Abstract
Molecular mechanics studies have been carried out on “B-DNA-like” structures of [d(C-G-C-G-A-A-T-T-C-G-C-G)]2 and [d(A)]12·[d(T)]12. Each of the backbone torsion angles (ψ, φ, ω, ω′, φ′) has been “forced” to alternative values from the normal B-DNA values (g+, t, g-, g-, t conformations). Compensating torsion angle changes preserve most of the base stacking energy in the double helix. In a second part of the study, one purine N3-pyrimidine N1 distance at a time has been forced to a value of 6 Å in an attempt to simulate the base opening motions required to rationalize proton exchange data for DNA. When the 6-Å constraint is removed, many of the structures revert to the normal Watson-Crick hydrogen-bonded structure, but a number are trapped in structures ≈5 kcal/mol higher in energy than the starting B-DNA structure. The relative energy of these structures, some of which involve a non-Watson-Crick thymine C2(carbonyl)[unk]adenine 6NH2 hydrogen bond, are qualitatively consistent with the ΔH for a “base pair-open state” suggested by Mandal et al. of 4-6 kcal/mol [Mandal, C., Kallenbach, N. R. & Englander, S. W. (1979) J. Mol. Biol. 135, 391-411]. The picture of DNA flexibility emerging from this study depicts the backbone as undergoing rapid motion between local torsional minima on a nanosecond time scale. Backbone motion is mainly localized within a dinucleoside segment and generally not conformationally coupled along the chain or across the base pairs. Base motions are much smaller in magnitude than backbone motions. Base sliding allows imino N—H exchange, but it is localized, and only a small fraction of the N—H groups is exposed at any one time. Stacking and hydrogen bonding cause a rigid core of bases in the center of the molecule accounting for the hydrodynamic properties of DNA.
Keywords: computer modeling, base stacking, motions
Full text
PDF
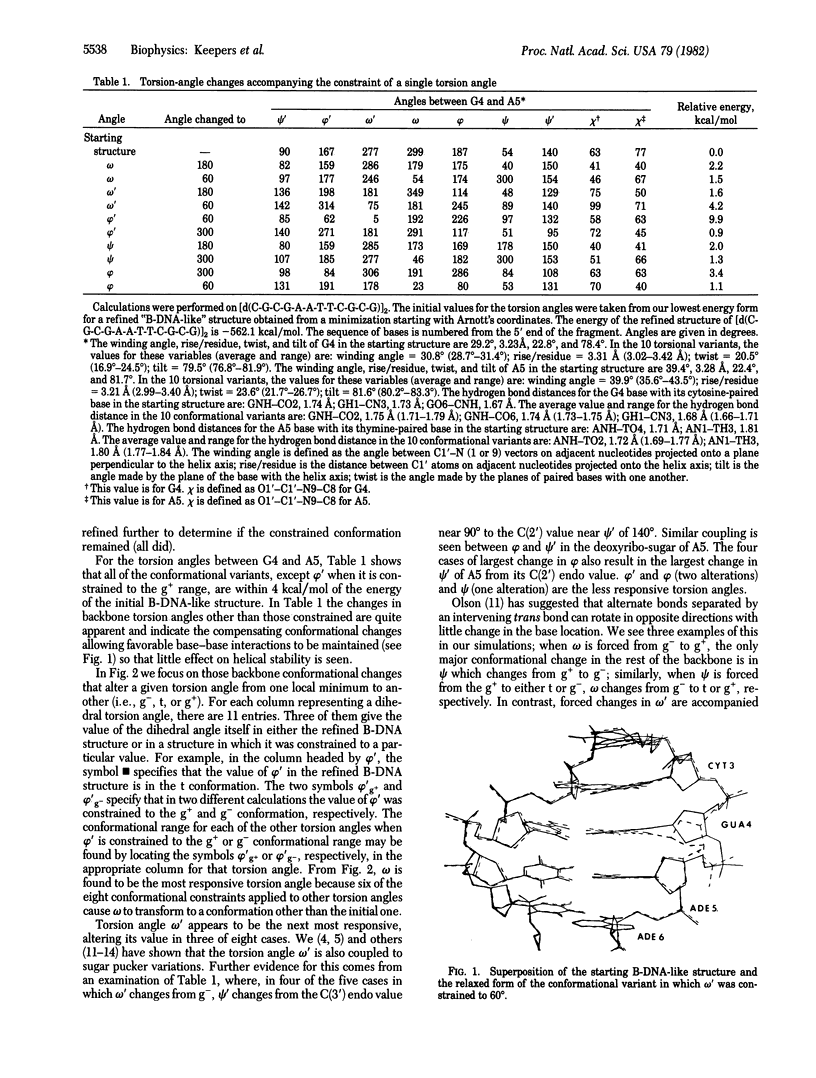
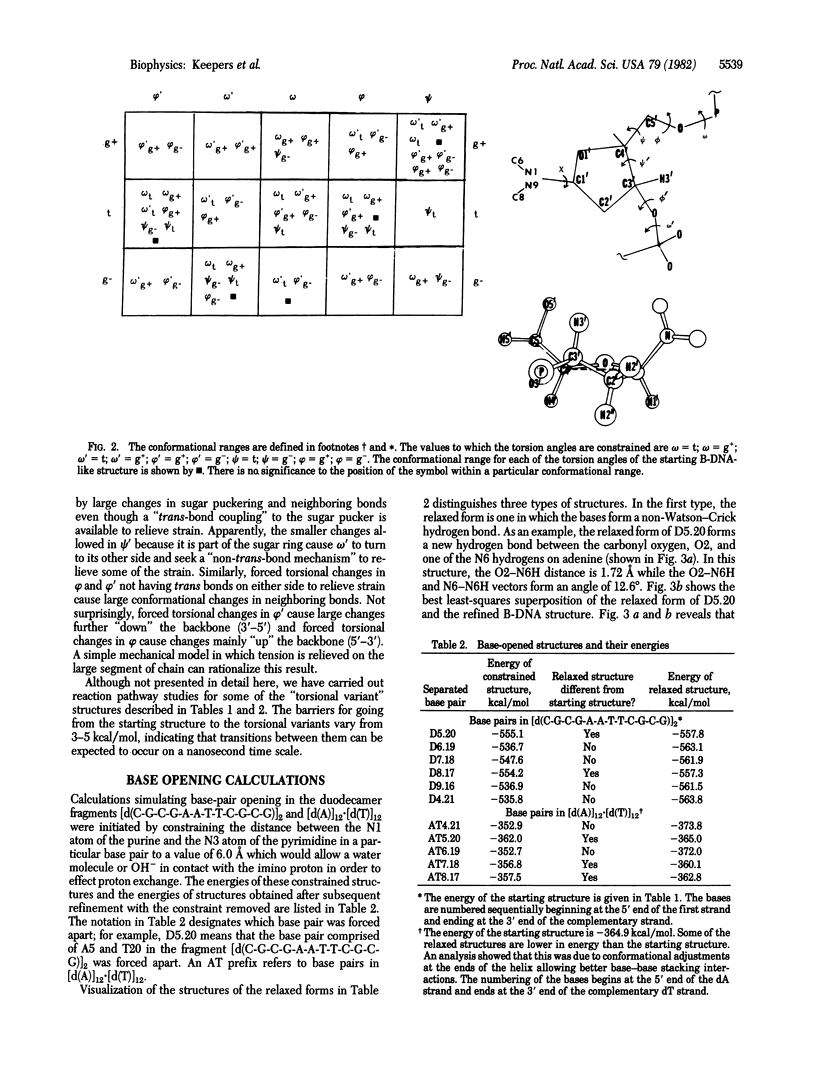


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolton P. H., James T. L. Conformational mobility of deoxyribonucleic acid, transfer ribonucleic acid, and poly(adenylic acid) as monitored by carbon-13 nuclear magnetic resonance relaxation. Biochemistry. 1980 Apr 1;19(7):1388–1392. doi: 10.1021/bi00548a019. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R., Heeger A. J., Krumhansl J. A., Litwin S. Nature of the open state in long polynucleotide double helices: possibility of soliton excitations. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7222–7226. doi: 10.1073/pnas.77.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman P. A., Weiner P. K., Dearing A. Studies of nucleotide conformations and interactions. The relative stabilities of double-helical B-DNA sequence isomers. Biopolymers. 1981 Dec;20(12):2583–2621. doi: 10.1002/bip.1981.360201208. [DOI] [PubMed] [Google Scholar]
- Mandal C., Kallenbach N. R., Englander S. W. Base-pair opening and closing reactions in the double helix. A stopped-flow hydrogen exchange study in poly(rA).poly(rU). J Mol Biol. 1979 Dec 5;135(2):391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- Olson W. K. The flexible DNA double helix. II. Superhelix formation. Biopolymers. 1979 May;18(5):1235–1260. doi: 10.1002/bip.1979.360180515. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Broka C., Rice J. A., Itakura K., Breslauer K. J. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982 Feb 2;21(3):428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Woodbury C. P., Inman R. B. Characterization of rodlike RNA fragments. Biopolymers. 1975 Feb;14(2):393–408. doi: 10.1002/bip.1975.360140212. [DOI] [PubMed] [Google Scholar]
- Sundaralingam M., Westhof E. Correlated motions in DNA. Biophys J. 1980 Oct;32(1):250–252. doi: 10.1016/S0006-3495(80)84952-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- de Santis P., Morosetti S., Palleschi A., Savino M. Conformational analysis of double-stranded B-type DNA structures. Biopolymers. 1981 Aug;20(8):1727–1739. doi: 10.1002/bip.1981.360200812. [DOI] [PubMed] [Google Scholar]


