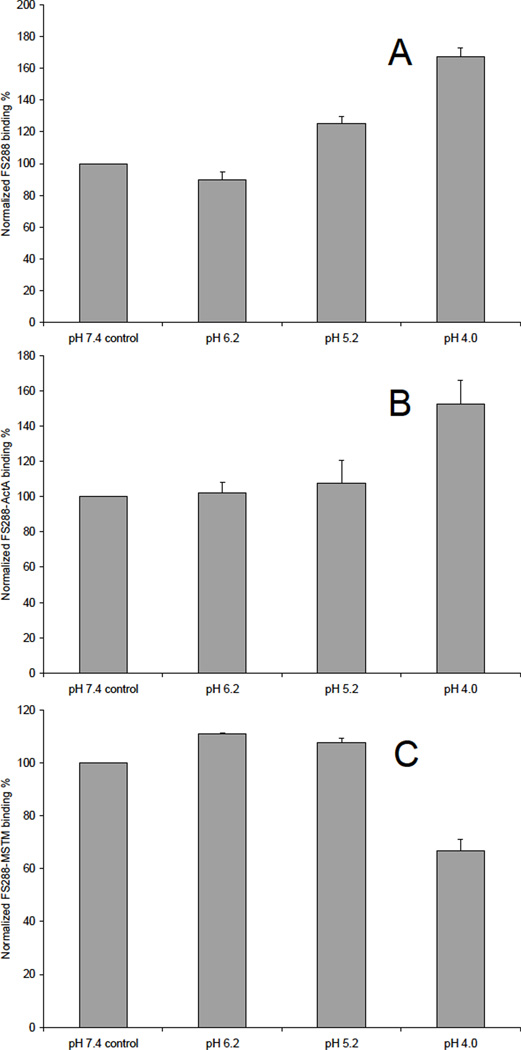
A. Binding preference of FS288 in various pH buffers. Bar graphs (based on triplicate experiments) displaying the FS288 binding to immobilized heparin (on chip surface). B. Binding preference of FS288-ActA complex in various pH buffers. Bar graphs (based on triplicate experiments) displaying the FS288-ActA complex binding to immobilized heparin (on chip surface). C. Binding preference of FS288-Mstn complex in various pH buffers. Bar graphs (based on triplicate experiments) displaying the FS288-Mstn complex binding to immobilized heparin (on chip surface).
The impact of pH on these interactions was also assessed. The results showed that acidic pH improved FS288 and FS288-ActA binding. Lower pH values cause more amino acids to become positively charged. For example, at pH values below 6 histidines become protonated and hence carry a positive charge. This favors electrostatic interactions with the negatively charged heparin.30 For FS288-Mstn, the results showed that slightly acidic pH values (pH 6.2 and 5.2) improved its binding to heparin, but at lower pH value (pH 4), the binding was inhibited. This could result from the protonation of carboxyl groups on heparin required for protein interaction or alteration of protein confirmation. There are a number of heparin/HS–protein interactions that are regulated by pH, such as β-amyloid peptide (Aβ),31 selenoprotein P.32 the stromal cell-derived factor-1 (SDF-1),33 although some of these effects might be attributable to proteins localized in acidic cell compartments like the lysozymes.
FS plays an important role in the bioneutralization of ActA and Mstn. This is partially a result of FS mediating ligand degradation through heparin dependent cell surface binding events. Previously, we identified heparin-binding differences when FS was in complex Mstn. Here we further define these differences and provide a quantitative analysis of heparin interactions with FS288 and FS288 complexes. Overall, knowledge of these differences might augment efforts to further modify follistatin to specifically antagonize Mstn therapeutically.
Acknowledgments
Funding
This work was supported by grants from the National Institutes of Health: GM-38060 to R.J.L and GM084186 to T.B.T.
ABBREVIATIONS
- FS
follistatin
- Mstn
myostatin
- ActA
Activin A
- SPR
surface plasmon resonance
- GAG
glycosaminoglycan
- HS
heparan sulfate
- HP
heparin
- LMWH
low molecular weight heparin
- Mstn
Myostatin
- FS
follistatin
- ActA
activin A
- TGF-β
transforming growth factor-β
- FGFs
fibroblast growth factors
- Hh
hedgehog
- ECM
extracellular matrix
- SA
streptavidin
- FC
flow-cell
- RU
resonance unit
References
- 1.Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv. Carbohydr. Chem. Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- 2.Linhardt RJ. Heparin: Structure and activity. J. Med. Chem. 2003;46:2551–2554. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 3.Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 4.Capila I, Linhardt RJ. Heparin-protein interactions. Angewandte Chemie-Intern. Ed. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nature Reviews Molecular Cell Biology. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 6.Parish CR. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 7.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 8.Roberts R, Gallagher J, Spooncer E, Allen TD, Bloomfield F, Dexter TM. Heparan sulphate bound growth factors: a mechanism for stromal cell mediated haemopoiesis. Nature. 1988;332:376–378. doi: 10.1038/332376a0. [DOI] [PubMed] [Google Scholar]
- 9.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annu. Rev. Biomed. Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 10.Lin X. Functions of Heparan Sulfate Proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 11.Wu ZLL, Zhang LJ, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J. Biol. Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 12.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 13.Turnbull JE, Fernig DG, Ke YQ, Wilkinson MC, Gallagher JT. Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. J. Biol. Chem. 1992;267:10337–10341. [PubMed] [Google Scholar]
- 14.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & Development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 15.Zhang FM, McLellan JS, Ayala AM, Leahy DJ, Linhardt RJ. Kinetic and structural studies on interactions between heparin or heparan sulfate and proteins of the hedgehog signaling pathway. Biochemistry. 2007;46:3933–3941. doi: 10.1021/bi6025424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derynck R, Miyazono K. The TGF-b Family. Woodbury, N.Y.: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 17.Feng XH, Derynck R. Specificity and versatility in TGF-b signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 18.Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell. Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 19.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell. Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 20.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: insights into receptor utilization and heparin binding. EMBO J. 2009;28:2662–2676. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innis CA, Hyvonen M. Crystal structures of the heparan sulfate-binding domain of follistatin. Insights into ligand binding. J. Biol. Chem. 2003;278:39969–39977. doi: 10.1074/jbc.M211284200. [DOI] [PubMed] [Google Scholar]
- 24.Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev. Cell. 2005;9:535–543. doi: 10.1016/j.devcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Pervin A, Gallo C, Jandik KA, Han X-J, Linhardt RJ. Preparation and Structural Characterization of Large Heparin-Derived Oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 26.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem. Biophys. Res. Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 27.Sidis Y, Schneyer AL, Keutmann HT. Heparin and activin binding determinants in follistatin and FSL3. Endocrinology. 2005;146:130–136. doi: 10.1210/en.2004-1041. [DOI] [PubMed] [Google Scholar]
- 28.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 29.Lerch TF, Shimasaki S, Woodruff TK, Jardetzky TS. Structural and Biophysical Coupling of Heparin and Activin Binding to Follistatin Isoform Functions. J. Biol. Chem. 2007;282:15930–15939. doi: 10.1074/jbc.M700737200. [DOI] [PubMed] [Google Scholar]
- 30.Gandhi NS, Mancera RL. The Structure of Glycosaminoglycans and their Interactions with Proteins. Chem. Biol. Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 31.Fraser PE, Nguyen JT, Surewicz WK, Kirschner DA. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys J. 1991;60:1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arteel GE, Franken S, Kappler J, Sies H. Binding of Selenoprotein P to heparin: characterization with surface plasmon resonance. Biol. Chem. 2000;381:265–268. doi: 10.1515/BC.2000.034. [DOI] [PubMed] [Google Scholar]
- 33.Veldkamp CT, Peterson FC, Pelzek AJ, Volkman BF. The monomer–dimer equilibrium of stromal cell-derived factor- 1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 2005;14:1071–1081. doi: 10.1110/ps.041219505. [DOI] [PMC free article] [PubMed] [Google Scholar]