Abstract
Killer-cell immunoglobulin-like receptor (KIR) proteins are expressed on natural killer (NK) cells and appear important in innate and adaptive immunity. There are about 14 KIR genes on chromosome 19q13.4, composed of those that inhibit and those that activate NK cell killing. Haplotypes have different combinations of these genes meaning that not all genes are present in a subject. There are two main classes of cognate human leukocyte antigen (HLA) ligands (HLA-Bw4 and HLA-C1/C2) that bind to the inhibitory/activating receptors. As a general rule, the inhibitory state is maintained except when virally infected or tumor cells are encountered; however, both increased activation and inhibition states have been associated with susceptibility and protection against numerous disease states including cancer, arthritis, and psoriasis.
Utilizing DNA from 158 Caucasian subjects with autism and 176 KIR control subjects we show for the first time a highly significant increase in four activating KIR genes (2DS5, 3DS1, 2DS1 and 2DS4) as measured by chi square values and odds ratios. In addition, our data suggests a highly significant increase in the activating KIR gene 2DS1 and its cognate HLA-C2 ligand (2DS1+C2; p=0.00003 [Odds Ratio=2.87]). This information ties together two major immune gene complexes, the Human Leukocyte Complex and the Leukocyte Receptor Complex, and may partially explain immune abnormalities observed in many subjects with autism.
Keywords: killer-cell immunoglobulin-like receptor, KIR genes, KIR haplotypes, human leukocyte antigen, HLA ligands, leukocyte receptor complex, autism, immune dysfunction, natural killer cells
Introduction
Autism Spectrum Disorder (ASD) is a term used for a complex group of neurodevelopmental disorders characterized by deficits in communication and social skills and the presence of restricted and repetitive stereotyped behaviors. The latest statistics released by the U.S. Centers for Disease Control and Prevention estimate that the incidence of ASDs increased 23% from 2006 to 2008 to 1 in 88 children (CDC, 2012).
Years ago familial clustering and twin studies indicated a strong genetic component to predisposition; however, after extensive genetic research, only a small number of cases can be associated with specific genes (McClellan and King, 2010). There is considerable evidence that suggests involvement of the immune system in the etiology of autism (Westover et al., 2011).
Both cellular and humoral immunological changes have been reported in children with autism. Those abnormalities include changes in certain immune cell functions (Ashwood et al., 2010), an increase in the C4B null allele (Warren et al., 1991; Odell et al., 2005; Mostafa and Shehab, 2010), association of certain human leukocyte antigen (HLA) alleles (Torres et al., 2002; Torres et al., 2006), and an increase in certain ancestral HLA haplotypes (Daniels et al., 1995). These last three papers, published by our research group, have all described typical HLA Class I and Class II HLA antigen-presenting alleles which involve different protein binding sites (T-cell receptor) outside the KIR binding sites. Additional immune associations include imbalances in antibody levels (Croonenberghs et al., 2002; Heuer et al., 2008; Enstrom et al., 2009b), an increase in autoantibodies to neural tissue (Cabanlit et al., 2007; Wills et al., 2009; Rosenspire et al., 2011), altered cytokine levels (Molloy et al., 2006; Ashwood et al., 2011), changes in lymphocyte subsets (Furlano et al., 2001), a family history of autoimmune diseases (Atladóttir et al., 2009), and reduced natural killer (NK) cell activity (Warren et al., 1987; Enstrom et al., 2009a).
The HLA region on chromosome 6, the most complex region in the human genome, is central to many immunological reactions. HLA cell surface proteins interact with receptors on various T-cells and thus play an important role in inflammation, the complement cascade, and the innate (inborn) and adaptive (acquired) immune responses (Shiina et al., 2009).
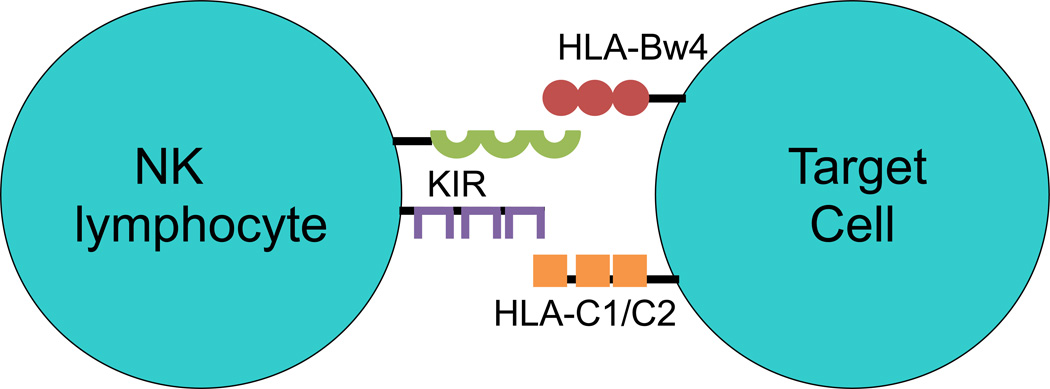
NK cells are a subset of lymphocytes with the innate ability to produce cytokines and kill target cells without prior sensitization and are essential for self-tolerance (Vivier et al., 2008). NK cells thus participate in early responses against virally infected and transformed cells by recognizing the lack of HLA class I proteins on the cell surface (“missing self”). NK cell function is partially regulated by inhibitory and stimulatory signals generated by ligand-receptor binding at the cell surface. Among these cell surface receptors are killer-cell immunoglobulin-like receptors (KIR) (Kulkarni et al., 2008). The ligands for these KIR inhibitory and activating receptors are amino acid epitopes contained in HLA-B, & C and rarely HLA-A proteins. The HLA-Bw4, HLA-C1 and HLA-C2 protein epitopes (serological alleles) are binding ligands for KIR proteins and do not present peptide antigens to T-cells. These serological alleles were detected by HLA-antibodies years before antigen-presenting alleles were delineated. All of the antigen-presenting HLA-B alleles can be divided into KIR binding (Bw4) or non-KIR binding (Bw6) serological alleles and all of the antigen-presenting HLA-C alleles can be placed into HLA-C1 or -C2 KIR ligand alleles. The bindings of these HLA cell surface ligands to KIR cell surface receptors thus play an important role in the regulation of the innate and adaptive immune response of NK cells (Fig. 1). In this report it is suggested that the frequencies of certain KIR activating genes and their HLA ligands are increased in autism.
Figure 1.
NK lymphocytes have several receptors on their surface including killer-cell immunoglobulin-like receptors (KIR). The ligands for the KIR are Class I HLA molecules, in particular, HLA-C1/C2 and HLA-Bw4.
Materials and Methods
Two sample cohorts have been used in this study. The first is from a Utah/Oregon autism study that consisted of 70 Caucasian subjects from 70 different families with a single autistic child (7 females, 63 males; 1:9 female:male ratio). Of the total 70 subjects, 51 subjects were obtained from the Utah Autism Project and 19 subjects from the Oregon Health and Science University Autism Clinic (Torres et al., 2006). The second family cohort sample was obtained from the Autism Genetics Resource Exchange (AGRE) which has samples from over 2000 families, many with 2 or more autistic children, which have been collected throughout the USA since 1997. These samples have been utilized by more than 150 research groups worldwide in the search for genetic markers and/or risk factors for autism. In our study we utilized a subset of Caucasian families (88 subjects, 16 females, 72 males; 1:4.5 female:male ratio) and we used DNA from only a single child with autism in any family. All subjects were diagnosed with autism by psychologists or pediatric psychiatrists using the Autism Diagnostic Inventory-Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS) (Lord et al., 1994; Lord et al., 1989). The severity of the disease in individuals can vary significantly within the Autism Spectrum and the degree of phenotypic and genetic heterogeneity in the two cohorts, even if fully known, would be extremely difficult to compare in a meaningful manner.
DNA
Genomic DNA was purified with Qiagen kits (QIAmp 96 DNA Blood Kit) from the Utah/Oregon (UT/OR) population whereas purified DNA was obtained from AGRE for the AGRE samples.
Whole genome amplification
About 100ng of genomic DNA from each individual was subjected to whole genome amplification following the multiple displacement amplification (MDA) procedure of Dean et al. (2002) using random hexamers. Phi29 polymerase and dNTPs were purchased from Epicentre Inc. The quantity of MDA-DNA was determined with the PicoGreen dsDNA kit from Molecular Probes (Eugene, Oregon).
HLA genotyping
The HLA genotyping was done using about 2µg of MDA-DNA using Taq polymerase (GenScript; cat.#E00007) with SSP UniTray low resolution HLA kits (Invitrogen). The HLA kits can distinguish between 23 HLA-A alleles, 49 HLA-B alleles, and 18 HLA-C alleles. The HLA-A and HLA-B allotypes for the UT/OR and AGRE subjects were used to determine Bw4/Bw6 serological alleles (http://hla.alleles.org/antigens/bw46.html). Bw4 alleles which have an isoleucine or threonine at the 80 position (80I & 80T respectively) are ligands for KIR receptors (Carrington and Norman, 2003) whereas Bw6 serological alleles are not ligands for KIR receptors. HLA-A3 & A11 are Bw4 ligands for 3DL2, but only when specific Epstein Barr virus peptides are present (Hansasuta et al., 2004). The HLA-C alleles were placed into KIR ligand binding specificities (C1 and C2) according to the amino acid sequence at positions 77 and 80 (Winter and Long, 1997).
The frequencies of the Bw4 and C1/C2 ligands were analyzed by a case-control study design using normal HLA gene frequencies from the Centre d’Etude Polymorphisms Human (CEPH) (http://www.cephb.fre/en/cephdb/). The CEPH control HLA population had 164 unrelated individuals.
KIR genotyping
The KIR genotyping was done on SSP kits (Invitrogen; cata#78930-3) using about 2µg of MDA-DNA as described above. The KIR kits can distinguish between the 14 KIR inhibitory/activating genes (2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3, 2DS1) used in this study. The inhibitory and activating KIR genes for NK killing activity have L and S in their names, respectively.
The KIR frequency genotyping data for the UT/OR and AGRE subjects were also analyzed by a case-control study using normal KIR gene frequencies from the Centre d’Etude Polymorphisms Human (CEPH) (http://www.cephb.fre/en/cephdb/). The CEPH control KIR gene population had 176 unrelated individuals (Martin et al., 2008) (Table 4) whereas the HLA comparisons used the CEPH control population with 164 individuals.
Table 4.
Comparison of the frequencies between the CEPH control population and the UT/OR and AGRE populations for the KIR genes plus the corresponding HLA Bw4 and C1/C2 ligands. HLA-A3/11 are Bw4 ligands.
| CEPH Controls |
UT/OR(n=70) | AGRE(n=88) | Combined UT/OR & AGRE(n=158) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KIR+HLA | UT/OR | OR | 95% CI | p-value | AGRE | OR | 95% CI | p- value |
Both | OR | 95% CI | p-value | |
| 2DL2+C1 | 61 | 31 | 1.35 | (0.76, 2.40) | 0.311 | 41 | 1.40 | (0.83, 2.38) | 0.209 | 72 | 1.38 | (0.88, 2.16) | 0.162 |
| 2DL3+C1 | 113 | 54 | 1.27 | (0.65, 2.52) | 0.482 | 65 | 1.00 | (0.55, 1.82) | 0.999 | 119 | 1.11 | (0.66, 1.86) | 0.694 |
| 2DL1+C2 | 90 | 49 | 1.74 | (0.95, 3.18) | 0.072 | 49 | 0.94 | (0.55, 1.59) | 0.803 | 98 | 1.22 | (0.77, 1.91) | 0.395 |
| 3DL1+Bw4 | 108 | 52 | 1.90 | (1.02, 3.52) | 0.041 | 56 | 1.15 | (0.68, 1.95) | 0.603 | 108 | 1.42 | (0.90, 2.23) | 0.126 |
| 3DL2+A3/11 | 47 | 19 | 1.12 | (0.60, 2.09) | 0.726 | 25 | 1.19 | (0.67, 2.11) | 0.548 | 44 | 1.16 | (0.72, 1.87) | 0.549 |
| 2DS2+C1 | 63 | 31 | 1.20 | (0.68, 2.12) | 0.533 | 44 | 1.51 | (0.89, 2.56) | 0.125 | 75 | 1.36 | (0.87, 2.13) | 0.174 |
| 3DS1+Bw4 | 38 | 34 | 3.50 | (1.94, 6.34) |
0.00003 0.003* |
30 | 1.92 | (1.08, 3.40) | 0.024 | 64 | 2.53 | (1.56, 4.08) |
0.0001 0.008* |
| 2DS1+C2 | 33 | 32 | 3.55 | (1.93, 6.51) |
0.00004 0.003* |
32 | 2.41 | (1.35, 4.30) | 0.003 | 64 | 2.87 | (1.75, 4.71) |
0.00003 0.003* |
| 2DS4+Bw4 | 24 | 13 | 1.27 | (0.60, 2.69) | 0.524 | 11 | 0.80 | (0.37, 1.72) | 0.563 | 24 | 1.00 | (0.54, 1.85) | >0.999 |
Bold type p-values are significant at the p≤0.05 level. Bonferroni allelic corrected p-values significant at the p≤0.05 are listed below in Bold type*.
Comparison of two published KIR control Caucasian populations (Du et al., 2008; Hollenbach et al., 2010) with the CEPH control population showed no significant differences in the p-values for KIR inhibitory and activating gene frequencies. This indicates that the Caucasian KIR gene frequencies are very similar in the USA. Although any of the 3 control populations could have been used, it was decided that the CEPH population would be used as this population is included in many genetic studies around the world.
Statistical Analysis
Odds ratios and p-values were calculated by multiple logistic regression, with all relevant alleles included in each statistical model. With the significance level set at p≤0.05 there is a 5% chance that each factor may be erroneously found to be significant. Multiple testing correction methods are available that maintain the overall, study-wide error rate to less than or equal to the user-specified p-value cutoff. One of the most stringent methods is the Bonferroni correction and it offers a very conservative approach to control for false positive results. A Bonferroni allelic correction for 42 tests in two cohorts (84 total tests) was used for a statistical significance threshold. We have provided the p-values for the initial testing and a Bonferroni significance threshold for ultimate determination of statistical significance. In this study a total of 84 pairwise comparisons were made, so an individual p-value of 0.0006 or less is necessary for statistical significance to maintain a study-wide error rate of p=0.05 (0.0006 × 84=0.05). The KIR gene frequencies from the CEPH control population were compared to the UT/OR and AGRE autism populations separately and combined (Table 2).
Table 2.
Comparison of KIR gene frequencies for subjects with autism from the UT/OR and AGRE populations against the CEPH control population. The two autistic populations were compared separately and then combined when compared to the control population.
| CEPH Controls |
UT/OR(n=70) | AGRE(n=88) | Combined UT/OR & AGRE(n=158) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KIR | UT/OR | OR | 95% CI | p-value | AGRE | OR | 95% CI | p-value | Both | OR | 95% CI | p-value | |
| 2DS2 | 97 | 37 | 0.89 | (0.51, 1.56) | 0.682 | 53 | 1.20 | (0.71, 2.03) | 0.489 | 90 | 1.05 | (0.68, 1.62) | 0.824 |
| 2DS3 | 46 | 24 | 1.47 | (0.81, 2.69) | 0.203 | 30 | 1.46 | (0.84, 2.55) | 0.180 | 54 | 1.47 | (0.92, 2.35) | 0.110 |
| 2DS5 | 53 | 33 | 2.04 | (1.15, 3.61) | 0.014 | 35 | 1.51 | (0.88, 2.58) | 0.133 | 68 | 1.72 | (1.10, 2.71) | 0.018 |
| 3DS1 | 65 | 41 | 2.41 | (1.37, 4.26) | 0.002 | 42 | 1.56 | (0.93, 2.62) | 0.093 | 83 | 1.89 | (1.22, 2.93) | 0.004 |
| 2DS1 | 67 | 45 | 2.93 | (1.64, 5.22) |
0.0003 0.025* |
50 | 2.14 | (1.27, 3.61) | 0.004 | 95 | 2.45 | (1.58, 3.82) |
0.00007 0.006* |
| 2DS4 | 160 | 68 | 3.40 | (0.76, 15.3) | 0.109 | 86 | 8.60 | (1.11, 66.6) | 0.038 | 154 | 5.13 | (1.45, 18.1) | 0.010 |
| 2DL2 | 96 | 38 | 1.06 | (0.60, 1.86) | 0.851 | 50 | 1.10 | (0.65, 1.84) | 0.726 | 88 | 1.08 | (0.70, 1.67) | 0.733 |
| 2DL3 | 162 | 66 | 1.90 | (0.53, 6.88) | 0.325 | 78 | 0.67 | (0.29, 1.59) | 0.366 | 144 | 0.96 | (0.43, 2.11) | 0.913 |
| 2DL1 | 172 | 68 | 0.79 | (0.14, 4.46) | 0.789 | 85 | 0.66 | (0.14, 3.03) | 0.590 | 153 | 0.71 | (0.19, 2.71) | 0.617 |
| 3DL1 | 174 | 64 | 0.12 | (0.02, 0.63) | 0.011 | 84 | 0.24 | (0.04, 1.36) | 0.105 | 148 | 0.17 | (0.04, 0.79) | 0.024 |
| 2DL5A | 63 | 39 | 2.16 | (1.22, 3.80) | 0.008 | 41 | 1.50 | (0.89, 2.53) | 0.130 | 80 | 1.75 | (1.30, 2.74) | 0.012 |
Bold type p-values significant at the p≤0.05 level. Bonferroni allelic corrected p-values significant at the p≤0.05 are listed below in Bold type*.
Results
The comparison of the HLA Bw4/Bw6 allelic frequencies of the UT/OR and AGRE populations suggests that there is a decrease in the Bw4 allele in the AGRE autism population, but not the UT/OR combined population (Table 1). This is in agreement with data that suggests a decrease in the Bw4 80I and Bw4 80T frequencies in the AGRE subjects or combined population (Table 1).
Table 1.
Comparison of CEPH HLA Bw4/Bw6 and HLA C1/C2 allelic control population to the UT/OR and AGRE allele frequencies.
| CEPH Controls |
UT/OR(n=70) | AGRE(n=88) | Combined UT/OR & AGRE(n=158) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UT/OR | OR | 95% CI | p- value |
AGRE | OR | 95% CI | p- value |
Both | OR | 95% CI | p- value |
||
| no Bw4 | 29 | 11 | 1 | 21 | 1 | 32 | 1 | ||||||
| Bw4 | 123 | 56 | 1.2 | (0.56, 2.58) | 0.639 | 60 | 0.67 | (0.35, 1.28) | 0.227 | 116 | 0.85 | (0.49, 1.50) | 0.585 |
| B15 | 9 | 3 | 0.88 | (0.2, 3.89) | 0.864 | 7 | 1.07 | (0.34, 3.36) | 0.902 | 10 | 1.01 | (0.36, 2.84) | 0.999 |
| no Bw4 | 29 | 11 | 1 | 21 | 1 | 32 | 1 | ||||||
| Bw4 80I | 107 | 33 | 0.69 | (0.39, 1.21) | 0.192 | 36 | 0.55 | (0.32, 0.93) | 0.026 | 69 | 0.60 | (0.39, 0.93) | 0.021 |
| Bw4 80T | 102 | 30 | 0.64 | (0.36, 1.14) | 0.124 | 29 | 0.45 | (0.26, 0.78) | 0.004 | 59 | 0.52 | (0.33, 0.81) | 0.004 |
| B15 or B27 | 31 | 12 | 0.87 | (0.40, 1.87) | 0.715 | 23 | 1.43 | (0.75, 2.71) | 0.270 | 35 | 1.17 | (0.67, 2.06) | 0.573 |
| no C1 | 30 | 12 | 1 | 13 | 1 | 25 | 1 | ||||||
| C1 | 134 | 58 | 1.08 | (0.52, 2.27) | 0.834 | 75 | 1.29 | (0.63, 2.63) | 0.480 | 133 | 1.19 | (0.66, 2.14) | 0.556 |
| no C2 | 64 | 19 | 1 | 38 | 1 | 57 | 1 | ||||||
| C2 | 100 | 51 | 1.72 | (0.93, 3.18) | 0.084 | 50 | 0.84 | (0.5, 1.43) | 0.522 | 101 | 1.13 | (0.72, 1.78) | 0.585 |
| Bw6/Bw6 | 29 | 11 | 1 | 21 | 1 | 32 | 1 | ||||||
| Bw4/Bw6 | 92 | 40 | 1.15 | (0.52, 2.53) | 0.734 | 44 | 0.66 | (0.34, 1.29) | 0.223 | 84 | 0.83 | (0.46, 1.49) | 0.524 |
| Bw4/Bw4 | 31 | 16 | 1.36 | (0.54, 3.43) | 0.512 | 16 | 0.71 | (0.31, 1.63) | 0.421 | 32 | 0.94 | (0.46, 1.90) | 0.853 |
| Bw6/B15 | 9 | 3 | 0.88 | (0.20, 3.89) | 0.864 | 7 | 1.07 | (0.34, 3.36) | 0.902 | 10 | 1.01 | (0.36, 2.84) | 0.999 |
| C1/C1 | 64 | 19 | 1 | 38 | 1 | 57 | 1 | ||||||
| C2/C1 | 70 | 39 | 1.88 | (0.98, 3.59) | 0.056 | 37 | 0.89 | (0.50, 1.57) | 0.687 | 76 | 1.22 | (0.75, 1.98) | 0.421 |
| C2/C2 | 30 | 12 | 1.35 | (0.58, 3.14) | 0.488 | 13 | 0.73 | (0.34, 1.57) | 0.419 | 25 | 0.94 | (0.49, 1.78) | 0.839 |
Bold type p-values significant at the p≤0.05 level.
No differences between the C1 and C2 alleles in the autism and control populations were noted (Table 1). Likewise, examination of individuals for diploid HLA Bw4/Bw6 and C1/C2 alleles did not indicate any significant differences (Table 1). Overall, the HLA ligand data is unremarkable by itself and only becomes important when examined with their cognate KIR receptors (Tables 3,4).
Table 3.
Comparison of inhibitory (i) and activating (a) KIR genes between the CEPH control population and the UT/OR and AGRE autistic populations separately and together.
| CEPH Controls |
UT/OR(n=70) | AGRE(n=88) | Combined UT/OR & AGRE(n=158) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UT/OR | OR | 95% CI | p- value |
AGRE | OR | 95% CI | p- value |
Both | OR | 95% CI | p- value |
||
| iKIR | 176 | 67 | 0.71 | (0.43, 1.19) | 0.194 | 88 | 0.72 | (0.44, 1.17) | 0.188 | 155 | 0.71 | (0.47, 1.07) | 0.097 |
| aKIR | 128 | 54 | 1.41 | (1.13, 1.76) | 0.002 | 72 | 1.32 | (1.08, 1.61) | 0.007 | 126 | 1.36 | (1.15, 1.61) |
0.0004 0.034* |
| iKIR + HLA | 147 | 67 | 0.81 | (0.57, 1.15) | 0.244 | 88 | 0.69 | (0.50, 0.97) | 0.033 | 155 | 0.76 | (0.58, 1.01) | 0.062 |
| aKIR + HLA | 104 | 49 | 1.74 | (1.25, 2.40) | 0.001 | 68 | 1.48 | (1.09, 2.01) | 0.012 | 117 | 1.56 | (1.21, 2.02) |
0.0006 0.050* |
Bold type p-values are significant at the p≤0.05 level. Bonferroni allelic corrected p-values significant at the p≤0.05 are listed below in Bold type*.
Table 2 shows the p-values and odds ratios of the KIR gene frequencies of the CEPH control population compared against the UT/OR and AGRE autism populations separately and together. One can see that the gene frequencies are similar in the two populations; therefore, we will only discuss the combined UT/OR and AGRE statistics. Four activating genes frequencies (2DS5, 3DS1, 2DS1, and 2DS4) and one inhibitory gene frequency (2DL5A) are increased in the autism samples and one inhibitory gene is decreased (3DL1). The 2DL5A gene is usually paired with activating genes. There is either a 3DL1 or a 3DS1 at the 3D location so an increase in one would show a decrease in the other except in rare instances that contain both so the decrease in 3DL1 is expected. The most important observation is the highly significant increase in 2DS1 (p=0.00007) (Bonferroni correction p=0.00588) (odds ratio= 2.45). Although the chi square value for 2DS4 is a modest p=0.010, it has an odds ratio of 5.13 suggesting that this gene also has a moderate to strong association with autism.
Comparing the total of the activating gene frequencies (actKIR) against the total of the inhibitory gene frequencies (inhKIR) also suggests a highly significantly increased in activating genes in the autistic populations (p=0.0004) (Bonferroni correction p=0.0336) (Table 3). Examining the frequencies of HLA ligands and activation KIR genes together is also highly significant (p=0.0006) (Bonferroni correction p=0.0504).
The last evaluation involves the examination of particular HLA ligands with KIR activating genes. The Bw4 ligand interacts with the 3DS1 receptor and together they are increased compared to controls (odds ratio of 2.53; p=0.0001) (Bonferroni correction p=0.0084). The most significant result is the 2DS1+C2 combination (odds ratio of 2.87; p=0.00003) (Bonferroni correction p=0.0025) (Table 4). The combined statistical evaluation of the HLA and KIR genotyping data indicates a remarkable autism association. It is important to note that the chi square evaluation of the HLA alleles is rather unremarkable (Table 1) until combining the cognate HLA ligand with the binding KIR receptor (Table 4).
Discussion
The main purpose of the HLA locus is to protect the individual against infectious agents and to eliminate damaged, dying or infected cells and tissue. The extraordinarily high level of genetic polymorphisms in the HLA region allows a selective advantage for the immune system in combating microorganisms. However, this high level of genetic polymorphisms adds a risk of creating autoimmune and genetic diseases. Besides antigen presentation to T-cells and B-cells, it is well known that HLA proteins interact with proteins encoded in genes outside the HLA region. For example, C4 proteins from the HLA class III region interact with proteins encoded on chromosome 1q32 in the regulators of complement activation block (RCAa) and Cystatin C on chromosome 20p11 (Shiina et al., 2009). HLA proteins influence cellular behavior by presenting antigen (self and non-self) to T-cell and B-cell receptors. NK cells are largely controlled by HLA ligands that bind to cell surface receptor proteins encoded in the leukocyte receptor complex (LRC) on chromosome 19q13.4. There are six important receptor complexes including the KIR that help control NK behavior. HLA class I molecules serve as ligands for specific KIR receptor proteins and help activate or inhibit effector function and cytokine production. It should be clear that this ligand-receptor interaction does not involve antigen presentation.
Specific HLA class I ligand and KIR receptor combinations have been associated with autoimmunity, viral infections, pregnancy-related disorders and cancer (Kulkarni et al., 2008). The surveillance of self must be tempered to block self-destruction, therefore, the pairing of inhibitory and activating effects on NK cells must be somewhat balanced. Our data suggests that HLA-KIR interactions may be of importance in autism, as has been observed in other disease pathologies. The activation and inhibition of NK cells also include cell surface receptors that interact with soluble ligands like cytokines and chemokines and soluble HLA molecules. The balance of activation and inhibition of NK cells is thus an extraordinarily complicated process.
Warren et al. (1987) reported decreased NK killing in cells from ASD subjects upon stimulation with K562 target cells. In an elegant study using RNA expression microarray and cell culture assays, Enstrom et al. (2009) also noted decreased NK killing upon stimulation with K562 cells. The microarray experiment showed the increased expression of 11 probes for KIR receptors with 9 of these probes being for inhibitory KIR genes in ASD subjects. However, under resting conditions, NK cells from ASD subjects demonstrated an increase in cytolytic capacity as determined by higher levels of perforin, granzyme B, and interferon γ. Upon K562 target cell stimulation, these three markers were lower in the ASD NK cells as would be expected from the decreased killing. The increase in the activating KIR-HLA suggests an increase in NK activation as noted by Enstrom et al. (2009) in their resting NK experiment. However, NK cells have an array of receptors that can either stimulate or dampen cell activity (Vivier et al., 2011) and although KIR genes may be the most important genes for NK activity they represent only one gene cluster in the LRC on chromosome 19q13.4 (Barrow and Trowsdale, 2008; Carrington and Norman, 2003). Although previous research has suggested a role for NK cells in autism, this is the first report describing the association of HLA ligands and an increase in the activating KIR receptors. The observation that there is an increase in the activating KIR genes is very strong with highly significant chi square values even after Bonferroni allelic correction.
There is still debate about how to best define autism and the reported increase in autism prevalence rates over the years may be partially due to changes in diagnostic practices (Desoto and Hitlan, 2010; King and Bearman 2009; Hertz-Picciotto, 2009). Latif and Williams (2007) reported that classical Kanner autism did not increase over time when the same diagnostic criteria were employed and Altevogt et al., (2008) has suggested that there may be more than one type of autism.
This broadening of the diagnosis may be beneficial for clinical practice, however, it lumps individuals from different regions on the autism spectrum together and complicates genetic studies. Although we cannot compare diagnostic heterogeneity of these two cohorts, the fact that both families show statistically significant increases in specific activating KIR genes and their cognate HLA ligands suggests that diagnostic heterogeneity may not be a confounding factor.
To better answer these questions, a new population of autism subjects that has much better clinical data is currently being HLA-KIR genotyped. This new population has three racial groups (African-American, Hispanic and Caucasian) which will allow us to confirm or not confirm these results in each of the three populations and compare with detailed diagnostic criteria.
Increased KIR-related NK activation has been associated with several autoimmune diseases like psoriatic arthritis, scleroderma, and rheumatoid vasculitis as well as numerous cancers including cervical cancer and nasopharyngeal carcinoma. On the other hand, increased KIR-related activation slows down HIV progression. The increase of activating KIR genes in autism may partially explain the association of autoimmune diseases with autism (Sweeten et al., 2003; Altevogt et al., 2008). It is important to note that KIR2DS1, which has the highest association with autism in this report, has been associated with psoriasis (Holm et al., 2005; Luszczek et al., 2004) and an increased incidence of psoriasis has been detected in mothers of children with autism (Croen et al., 2005).
The new data presented in this study about an increase in activating KIR genes in autism combined with the information about HLA ligand interactions with KIR receptors could be important in how immune dysfunction may be involved in the etiology of autism. The data also opens another avenue for further research on the etiology of autism since HLA ligand-KIR receptor interactions are important in histocompatibility and restricted cellular interactions which influence central nervous system development and plasticity, neurological cell interactions, synaptic function, as well as neurological and psychiatric disorders (Shiina et al., 2009).
The KIR gene complex is extraordinarily complicated with 14 genes comprised of structural, inhibitory, and activating genes that are inherited together as haplotypes. There is one inhibitory haplotype comprised of 5 inhibitory genes and one activating gene (2DS4). The inhibitory haplotype is the most common with 50% to 60% of individuals having one inhibitory haplotype (Pyo et al., 2010) and about 27% of individuals having two inhibitory haplotypes (Martin et al., 2008). The activating haplotypes contain a complex mixture of inhibitory and activating genes (2DS1, 2DS2, 2DS3, 2DS4, 2DS5, and 3DS1) in about 40 different combinations.
We decided to compare the number of inhibitory genes to activating genes for the autism and control populations instead of constructing incomplete haplotypes. Essentially all inhibitory haplotypes have a 2DS4 gene, but only about half of the activating haplotypes have the 2DS4 gene. This suggests that activating haplotypes which have the 2DS4 gene are increased in the subjects with autism. Our current goal is to extend our research to delineate KIR haplotypes in the UT/OR and AGRE populations. Newer genotyping methods are becoming available that allow for more accurate determination of the complicated KIR haplotypes (Pyo et al., 2010); this should help further define the genetic understanding of the KIR gene families in the etiology of autism.
Research Highlight.
The data is highly significant as it suggests a strong genetic connection between two immune complexes: the human leukocyte complex and the leukocyte receptor complex.
Acknowledgments
We gratefully acknowledge the resources provided by the AGRE Consortium and the participating AGRE families. Funding was provided [in part] by the Early Markers for Autism NIH grant RO1ESO16669, the Utah Science Technology and Research (USTAR) initiative at Utah State University, and the Utah Autism Foundation (SLC, Utah). This project also received Federal funding [in part] from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was also supported [in part] by the Intramural Research Program of NIH, Frederick National Laboratory, Center for Cancer Research. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does its mention of trade names, commercial products or organizations imply endorsement of the US Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: All authors declare that there are no conflicts of interest.
References
- Altevogt BM, Hanson SL, Leshner AI. Autism and the environment: challenges and opportunities for research. Pediatrics. 2008;121:1225–1229. doi: 10.1542/peds.2007-3000. [DOI] [PubMed] [Google Scholar]
- Ashwood P, Krakowiak P, Hertz-Picciotto I, Pessah IN, Van de Water J. Altered T cell responses in children with autism. Brain Behav Immun. 2010;25:840–849. doi: 10.1016/j.bbi.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah I, Van de Water J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun. 2011;25:40–45. doi: 10.1016/j.bbi.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atladóttir HÓ, Pedersen MG, Thorsen P, Mortensen PB, Deleuran B, Eaton WW, et al. Association of family history of autoimmune diseases and autism spectrum disorders. Pediatrics. 2009;124:687–694. doi: 10.1542/peds.2008-2445. [DOI] [PubMed] [Google Scholar]
- Barrow AD, Trowsdale J. The extended human leukocyte receptor complex:diverse ways of modulating immune response. Immunological Reviews. 2008;224:99–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- Cabanlit M, Wills S, Goines P, Ashwood P, Van de Water J. Brain-specific autoantibodies in the plasma of subjects with autistic spectrum disorder. Ann NY Acad Sci. 2007;1107:92–103. doi: 10.1196/annals.1381.010. [DOI] [PubMed] [Google Scholar]
- Carrington M, Norman P. The KIR Gene Cluster. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information (US); 2003. May 28, pp. 1–48. [Google Scholar]
- Centers for Disease Control and Prevention. Prevalence of Autism Spectrum Disorders - Autism and Developmental Disabilities Monitoring Network, 14 Sites, United States. MMWR. 2012;61:1–19. [PubMed] [Google Scholar]
- Croen LA, Grether JK, Yoshida CK, Odouli R, Van de Water J. Maternal autoimmune diseases, asthma and allergies, and childhood autism spectrum disorders: a case-control study. Arch Pediatr Adolesc Med. 2005;159:151–157. doi: 10.1001/archpedi.159.2.151. [DOI] [PubMed] [Google Scholar]
- Croonenberghs J, Wauters A, Devreese K, Verkerk R, Scharpe S, Bosmans E, et al. Increased serum albumin, gamma globulin, immunoglobulin IgG, and IgG2 and IgG4 in autism. Psychol Med. 2002;32:1457–1463. doi: 10.1017/s0033291702006037. [DOI] [PubMed] [Google Scholar]
- Daniels WW, Warren RP, Odell JD, Maciulis A, Burger RA, Warren WL, Torres AR. Increased frequency of the extended or ancestral haplotype B44-SC30-DR4 in autism. Neuropsychobiology. 1995;32:120–123. doi: 10.1159/000119223. [DOI] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive Human Genome Amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoto MC, Hitlan RT. Sorting out the spinning of autism: heavy metals and the question of incidence. Acta Neurobiol Exp. 2010;70:165–176. doi: 10.55782/ane-2010-1788. [DOI] [PubMed] [Google Scholar]
- Du Z, Sharma SK, Spellman S, Reed EF, Rajalingam R. KIR2DL5 alleles mark certain combination of activating KIR genes. Genes Immun. 2008;9:470–480. doi: 10.1038/gene.2008.39. [DOI] [PubMed] [Google Scholar]
- Enstrom AM, Lit L, Onore CE, Gregg JP, Hansen RL, Pessah IN, et al. Altered gene expression and function of peripheral blood natural killer cells in children with autism. Brain Behav Immun. 2009a;23:24–33. doi: 10.1016/j.bbi.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enstrom A, Krakowiak P, Onore C, Pessah IN, Hertz-Picciotto I, Hansen RL, et al. Increased IgG4 levels in children with autism disorder. Brain Behav Immun. 2009b;23:389–395. doi: 10.1016/j.bbi.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlano RI, Anthony A, Day R, Brown A, McGarvey L, Thomson MA, et al. Colonic CD8 and gamma delta T-cell infiltration with epithelial damage in children with autism. J Pediatr. 2001;138:366–372. doi: 10.1067/mpd.2001.111323. [DOI] [PubMed] [Google Scholar]
- Hansasuta P, Dong T, Thananchai H, Weekes M, Wilberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- Hertz-Picciotto I. Commentary: Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38:1239–1241. doi: 10.1093/ije/dyp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer L, Ashwood P, Schauer J, Goines P, Krakowiak P, Hertz-Picciotto I, et al. Reduced levels of immunoglobulin in children with autism correlates with behavioral symptoms. Autism Res. 2008;1:275–283. doi: 10.1002/aur.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach JA, Meenagh A, Sleator C, Alaez C, Bengoche M, Canossi A, et al. Report from the killer immunoglobulin-like receptor (KIR) anthropology component of the 15th International Histocompatibility Workshop: worldwide variation in the KIR loci and further evidence for the co-evolution of KIR and HLA. Tissue Antigens. 2010;76:9–17. doi: 10.1111/j.1399-0039.2010.01459.x. [DOI] [PubMed] [Google Scholar]
- Holm SJ, Sakuraba K, Mallbris L, Wolk K, Stahle M, Sanchez FO. Distinct HLA-C/KIR genotype profile associates with guttate psoriasis. J Invest Dermatol. 2005;125:721–730. doi: 10.1111/j.0022-202X.2005.23879.x. [DOI] [PubMed] [Google Scholar]
- King M, Bearman P. Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38:1224–1234. doi: 10.1093/ije/dyp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. The Ying and Yang of HLA and KIR in human disease. Seminars in Immunology. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif AH, Williams WR. Diagnostic trends in autism spectrum disorder in the South Wales valleys. Autism. 2007;11:479–487. doi: 10.1177/1362361307083256. [DOI] [PubMed] [Google Scholar]
- Luszczek W, Manczak M, Cislo M, Nockowski P, Wisniewski A, Jasek M, et al. Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol. 2004;65:758–766. doi: 10.1016/j.humimm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Martin MP, Single RM, Wilson MJ, Trowsdale J, Carrington M. KIR haplotypes defined by segregation analysis in 59 Centre d’Etude Polymorphisme Humain (CEPH) families. Immunogenetics. 2008;60:767–774. doi: 10.1007/s00251-008-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Molloy CA, Morrow AL, Meinzen-Derr J, Schleifer K, Dienger K, Manning-Courtney P, et al. Elevated cytokine levels in children with autism spectrum disorder. J Neuroimmunol. 2006;172:198–205. doi: 10.1016/j.jneuroim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Mostafa GA, Shehab AA. The link of C4B null allele to autism and to a family history of autoimmunity in Egyptian autistic children. J Neuroimmunol. 2010;223:115–119. doi: 10.1016/j.jneuroim.2010.03.025. [DOI] [PubMed] [Google Scholar]
- Odell D, Maciulis A, Cutler A, Warren L, McMahon WM, Coon H, et al. Confirmation of the association of the C4B null allele in autism. Hum Immunol. 2005;66:140–145. doi: 10.1016/j.humimm.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. 2010;5:e15115, 1–14. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenspire A, Yoo W, Menard S, Torres AR. Autism spectrum disorders are associated with an elevated autoantibody response to tissue transglutaminase-2. Autism Res. 2011;4:242–249. doi: 10.1002/aur.194. [DOI] [PubMed] [Google Scholar]
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- Sweeten TL, Bowyer SL, Posey DJ, Halberstadt GM, McDougle CJ. Increased prevalence of familial autoimmunity in probands with pervasive developmental disorders. Pediatrics. 2003;112:e420–e424. doi: 10.1542/peds.112.5.e420. [DOI] [PubMed] [Google Scholar]
- Torres AR, Maciulis A, Stubbs EG, Cutler A, Odell D. The transmission disequilibrium test suggests that HLA-DR4 and DR13 are linked to autism spectrum disorder. Hum Immunol. 2002;63:311–316. doi: 10.1016/s0198-8859(02)00374-9. [DOI] [PubMed] [Google Scholar]
- Torres AR, Sweeten TL, Cutler A, Bedke BJ, Fillmore M, Stubbs EG, et al. The association and linkage of the HLA-A2 class I allele with autism. Hum Immunol. 2006;67:346–351. doi: 10.1016/j.humimm.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nature Immunology. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RP, Foster A, Magaretten NC. Reduced natural killer cell activity in autism. J Am Acad Child Adolesc Psychiatry. 1987;26:333–335. doi: 10.1097/00004583-198705000-00008. [DOI] [PubMed] [Google Scholar]
- Warren RP, Singh VK, Cole P, Odell JD, Pingree CB, Warren WL, et al. Increased frequency of the null allele at the complement C4b locus in autism. Clin Exp Immunol. 1991;83:438–440. doi: 10.1111/j.1365-2249.1991.tb05657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills S, Cabanlit M, Bennett J, Ashwood P, Amaral DG, Van de Water J. Detection of autoantibodies to neural cells of the cerebellum in the plasma of subjects with autism spectrum disorders. Brain Behav Immun. 2009;23:64–74. doi: 10.1016/j.bbi.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CC, Long EO. A single amino acid in p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- Westover JB, Sweeten TL, Benson M, Bray-Ward P, Torres AR. Immune dysfunction in autism spectrum disorder. In: Valsamma Eapen., editor. Autism - A Neurodevelopmental Journey from Genes to Behaviour. 2011. InTech, http://www.intechopen.com/articles/show/title/immune-dysfunction-in-autism-spectrum-disorder. [Google Scholar]