Abstract
The intracellular bacterial pathogen Listeria monocytogenes secretes a broad-range phospholipase C enzyme called PC-PLC (phosphatidylcholine phospholipase C) whose compartmentalization and enzymatic activity is regulated by a 24-amino-acid propeptide (Cys28–Ser51). During intracytosolic multiplication, bacteria accumulate the proform of PC-PLC at their membrane–cell-wall interface, whereas during cell-to-cell spread vacuolar acidification leads to maturation and rapid translocation of PC-PLC across the cell wall in a manner that is dependent on Mpl, the metalloprotease of Listeria. In the present study, we generated a series of propeptide mutants to determine the minimal requirement to prevent PC-PLC enzymatic activity and to identify residues regulating compartmentalization and maturation. We found that a single residue at position P1 (Ser51) of the cleavage site is sufficient to prevent enzymatic activity, which is consistent with P1′ (Trp52) being located within the active-site pocket. We observed that mutants with deletions at the N-terminus, but not the C-terminus, of the propeptide are translocated across the cell wall more effectively than wild-type PC-PLC at a physiological pH, and that individual amino acid residues within the N-terminus influence Mpl-mediated maturation of PC-PLC at acidic pH. However, deletion of more than 75% of the propeptide was required to completely prevent Mpl-mediated maturation of PC-PLC. These results indicate that the N-terminus of the propeptide regulates PC-PLC compartmentalization and that specific residues within the N-terminus influence the ability of Mpl to mediate PC-PLC maturation, although a six-residue propeptide is sufficient for Mpl to mediate PC-PLC maturation.
Keywords: bacterial virulence factor, Gram-positive bacterium, Listeria monocytogenes, metalloprotease, phosphatidylcholine phospholipase C (PC-PLC), propeptide function
INTRODUCTION
Listeria monocytogenes is the aetiological agent of listeriosis, a foodborne disease primarily affecting humans and ruminants [1,2]. As a foodborne pathogen, L. monocytogenes initially invades through the intestinal epithelium and Peyer's patches, potentially causing a transient febrile gastroenteritis. Bacteria then translocate to the liver and multiply, primarily in hepatocytes, causing a subclinical hepatitis. For most individuals, the infection is abrogated at this stage. If the immune system fails to control bacterial growth, a more invasive infection will develop within 3–4 weeks of infection. At that point, human listeriosis will manifest itself primarily as a meningoencephalitis or it will cause abortion in pregnant women.
During infection, L. monocytogenes is found in professional and non-professional phagocytic cells [3–5]. Although neutrophils and activated macrophages are very efficient at controlling infection, L. monocytogenes is capable of multiplying in non-activated macrophages, as well as in intestinal epithelial cells, hepatocytes and trophoblasts. The intracellular life cycle of L. monocytogenes involves bacterium-mediated lysis of vacuoles, growth in the host cytosol and an actin-based mechanism of motility that enables bacteria to move from cell to cell without exiting the intracellular milieu [6]. Remarkably, L. monocytogenes can grow to a high density in cells with very low cytotoxic effects, indicating that it tightly regulates the activity of its membrane-damaging factors.
Among the factors responsible for lysing vacuoles is a broad-range phospholipase C called PC-PLC (phosphatidylcholine phospholipase C) [7]. PC-PLC is made as a proenzyme with a 24-amino-acid residue propeptide, whose proteolytic cleavage is mediated by Mpl, the metalloprotease of Listeria [8,9]. The propeptide of PC-PLC assumes at least two functions. First, it inhibits enzyme activity as determined by the requirement for propeptide cleavage to generate an active enzyme [10]. Secondly, it influences the compartmentalization of the enzyme. During intracellular infection, the proform of PC-PLC is secreted into the cytosol of the host cell [10]. Nonetheless, the rate of translocation of the proform of PC-PLC across the bacterial cell wall is slower than the rate of secretion across the cytoplasmic membrane resulting in the accumulation of a protein pool at the membrane–cell-wall interface [11,12]. Rapid translocation of bacterium-associated PC-PLC occurs in vacuoles formed upon cell-to-cell spread and is dependent on a decrease in pH. Acidic pH-mediated translocation of PC-PLC across the bacterium cell wall is associated with cleavage of the propeptide in an Mpl-dependent manner. In the absence of Mpl, the pool of bacterium-associated PC-PLC does not respond to a decrease in pH; however, a PC-PLC mutant that is synthesized in absence of its propeptide (plcB Δpro) translocates efficiently across the cell wall in an Mpl- and pH-independent manner [13]. These observations suggested that the propeptide defines the localization of PC-PLC. However, studies with a PC-PLC cleavage-site mutant showed that cleavage of the propeptide is not necessary for rapid translocation of PC-PLC across the cell wall in response to a decrease in pH, although Mpl is required [13]. Therefore Mpl regulates the compartmentalization of PC-PLC by two independent means, both of which are pH-regulated: it mediates (i) the rapid translocation of PC-PLC across the cell wall; and (ii) the maturation of PC-PLC. Both functions of Mpl are dependent on the PC-PLC propeptide.
In the present study, we tested the hypothesis that different parts of the propeptide control PC-PLC activity and localization. To test this hypothesis, we generated a series of propeptide deletion and substitution mutants, and characterized the phenotype of these mutants. Our results indicate that inhibition of PC-PLC activity requires only a single C-terminal propeptide residue, whereas the ability of PC-PLC to remain bacterium-associated is conferred by the N-terminus of the propeptide. In addition, specific amino acid residues located within the N-terminus of the propeptide are required for efficient Mpl-mediated maturation of PC-PLC, but complete inhibition of PC-PLC maturation requires deletion of more than 75% of the propeptide.
EXPERIMENTAL
Bacterial strains and growth conditions
The L. monocytogenes strains used in the present study are listed in Table 1. Strains 10403S, DP-L1935, and DP-L2787 have been described previously [10,14,15]. All of the other L. monocytogenes strains were generated during the present study. L. monocytogenes was routinely cultured in brain-heart infusion broth. Escherichia coli strains carrying pPL2-derived plasmids were cultured in LB (Luria–Bertani) broth supplemented with 25 μg/ml chloramphenicol [16]. L. monocytogenes pPL2 integrants were selected with 10 μg/ml chloramphenicol.
Table 1. L. monocytogenes strains, plasmids and primers used in the present study.
Strains 10403S, DP-L1935 and DP-L2787 have been described previously [10,14,15]. All of the other strains and plasmids were generated for the present study. List of plasmids refer to pPL2 constructs that were made to generate the chromosomal integrants expressing wild-type PC-PLC and PC-PLC propeptide mutants. Primer numbers refer to primers used to generate each construct. For primer sequences, see Supplementary Table S1 at http://www.BiochemJ.org/bj/432/bj4320557add.htm.
| Strain | Genotype and relevant features | Plasmid | Primer |
|---|---|---|---|
| 10403S | Wild-type strain | ||
| DP-L1935 | Internal in-frame deletion of plcB in 10403S background | ||
| DP-L2787 | Internal in-frame deletions of mpl and plcB in 10403S background | ||
| HEL-1031 | DP-L1935 pPL2:PactA–plcB | pERS1018 | 512–515 |
| HEL-1034 | DP-L1935 pPL2:PactA–plcB C28A C29A D30A | pERS1025 | 516, 517 |
| HEL-1037 | DP-L1935 pPL2:PactA–plcB E31A Y32A L33A | PERS1026 | 518, 519 |
| HEL-1040 | DP-L1935 pPL2:PactA–plcB P39A H40A D41A | pERS1027 | 526, 527 |
| HEL-1043 | DP-L1935 pPL2:PactA–plcB ΔC28–P39 | pERS1028 | 522, 523 |
| HEL-1046 | DP-L1935 pPL2:PactA–plcB ΔH40–P47 | pERS1029 | 524, 525 |
| HEL-1056 | DP-L1935 pPL2:PactA–plcB Q34A T35A P36A | pAB1049 | 520, 521 |
| HEL-1058 | DP-L1935 pPL2:PactA–plcB A37Q A38Q | pAB1050 | 528, 529 |
| HEL-1060 | DP-L1935 pPL2:PactA–plcB I42A D43A S44A | pAB1051 | 530, 531 |
| HEL-1062 | DP-L1935 pPL2:PactA–plcB K45A L46A P47A | pAB1052 | 532, 533 |
| HEL-1064 | DP-L1935 pPL2:PactA–plcB H48A K49A | pAB1053 | 534, 535 |
| HEL-1066 | DP-L1935 pPL2:PactA–plcB L50A S51A | pAB1054 | 536, 537 |
| HEL-1068 | DP-L1935 pPL2:PactA–plcB Q34K T35P | pERS1055 | 538, 539 |
| HEL-1070 | DP-L2787 pPL2:PactA–plcB | pERS1018 | 513, 514 |
| HEL-1074 | DP-L2787 pPL2:PactA–plcB ΔC28–P39 | pERS1028 | 522, 523 |
| HEL-1106 | DP-L1935 pPL2:PactA–plcB D30A E31A | pAPB1101 | 555, 556 |
| HEL-1112 | DP-L1935 pPL2:PactA–plcB ΔC28–L33 | pAB1087 | 549, 550 |
| HEL-1114 | DP-L1935 pPL2:PactA–plcB ΔE31–P36 | pAB1089 | 553, 554 |
| HEL-1116 | DP-L1935 pPL2:PactA–plcB ΔQ34–P39 | pAB1088 | 551, 552 |
| HEL-1134 | DP-L1935 pPL2:PactA–plcB ΔC28–T35 | pERS1123 | 570, 571 |
| HEL-1136 | DP-L1935 pPL2:PactA–plcB ΔC28–A37 | pERS1124 | 572, 573 |
| HEL-1138 | DP-L1935 pPL2:PactA–plcB ΔA38–K49 | pERS1125 | 574, 575 |
| HEL-1140 | DP-L1935 pPL2:PactA–plcB ΔC28–D41 | pERS1126 | 576, 577 |
| HEL-1142 | DP-L1935 pPL2:PactA–plcB ΔC28–D43 | pERS1127 | 578, 579 |
| HEL-1144 | DP-L1935 pPL2:PactA–plcB ΔC28–K45 | pERS1128 | 580, 581 |
| HEL-1146 | DP-L1935 pPL2:PactA–plcB ΔC28–S51 | pERS1129 | 582, 583 |
| HEL-1161 | DP-L1935 pPL2:PactA–plcB Q34A T35A | pERS1157 | 597, 598 |
| HEL-1164 | DP-L1935 pPL2:PactA–plcB P36A | pERS1158 | 599, 600 |
| HEL-1166 | DP-L1935 pPL2:PactA–plcB Y32A | pERS1159 | 601, 602 |
| HEL-1167 | DP-L1935 pPL2:PactA–plcB L33A | pERS1160 | 603, 604 |
| HEL-1169 | DP-L2787 pPL2:PactA–plcB ΔC28–L33 | pAB1087 | 549, 550 |
| HEL-1176 | DP-L2787 pPL2:PactA–plcB ΔC28–T35 | pERS1123 | 570, 571 |
| HEL-1177 | DP-L2787 pPL2:PactA–plcB ΔC28–A37 | pERS1124 | 572, 573 |
| HEL-1182 | DP-L2787 pPL2:PactA–plcB ΔC28–D41 | pERS1126 | 576, 577 |
| HEL-1184 | DP-L2787 pPL2:PactA–plcB ΔC28–D43 | pERS1127 | 578, 579 |
| HEL-1185 | DP-L2787 pPL2:PactA–plcB ΔC28–K45 | pERS1128 | 580, 581 |
| HEL-1187 | DP-L2787 pPL2:PactA–plcB ΔC28–S51 | pERS1129 | 582, 583 |
| HEL-1204 | DP-L2787 pPL2:PactA–plcB ΔC28–H48 | pERS1189 | 609, 610 |
| HEL-1206 | DP-L2787 pPL2:PactA–plcB ΔC28–L50 | pERS1190 | 611, 612 |
| HEL-1223 | DP-L1935 pPL2:PactA–plcB E31A | pAB1217 | 595, 596 |
| HEL-1320 | DP-L1935 pPL2:PactA–plcB ΔC28–H48 | pERS1189 | 609, 610 |
| HEL-1321 | DP-L1935 pPL2:PactA–plcB ΔC28–L50 | pERS1190 | 611, 612 |
Construction of PC-PLC propeptide mutants
To facilitate the generation of a large number of PC-PLC propeptide mutants in L. monocytogenes, the actA promoter and the plcB gene (which encodes PC-PLC) were fused by SOEing (site-directed mutagenesis with overlap extension) and cloned into the site-specific shuttle integration vector pPL2 [16,17]. The actA promoter is the natural promoter for expression of the plcB gene, which is part of an operon that comprises actA and plcB [18]. The actA promoter and the plcB gene, including the plcB terminator, were amplified individually by PCR using genomic DNA from strain 10403S and primer pairs Marq 512/514 and Marq 513/515 respectively (Table 1 and Supplementary Table S1 at http://www.BiochemJ.org/bj/432/bj4320557add.htm). The products were purified and used in a second PCR with primer pair Marq 512/515 to generate the PactA-plcB product. The purified PactA–plcB product was digested with PstI and XhoI, and ligated into pPL2, generating pERS1018 (Table 1). Sequence integrity was verified by sequencing and pERS1018 was integrated into the chromosome of L. monocytogenes strains DP-L1935 (10403S ΔplcB) and DP-L2787 (10403S ΔplcB Δmpl), as described previously [19]. Integrants were screened for chloramphenicol resistance and by PCR using primers PL95 and NC16 [16] to amplify the site of pPL2 integration on the chromosome. The resulting strains were named HEL-1031 (DP-L1935 pPL2:PactA–plcB) and HEL-1070 (DP-L2787 pPL2:PactA–plcB) (Table 1).
Propeptide deletion and substitution mutants were generated by SOEing PCR using pERS1018 as the DNA template, and cloned into pPL2 as described above. All plasmid constructs and mutant strains generated are listed in Table 1.
Phospholipase assay and detection of PC-PLC on protein gels
Bacteria were grown overnight at 37°C in LB with 50 mM Mops, pH 7.3, 25 mM glucose 1-phosphate and 0.2% activated charcoal, as described previously [12]. Bacterial supernatants were concentrated by trichloroacetic acid precipitation and the equivalent of 0.5 ml of culture at a D600 normalized to 1 was loaded in each lane of a protein gel. Proteins were resolved by SDS/PAGE (12% gels). The gel was processed as described previously [10]. Briefly, it was rinsed in 25% propan-2-ol, equilibrated in PBS, overlaid with egg-yolk agar and incubated at 37°C. PC-PLC activity was detected by the formation of a zone of opacity in the overlay above the mature form of PC-PLC. After scanning the overlay, the protein gel was recovered and stained with Coomassie Brilliant Blue.
Metabolic labelling and immunoprecipitation assay
These experiments were performed, as described previously [12], using a system in which the effect of pH on bacterium-associated PC-PLC is synchronized by preventing the formation of double membrane vacuoles and by manipulating the cytosolic pH of host cells to mimic either the cytosolic or vacuolar environment that is experienced by intracellular bacteria during the successive steps of infection. Briefly, infected mouse macrophage-like J774 cells or human epithelial HeLa cells were treated with cytochalasin D at approx. 3 h post-infection to prevent cell-to-cell spread. After 1 h, the cells were pulse-labelled for 5 min with [35S]methionine/cysteine in the presence of host protein synthesis and proteasome inhibitors, followed by a chase period of 5 min in the presence of chloramphenicol to block bacterial protein synthesis. A second 5-min chase in a potassium-based buffer, at pH 7.3 or pH 6.5, and supplemented with nigericin, was used to manipulate the intracellular pH of host cells. The host cells were lysed, cleared of bacteria and insoluble products by centrifugation, and PC-PLC was immunoprecipitated from the cleared host-cell lysates using rabbit anti-PC-PLC antibodies. Labelled proteins were resolved by SDS/PAGE and detected by autoradiography. Quantification of band intensities was performed with Image J (http://rsbweb.nih.gov/ij/). Half-areas were utilized for this calculation because the proform and mature forms of PC-PLC were not always completely separated. After background subtraction, the results were normalized for the number of methionine and cysteine residues in each band.
A control experiment was performed to determine the ratio of band intensity to protein concentration. Infected cells were pulselabelled and chased as described above and 2-fold serial dilutions of immunoprecipitated PC-PLC were fractionated by SDS/PAGE. The autoradiograph was analysed with Image J as described above and a regression curve was drawn (Supplementary Figure S1 at http://www.BiochemJ.org/bj/432/bj4320557add.htm). A third-order polynominal equation derived from this regression curve (y=[2.897×10−13]x3−[2.68×10−9]x +[4.176×10−5]x+0.03561) was used normalize to the ratio+ of band intensity to protein concentration for protein bands detected on the autoradiographs.
To evaluate levels of PC-PLC secretion and maturation, three protein bands were analysed: (A) the proform of PC-PLC secreted at pH 7.3, (B) the proform of PC-PLC secreted at pH 6.5; and (C) the mature form of PC-PLC secreted at pH 6.5. To normalize for potential differences in the amount of PC-PLC made by each individual mutant, we used the equations A/[B+C] to determine the efficacy of PC-PLC translocation at physiological pH, and C/[B+C] to determine the efficacy of PC-PLC maturation at pH 6.5.
Statistical analysis
Statistical analysis of the results was performed in consultation with Dr Shamil Sadigov from the Cornell University Statistical Consulting Unit. Data were analysed with StatXact®8 (Cytel Statistical Software & Services), using a non-parametric ANOVA test. Because of the small sample size (2–5 data points per mutant), the permutation test with two-sided exact Monte Carlo P-values was used to determine the degree of significance between the mutant and the reference wild-type strain (HEL-1031).
RESULTS
A single amino acid propeptide is sufficient to inhibit PC-PLC activity, whereas a six-residue propeptide is sufficient for Mpl to mediate the proteolytic maturation of PC-PLC
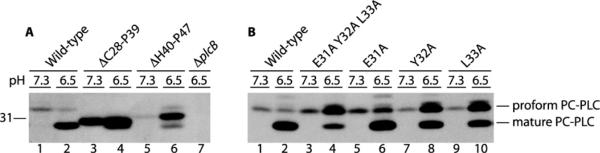
PC-PLC is made as a proenzyme with a 24-amino-acid propeptide that begins at Cys28 and ends at Ser51 (CCDEYLQTPAAPHDIDSKLPHKLS). The activity of PC-PLC is regulated by its propeptide, as loss of the propeptide either by proteolytic cleavage or by gene deletion generates an active enzyme [10,13]. To determine which part of the propeptide is necessary to inhibit PC-PLC activity and to enable Mpl-mediated maturation of PC-PLC, we generated a series of nested propeptide deletions in PC-PLC. All of the mutant proteins were synthesized (Figures 1A and 1C) and secreted in the supernatant of broth-grown bacteria, although the 16-(ΔC28–D43) and 21-(ΔC28–H48) residue deletion mutants were detected in smaller amounts (Figures 1A and 1C, lanes 8 and 10 respectively). The stained protein gel indicated that in broth culture the proform of wild-type PC-PLC predominates over the mature form (Figures 1A and 1C, lane 1), but it was not possible to distinguish the proform and mature form of PC-PLC from the deletion mutants (Figures 1A and 1C, lanes 3–12).
Figure 1. Detection of PC-PLC propeptide deletion mutants on protein gels and of PC-PLC activity by the egg-yolk overlay assay.
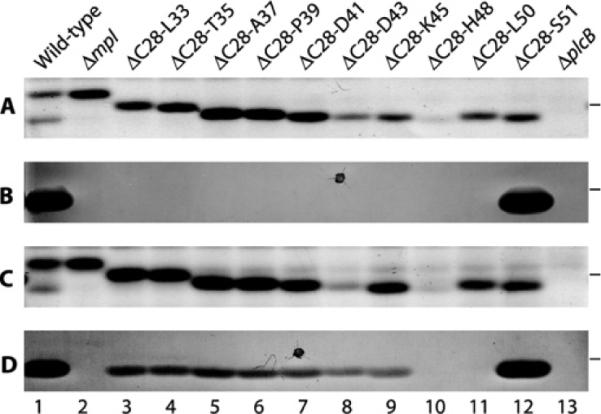
The indicated L. monocytogenes strains were grown in broth and concentrated supernatants were resolved by SDS/PAGE. The gel was overlaid with egg-yolk agar and incubated at 37°C for 2 h. PC-PLC activity was detected by a zone of opacity below the 31 kDa molecular-mass marker (the position of which is indicated at the right-hand side of each panel). The agar was scanned and the gel was recovered for Coomassie Blue staining. (A and C) Coomassie-Blue-stained gel. (B and D) Egg-yolk overlay agar. (A and B) All strains except the wild-type (HEL-1031) and ΔplcB (DP-L1935) in lanes 1 and 13 have a deletion of the mpl gene to assess which part of the propeptide is required to inhibit PC-PLC activity. Lane 1, HEL-1031; lane 2, HEL-1070; lane 3, HEL-1169; lane 4, HEL-1176; lane 5, HEL-1177; lane 6, HEL-1074; lane 7, HEL-1182; lane 8, HEL-1184; lane 9, HEL-1185; lane 10, HEL-1204; lane 11, HEL-1206; lane 12, HEL-1187; and lane 13, DP-L1935. (C and D) All strains except Δmpl (HEL-1070) in lane 2 express mpl to assess the functionality of the propeptide deletion mutants. Lane 1, HEL-1031; lane 2, HEL-1070; lane 3, HEL-1112; lane 4, HEL-1134; lane 5, HEL-1136; lane 6, HEL-1043; lane 7, HEL-1140; lane 8, HEL-1142; lane 9, HEL-1144; lane 10, HEL-1320; lane 11, HEL-1321; lane 12, HEL-1146; and lane 13, DP-L1935. The dark dots in (B) and (D) represent artefacts from the Petri dish.
The ability of the deletion mutants to become active PC-PLCs was assessed by the egg-yolk overlay assay, in which a protein gel loaded with supernatant from bacterial broth cultures is overlaid with agar containing egg yolk, which serves as a substrate for the enzyme. The positive control for this assay was an L. monocytogenes strain that synthesizes wild-type Mpl and PC-PLC (HEL-1031). As expected, a zone of opacity was detected below the 31 kDa protein standard, indicating PC-PLC activity (Figure 1B, lane 12). A second positive control consisted of a L. monocytogenes strain expressing a propeptide deletion form of PC-PLC (plcBΔpro) in an mpl-deletion background (HEL-1187). A zone of opacity indicating PC-PLC activity was detected in the lane loaded with supernatant from this second control strain (Figure 1B, lane 12). This is consistent with previous results indicating that the propeptide serves to inhibit PC-PLC activity and that Mpl does not contribute to the activity of mature PC-PLC [13]. The negative control was an L. monocytogenes strain that synthesizes wild-type PC-PLC in a Δmpl background (HEL-1070). No PC-PLC activity was detected with this strain, confirming that Mpl is required for the proteolytic activation of PC-PLC in broth culture (Figures 1B and 1D, lane 2). Lastly, we tested a ΔplcB deletion mutant of L. monocytogenes (DP-L1935) to confirm that the zone of opacity was a result of PC-PLC activity (Figures 1B and 1D, lane 13).
To determine the minimal propeptide requirement for inhibition of PC-PLC activity, we assessed the ability of the nested propeptide deletion mutants to generate active PC-PLC when expressed in an Δmpl background strain. No PC-PLC activity was detected from any of the partial propeptide deletion strains in the absence of Mpl (Figure 1B, lanes 3–11). To identify the minimum propeptide requirement for Mpl-mediated processing of PC-PLC, we assessed the ability of the nested propeptide deletion mutants to generate active PC-PLC when expressed in a wild-type Mpl background strain. The egg-yolk activity assay indicated that active PC-PLC was generated from every one of the propeptide deletion strains in the presence of Mpl, except for the two mutants with the largest deletions (HEL-1320, ΔC28-H48 and HEL-1321, ΔC28–L50) (Figure 1D). Although the lack of activity with the ΔC28–H48 deletion mutant could possibly be attributed to a decrease in protein concentration, this is not the case for the ΔC28–L50 deletion mutant (Figures 1C and 1D, lanes 10 and 11). These results are consistent with Mpl being required for efficient proteolytic maturation of PC-PLC. We concluded that a single amino acid propeptide is sufficient to inhibit PC-PLC activity, and that the presence of six residues in the propeptide is sufficient for Mpl to mediate the maturation of PC-PLC.
The N-terminus of the PC-PLC propeptide regulates the compartmentalization of PC-PLC
During intracellular infection, the proform of PC-PLC accumulates at the membrane–cell-wall interface [11,12]. Upon cell-to-cell spread, bacteria become entrapped in secondary vacuoles that acidify, leading to the proteolytic maturation of PC-PLC and rapid translocation of the active enzyme across the bacterial cell wall and into the vacuole; Figure 2 is a schematic diagram representing this process during infection. Although the regulation of PC-PLC activity during intracellular infection was studied using mouse macrophage-like J774 cells, similar results were obtained using human epithelial HeLa cells (Supplementary Figure S2 at http://www.BiochemJ.org/bj/432/bj4320557add.htm). When expressed in the absence of its propeptide, PC-PLC diffusion across the bacterial cell wall occurs independently of pH [13], affecting bacterial virulence [20]. To assess which part of the propeptide contributes to PC-PLC compartmentalization, we tested a series of PC-PLC propeptide mutants for their efficacy to translocate across the cell wall at physiological pH. For these experiments, pulse-labelled infected cells were chased in buffer at pH 7.3 or 6.5, which was supplemented with nigericin to enable equilibration of pH across cell membranes. PC-PLC was immunoprecipitated from host-cell lysates, which excludes bacterium-associated PC-PLC. The immunoprecipitates were resolved by SDS/PAGE and detected by autoradiography.
Figure 2. Site of PC-PLC maturation, translocation and activity.
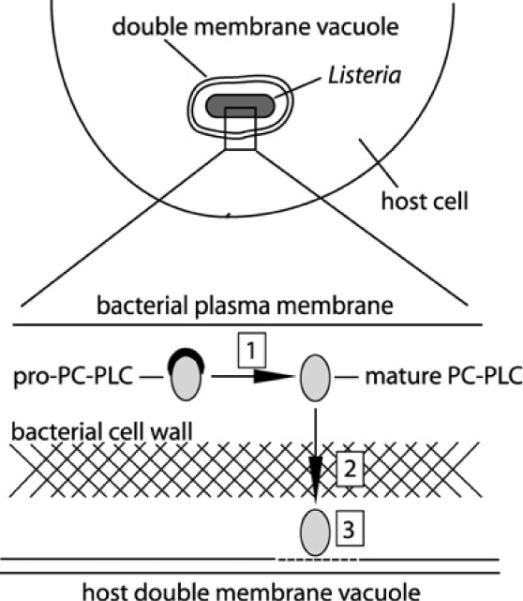
Overview of the location of PC-PLC maturation, translocation and activity with respect to host cell and bacterial membranes, and the bacterial cell wall. Boxed numbers represent the sites of: (1) PC-PLC maturation; (2) PC-PLC translocation across the bacterial cell wall; and (3) PC-PLC activity.
We first tested an N-terminal deletion mutant (HEL-1043, ΔC28–P39) and a C-terminal deletion mutant (HEL-1046, ΔH40–P47), and observed that deletion of the N-terminus of the propeptide, but not of the C-terminus, facilitates translocation of PC-PLC across the cell wall (Figure 3A, compare lanes 3 and 5 with lane 1). Next, we tested a series of deletion and substitution mutants and measured band intensities using Image J. To control for possible changes in secretion intrinsic to each mutant protein within each experiment, we divided the amount of protein translocated at pH 7.3 by the amount translocated at pH 6.5. We also calculated the ratio of band intensity to protein concentration in a control experiment and corrected the results accordingly. The efficacy of protein translocation is reported in reference to wild-type PC-PLC (Figure 4). PC-PLC translocation was increased by 5- and 2.5-fold at the physiological pH for the N-terminal deletion of 12 amino acid residues (HEL-1043, ΔC28–P39) and the entire propeptide deletion mutant (HEL-1146, ΔC28–S51) respectively. These differences were statistically significant when compared with wild-type PC-PLC. Two additional mutants showed an increase of approx. 2-fold in protein translocation at physiological pH: a 10-amino-acid N-terminal deletion (HEL-1136, ΔC28–A37) and a six-amino-acid internal deletion (HEL-1114, ΔE31–P36). However, this slight increase in translocation was not statistically significant. No other propeptide mutants behaved differently to wild-type PC-PLC in regard to translocation across the bacterial cell wall at physiological pH. Overall, these results indicate that the N-terminus of the propeptide impedes the translocation of PC-PLC across the bacterial cell wall.
Figure 3. Immunoprecipitation of PC-PLC secreted in cells infected with L. monocytogenes.
J774 cells were infected with the indicated strains of L. monocytogenes expressing either wild-type PC-PLC or a PC-PLC propeptide mutant. Pulse-labelled infected cells were perfused with nigericin-supplemented buffer at pH 7.3 or pH 6.5. Radiolabelled PC-PLC was immunoprecipitated from host cell lysates. The secreted protein was detected by autoradiography to determine the efficacy of translocation of PC-PLC at pH 7.3 and of maturation at pH 6.5. The position of a molecular-mass marker in kDa is indicated on the far left-hand side. Positions of the proform and mature form of wild-type PC-PLC are indicated on the far right-hand side. (A) Lanes 1 and 2, HEL-1031; lanes 3 and 4, HEL-1043; lanes 5 and 6, HEL-1046; and lane A 7, DP-L1935. (B) Lanes 1 and 2, HEL-1031; lanes 3 and 4, HEL-1037; lanes 5 and 6, HEL-1223; lanes 7 and 8, HEL-1166; and lanes 9 and 10, HEL-1167.
Figure 4. Quantification of translocation and maturation efficacy of PC-PLC propeptide mutants.
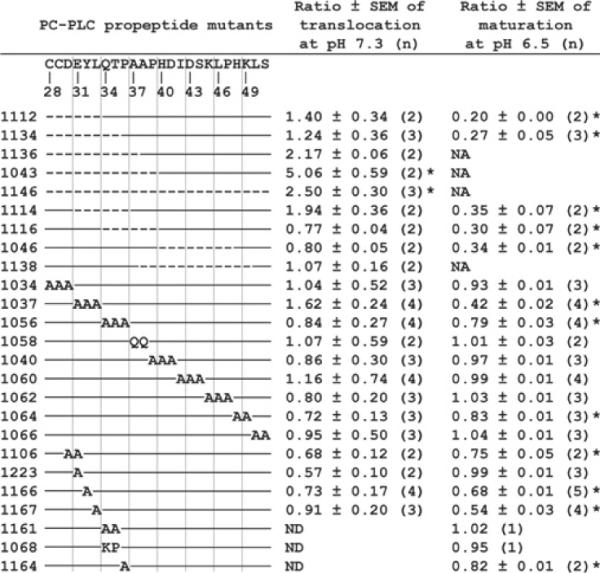
The strain numbers are indicated on the left. The propeptide sequence with the corresponding amino acid sequence numbers are listed above the schematic diagram representing each mutant. The broken line indicates that the corresponding amino acid residue was deleted and solid lines indicate propeptide sequences present within a mutant. Letters indicate the amino acids that were used for substitution mutants. The ratios were calculated in reference to the behaviour of wild-type PC-PLC expressed in HEL-1031 (the ratio for wild-type PC-PLC = 1). Wild-type PC-PLC was translocated with an efficacy of 16.4 ± 3.0% (means ± S.E.M. for 14 experiments). The efficacy of maturation of wild-type PC-PLC was 87.6 ± 0.8% (means ± S.E.M. for 15 experiments). (n) represents the number of immunoprecipitation experiments performed with each strain. NA, not applicable (the proform and mature form of PC-PLC could not be resolved by SDS/PAGE); ND, not determined. Statistical analysis of data was performed as described in the Experimental section. *P < 0.01.
Individual amino acid residues within the N-terminus of the PC-PLC propeptide regulate Mpl-mediated maturation of PC-PLC
Rapid translocation of PC-PLC across the bacterial cell wall is dependent on Mpl and a decrease in pH, and is associated with processing of the propeptide [11,12]. However, a PC-PLC cleavage-site mutant behaves similarly to wild-type PC-PLC, as its rapid translocation across the cell wall is still dependent on Mpl and a decrease in pH [13]. In contrast, the PC-PLC propeptide deletion mutant translocates across the bacterial cell wall independently of Mpl and pH. Therefore we hypothesized that Mpl interacts with the propeptide of PC-PLC when both proteins are at the membrane-cell-wall interface. To identify propeptide amino acids that contribute to the Mpl-mediated maturation of PC-PLC, we measured the efficacy of propeptide cleavage as a function of pH for all the propeptide mutants generated (except for those with large deletions whose pro and mature forms could not be resolved by SDS/PAGE). The experiments were performed as described above for measuring the rate of proform diffusion at physiological pH, except that we compared the amount of secreted mature form to the total amount of secreted protein (pro and mature) for cells chased at pH 6.5.
We observed that the efficacy of PC-PLC processing was reduced to 20% and 27% of wild-type PC-PLC when the first six and eight N-terminal amino acids were deleted respectively (HEL–1112, ΔC28–L33 and HEL–1134, ΔC28–T35) (Figure 4). Successive internal propeptide deletions of Glu31–Pro36 (HEL–1114, ΔE31–P36), of Gln34–Pro39 (HEL–1116, ΔQ34–P39) and of His40–Pro47 (HEL–1046, ΔH40–P47) also decreased the efficacy of PC-PLC processing to 30–35% of wild-type. Replacing the first three N-terminal amino acids (HEL–1034, C28A/C29A/D30A) had no effect on PC-PLC maturation, whereas D30A/E31A (HEL–1106), E31A/Y32A/L33A (HEL–1037), or Q34A/T35A/P36A (HEL–1056) alanine residue replacements decreased the efficacy of PC-PLC processing to 75%, 42% and 79% of wild-type respectively (Figure 4 and Figure 3B, compare lanes 4 and 2). Mutations E31A (HEL–1223), Q34A/T35A (HEL–1161) or Q34K/T35P (HEL–1068) had no effect on PC-PLC maturation, whereas mutations Y32A (HEL–1166), L33A (HEL–1167) or P36A (HEL–1164) decreased the efficacy of PC-PLC processing to 68%, 54% and 82% of wild-type respectively (Figure 4 and Figure 3B, compare lanes 2, 6, 8 and 10). The Q34K/T35P mutation was made because this substitution is frequently found in PC-PLC propeptides from other strains of L. monocytogenes. Overall these results indicate that three specific amino acid residues located at the N-terminus of the propeptide, Tyr32, Leu33 and Pro36, influence Mpl-mediated maturation of PC-PLC. In addition, the location of these three amino acids within the 24-residue propeptide is important, as internal in-frame deletions that do not eliminate these specific residues also cause a major defect in maturation of PC-PLC. Moreover, a negatively charged residue within the N-terminal of the propeptide is important for Mpl-mediated maturation of PC-PLC, as the double D30A/E31A mutant is deficient in maturation, whereas the triple C28A/C29A/D30A and the single E31A mutants are not deficient for maturation. Lastly, there was one C-terminal substitution mutant whose maturation efficacy was 83% of wild-type PC-PLC (HEL–1064, H48A/K49A), indicating that the third and fourth residues upstream of the cleavage site are important for Mpl-mediated maturation of PC-PLC (Figure 4).
DISCUSSION
The propeptide of PC-PLC is 24 amino acids long and serves two functions: it inhibits enzyme activity and influences the compartmentalization of PC-PLC. In the present study, we tested the hypothesis that different parts of the propeptide control PC-PLC activity and compartmentalization. Our results indicate that inhibition of PC-PLC activity requires the presence of only a single propeptide residue. The ability of PC-PLC to remain bacterium-associated is conferred by the N-terminus of the propeptide. Furthermore, four individual amino acid residues located between the third and ninth positions in the N-terminus of the propeptide influence the efficacy of Mpl-mediated maturation of PC-PLC, although a six-residue propeptide is sufficient for Mpl to mediate the proteolytic activation of PC-PLC.
The propeptide of PC-PLC serves to inhibit its enzymatic activity, as deletion of the entire propeptide generates an enzyme that is constitutively active [13]. However, deletion of 23 out of 24 amino acids did not compromise the ability of the propeptide to inhibit enzyme activity as shown in Figure 1. The crystal structure of the Bacillus cereus orthologue to PC-PLC (PLCBC) (PDB code 1AH7) indicates that the first N-terminal residue of the catalytic domain (P1′) is one of the nine zinc-co-ordinating residues, and that the following three residues (P2′–P4′) are located within the active site [21,22]. PC-PLC and PLCBC have identical P1′–P3′ residues and a similar P4′ residue [23]. PC-PLC and PLCBC are approx. 40% identical and the homology model of PC-PLC suggests that its structure is nearly identical with that of PLCBC (H. Marquis, unpublished work). Therefore it is reasonable that a single propeptide amino acid residue is sufficient to inhibit PC-PLC activity either by steric hindrance within the active-site pocket or by preventing native folding of the catalytic domain.
During intracellular infection, bacteria maintain a pool of PC-PLC at the membrane-cell-wall interface [12]. Upon cell-to-cell spread, bacteria sense a decrease in vacuolar pH and rapidly release this pool of bacterium-associated PC-PLC. The ability of PC-PLC to remain bacterium-associated is mediated by its propeptide. Our results indicate that deletion of the N-terminus of the propeptide, but not of the C-terminus, leads to an increase in protein translocation across the bacterial cell wall at physiological pH (Figures 3 and 4). Deleting the entire propeptide did not increase the efficacy of translocation above that of the N-terminal 12-amino-acid deletion (ΔC28–P39). It is not known how PC-PLC manages to remain bacterium-associated at physiological pH, because PC-PLC does not have a transmembrane domain or cell-wall-anchoring motif [24]. One possible hypothesis is that the propeptide contributes to the formation of a complex that immobilizes PC-PLC at the membrane-cell-wall interface. Alternatively or concomitantly, the propeptide may interfere with protein folding, slowing down its rate of translocation as the rate of protein translocation across the cell wall is, for some proteins, directly proportional to the rate of folding [25,26]. Either mechanism could be mediated by the N-terminus of the propeptide.
The most unexpected result from the present study was the importance of individual amino acid residues within the N-terminus of the propeptide in facilitating Mpl-mediated proteolytic maturation of PC-PLC. Deletion and substitution mutants encompassing Tyr32, Leu33 and Pro36 all showed a statistically significant PC-PLC maturation defect (Figures 3 and 4). In addition, the presence of a negatively charged residue, either Asp30 or Glu31, is important for Mpl-mediated maturation of PC-PLC, as the D30A/E31A substitution mutant was defective, whereas the C28A/C29A/D30A or the E31A substitution mutants were not. Considering that the residues influencing PC-PLC maturation are located between 22 and 16 residues upstream of the propeptide cleavage site, we propose that these residues interact with Mpl, leading to processing of the propeptide at Ser51. An additional deletion mutant (ΔH40–P47) that did not include Asp30, Glu31, Tyr32, Leu33 or Pro36 showed a major maturation defect. However, triple replacement mutants involving residues Pro39–Pro47 behaved like wild-type PC-PLC. It is possible that the defect associated with the ΔH40–P47 mutant indicates that the spatial location of the residues influencing PC-PLC maturation is imperative to the ability of PC-PLC to interact with Mpl.
Amino acid residues in proximity to the propeptide cleavage site are important for Mpl-mediated proteolytic maturation of PC-PLC. We had identified previously the cleavage-site mutant S51D/S53N whose maturation at pH 6.5 was greatly reduced [13]. However, the requirement for a serine residue at position 51 is not essential because a S51G mutant that we generated in that study behaved like wild-type PC-PLC. In the present study, the L50A/S51A mutant did not show a maturation defect, whereas the H48A/K49A mutant showed a small but statistically significant decrease in PC-PLC maturation. This result indicates that residues located at the P4 and/or P3 position in relation to the propeptide cleavage site also influence the ability of Mpl to mediate PC-PLC maturation. Interestingly, deletion of 18 residues (ΔC28–K45) did not prevent Mpl-mediated proteolytic activation of PC-PLC, but deletion of 21 residues (ΔC28–H48) completely abolished Mpl-mediated activation of PC-PLC. Thus a six-residue propeptide is sufficient for Mpl to mediate the maturation of PC-PLC, but a three-residue propeptide is not.
The regulation of PC-PLC activity during intracellular infection has been characterized using mouse macrophage-like J774 cells [11–13]. However, during infection, L. monocytogenes appears to multiply primarily in non-professional phagocytic cells, such as epithelial cells, hepatocytes and trophoblasts [3–5]. To address whether pH regulates PC-PLC activity in non-professional phagocytic cells, we used human epithelial HeLa cells as the infection host. In these cells, we observed that pH regulates PC-PLC compartmentalization and maturation in a manner similar to that observed in macrophages (Supplementary Figure S2), suggesting that the activity of PC-PLC is pH-regulated in both professional and non-professional cell types.
The results from the present study are compatible with the following model. The first 12 residues of the PC-PLC propeptide retard translocation of the protein across the bacterial cell wall by interacting with the cell wall or a protein complex, and/or by possibly interfering with native folding of the catalytic domain. Upon a decrease in pH, Mpl interacts with the N-terminus of the propeptide destabilizing the interaction of PC-PLC with the cell wall or protein complex. PC-PLC residues Tyr32, Leu33 and Pro36, which are located at the fifth, sixth and ninth positions within the N-terminus of the propeptide, along with a negatively charged residue located at the third or fourth position of the propeptide, stabilize the Mpl-PC-PLC interaction and promote native folding of the PC-PLC catalytic domain leading to rapid translocation of PC-PLC across the cell wall. It is possible, but not necessary, that Mpl concomitantly mediates the proteolytic maturation of PC-PLC. Future studies will aim to test this model.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shamil Sadigov from the Cornell University Statistical Consulting Unit for helping with the statistical analysis. We are grateful to Bryant Blank, Brian Forster and Gabriela Wagner for critical reading of the manuscript prior to submission.
FUNDING This work was supported by the Public Health Service of the National Institute of Allergy and Infectious Disease [grant number AI52154 (to H.M.)]
Abbreviations used
- LB
Luria–Bertani
- Mpl
metalloprotease
- PC-PLC
phosphatidylcholine phospholipase C
- PLCBC
Bacillus cereus PC-PLC
- SOEing
site-directed mutagenesis with overlap extension
Footnotes
AUTHOR CONTRIBUTION Emily Slepkov and Hélène Marquis designed the research; Emily Slepkov and Alan Pavinski Bitar performed the research; all authors analysed the results; Emily Slepkov and Hélène Marquis wrote the paper.
REFERENCES
- 1.Schlech WF., III Foodborne listeriosis. Clin. Infect. Dis. 2000;31:770–775. doi: 10.1086/314008. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wing EJ, Gregory SH. Listeria monocytogenes: clinical and experimental update. J. Infect. Dis. 2002;185:S18–S24. doi: 10.1086/338465. [DOI] [PubMed] [Google Scholar]
- 4.Drevets DA, Bronze MS. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 2008;53:151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- 5.Robbins JR, Skrzypczynska KM, Zeldovich VB, Kapidzic M, Bakardjiev AI. Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of Listeria monocytogenes. PLoS Pathog. 2010;6:e1000732. doi: 10.1371/journal.ppat.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfine H, Marquis H. Escape of Listeria monocytogenes from a vacuole. In: Goldfine H, Shen H, editors. Listeria monocytogenes: Pathogenesis and Host Response. Springer; New York: 2007. pp. 177–195. [Google Scholar]
- 8.Niebuhr K, Chakraborty T, Köllner P, Wehland J. Production of monoclonal antibodies to the phosphatidylcholine-specific phospholipase C of Listeria monocytogenes, a virulence factor for this species. Med. Microbiol. Lett. 1993;2:9–16. [Google Scholar]
- 9.Raveneau J, Geoffroy C, Beretti J-L, Gaillard J-L, Alouf JE, Berche P. Reduced virulence of a Listeria monocytogenes phospholipase-deficient mutant obtained by transposon insertion into the zinc metalloprotease gene. Infect. Immun. 1992;60:916–921. doi: 10.1128/iai.60.3.916-921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquis H, Goldfine H, Portnoy DA. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J. Cell Biol. 1997;137:1381–1392. doi: 10.1083/jcb.137.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquis H, Hager EJ. pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 2000;35:289–298. doi: 10.1046/j.1365-2958.2000.01708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder A, Marquis H. Restricted translocation across the cell wall regulates secretion of the broad-range phospholipase C of Listeria monocytogenes. J. Bacteriol. 2003;185:5953–5958. doi: 10.1128/JB.185.20.5953-5958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeung PS, Zagorski N, Marquis H. The metalloprotease of Listeria monocytogenes controls cell wall translocation of the broad-range phospholipase C. J. Bacteriol. 2005;187:2601–2608. doi: 10.1128/JB.187.8.2601-2608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop DK, Hinrichs DJ. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 1987;139:2005–2009. [PubMed] [Google Scholar]
- 15.Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauer P, Chow MYN, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Shetron-Rama LM, Marquis H, Bouwer HGA, Freitag NE. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 2002;70:1087–1096. doi: 10.1128/IAI.70.3.1087-1096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neil HS, Marquis H. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 2006;74:6675–6681. doi: 10.1128/IAI.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung PS, Na Y, Kreuder AJ, Marquis H. Compartmentalization of the broad-range phospholipase C activity to the spreading vacuole is critical for Listeria monocytogenes virulence. Infect. Immun. 2007;75:44–51. doi: 10.1128/IAI.01001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen S, Hough E, Svensson LA, Wong Y-L, Martin SF. Crystal structure of phospholipase C from Bacillus cereus complexed with a substrate analog. J. Mol. Biol. 1993;234:179–187. doi: 10.1006/jmbi.1993.1572. [DOI] [PubMed] [Google Scholar]
- 22.Hough E, Hansen LK, Birknes B, Jynge K, Hansen S, Hordvik A, Little C, Dodson E, Derewenda Z. High-resolution (1.5Å) crystal structure of phospholipase C from Bacillus cereus. Nature. 1989;338:357–360. doi: 10.1038/338357a0. [DOI] [PubMed] [Google Scholar]
- 23.Zuckert WR, Marquis H, Goldfine H. Modulation of enzymatic activity and biological function of Listeria monocytogenes broad-range phospholipase C by amino acid substitutions and by replacement with the Bacillus cereus ortholog. Infect. Immun. 1998;66:4823–4831. doi: 10.1128/iai.66.10.4823-4831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002;10:238–245. doi: 10.1016/s0966-842x(02)02342-9. [DOI] [PubMed] [Google Scholar]
- 25.Leloup L, Haddaoui el A, Chambert R, Petit-Glatron MF. Characterization of the rate-limiting step of the secretion of Bacillus subtilis α-amylase overproduced during the exponential phase of growth. Microbiol. 1997;143:3295–3303. doi: 10.1099/00221287-143-10-3295. [DOI] [PubMed] [Google Scholar]
- 26.Stephenson K, Carter NM, Harwood CR, Petit-Glatron MF, Chambert R. The influence of protein folding on late stages of the secretion of α-amylases from Bacillus subtilis. FEBS Lett. 1998;430:385–389. doi: 10.1016/s0014-5793(98)00698-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.