Abstract
Background
Yersinia enterocolitica is a gastrointestinal foodborne pathogen found worldwide and which especially affects infants and young children. While different bioserotypes have been associated with varying pathogenicity, research on Y. enterocolitica is mainly conducted on the highly virulent mouse-lethal strains of biotype 1B and serotype O:8. We demonstrate here that two Y. enterocolitica bioserotype 1B/O:8 strains, 8081 and WA-314, display different virulence and fitness properties in a mouse model. In vivo co-infection experiments revealed that strain WA-314 overcomes strain 8081 in the colonization of spleen and liver. To trace the reasons of this incongruity, we present here the first high-quality sequence of the whole genome of strain WA-314 and compare it to the published genome of strain 8081.
Results
Regions previously accepted as unique to strain 8081, like the YAPI and YGI-3 genomic islands, are absent from strain WA-314, confirming their strain-specificity. On the other hand, some fitness- and bacterial competition-associated features, such as a putative colicin cluster and a xenobiotic-acyltransferase-encoding gene, are unique to strain WA-314. Additional acquisitions of strain WA-314 are seven prophage-like regions. One of these prophages, the 28-kb P4-like prophage YWA-4, encodes a PilV-like protein that may be used for adhesion to and invasion of the intestinal cells. Furthermore, a putative autotransporter and two type 1 fimbrial proteins of strain WA-314 show a sequence similarity <50% with the orthologous proteins in strain 8081. The dissimilar sequences of these proteins indicate possible different functions or interaction modes, reflecting the specific adhesion properties of Y. enterocolitica strains 8081 and WA-314 and thus the different efficiency of host colonization. Further important differences were found in two pYV plasmid-encoded virulence factors, YopM and YscP. The impact of these differences on virulence is discussed.
Conclusions
Our study emphasizes that the virulence of pathogens can be increased, by acquiring new genes and/or improving the function of essential virulence proteins, resulting in permanently hyper-virulent strains. This work also highlights the importance of addressing genetic and phenotypic variations among closely related bacterial strains, even those belonging to the same bioserotype.
Keywords: Yersinia enterocolitica, Hyper-virulent, Genome comparison, Diversity, Host colonization, Virulence factors, YscP, YopM
Background
Yersinia enterocolitica is a globally disseminated gastrointestinal pathogen which is transmitted by the fecal-oral route, through ingestion of contaminated food or water [1]. Human clinical infections most commonly occur in young individuals and are associated with acute diarrhoea, terminal ileitis, mesenteric lymphadenitis and pseudo-appendicitis [2]. Critical elements for pathogenesis are the high pathogenicity island (HPI), carrying the siderophore-mediated iron uptake system named yersiniabactin, and the virulence plasmid pYV. While the HPI and pYV are absent in avirulent strains, they are both conserved among highly virulent strains of the three pathogenic Yersinia species, Y. pestis, Y. pseudotuberculosis and Y. enterocolitica. The 70-kb plasmid encodes a type III secretion system (T3SS) and a set of Yop effector proteins which, after injection by the T3SS into host cells, inhibit several host immune mechanisms which enable the bacteria to survive in the host environment [3].
Y. enterocolitica strains are heterogeneous and are classified into 6 biotypes (1A, 1B, 2, 3, 4 and 5) according to biochemical properties [4]. Biotype 1A strains, lacking both HPI and pYV plasmid, are considered as non-virulent in mice, whereas biotypes 2 to 5, which lack HPI, are low virulence (unable to kill mice) [5]. These five biotypes belong to the Y. enterocolitica subsp. palearctica and are generally isolated in Europe and Japan (termed “Old World” strains). Biotype 1B (subsp. enterocolitica), harboring both HPI and pYV plasmid, is highly virulent (mouse-lethal) and predominant in North America (the so called “New World” strains) [6]. More than 70 serotypes of Y. enterocolitica have been described; however only few of them are virulent with serotypes O:3, O:5,27, O:8, O:9, O:20 and O:13 being the most pathogenic to humans [2]. In the past, Y. enterocolitica bioserotype 1B/O:8 strains were predominant in the United States [7]. Nowadays these strains are also isolated in other countries; nevertheless, bioserotype 4/O:3 strains are the most commonly Y. enterocolitica strains found over the world [8], [9].
Many Yersinia research laboratories use in their studies two mouse-virulent Y. enterocolitica 1B/O:8 strains, named 8081 [10] and WA-314 [5]. Y. enterocolitica strain 8081 is an American isolate from a fatal-septicemia patient [10] and has been widely used in murine infection models. Y. enterocolitica strain WA-314 was isolated from the blood of a human patient and proved to be highly virulent for mice and rats [11]. Recently, in an attempt to identify chromosomal variations underlying the different properties of high versus low virulence strains, the genome of the highly virulent strain 8081 [12] has been compared to genomes of low virulence strains [13,14]. However, a complete comparative characterization of highly virulent 1B/O:8 strains with each other had not been performed. In this context, the sequence of the pYVWA-314 has been recently determined, showing global similarity to pYV plasmids of other Y. enterocolitica strains but with noticeable differences in the amino acid sequences of the T3SS proteins SycH, YopM, LcrV and YscP [15]. Phenotypic differences in autoagglutination experiments [15] and results from our pilot experiments with strains 8081 and WA-314, displaying differences in bacterial growth curves and optical density-cellular mass correlation in vitro, suggested that these two strains possessed distinct virulence and colonization properties.
In this work, we demonstrated that strain WA-314 overcame strain 8081 in the colonization of spleen and liver during in vivo co-infection experiments. Genome comparison between the genomic sequence of strain 8081 [12] and a high-quality whole-genome sequence of strain WA-314, presented in this study, allowed the identification of putative virulence factors which may account for the different in vivo phenotypic behavior of these two 1B/O:8 strains.
Results
Comparison of the infection ability of Y. enterocolitica strain 8081 versus strain WA-314 in mouse model
Y. enterocolitica strain WA-314 is a hyper-virulent strain with increased colonization in mice
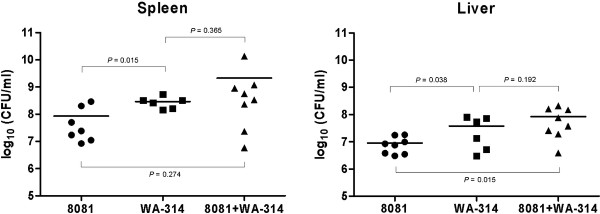
In order to directly compare the virulence efficiency of Y. enterocolitica strain 8081 and Y. enterocolitica strain WA-314, we infected 3 groups of mice with the same total CFU of strain 8081, strain WA-314, or a 1:1 mix of both. Intra-peritoneal infection was used to enable accurate and controlled injection of the dose of interest into a limited number of mice. Figure 1 indicates the bacterial loads in spleens and livers of the 3 groups of mice. Mice infected with strain 8081 contained significantly lower bacterial loads than mice infected with strain WA-314, both in the spleens (P = 0.01) and in the livers (P = 0.04). Mice co-infected with 1:1 mix of Y. enterocolitica strain 8081 and Y. enterocolitica strain WA-314 contained significantly more bacteria in liver samples, as compared to the livers of mice infected with strain 8081 alone (P = 0.01). However, no significant difference was shown between bacterial counts in livers of mice infected with strain WA-314 alone and with 1:1 mix of both strains 8081 and WA-314 (Figure 1). In addition, bacterial colonization in spleens of mice infected with individual versus combined strains was not significantly different.
Figure 1.
Mouse infection experiments. Bacterial counts in spleens and livers of mice infected with Y. enterocolitica strain 8081, Y. enterocolitica strain WA-314 and of co-infected mice with both strains are indicated. The means log10(CFU/ml) are displayed as error bars.
Digoxigenin-labeled probes specifically differentiate between Y. enterocolitica strain 8081 and Y. enterocolitica strain WA-314
Two strain-specific digoxigenin-labeled probes were designed for differentiating Y. enterocolitica 8081 and WA-314 (see Methods). The strain 8081-specific probe targets the putative hemolysin gene in the YAPI region, while the probe specific for strain WA-314 targets a region inside the colicin operon specifically acquired by strain WA-314 (see below). The two probes were tested on plates with cultivated single-strain and mixed-strain colonies. Two membranes were placed sequentially on each plate and then hybridized with probes Hem_8081 and Col_WA, respectively. As expected, colonies on membranes derived from strain 8081-plates were all detected by probe Hem_8081, whereas probe Col_WA gave no signal. Probe Hem_8081 did not detect any colonies on membranes replicated from strain WA-314-plates, while WA-314 colonies were all recognized by probe Col_WA. On mixed-strain membranes, probes Hem_8081 and Col_WA detected different colonies, all colonies gave a signal with the respective probe and no colonies were recognized by both probes, indicating sensitivity and specificity of the developed test (see Additional file 1).
Y. enterocolitica strain WA-314 overcomes 8081 in co-infection experiments
To investigate whether Y. enterocolitica strains 8081 and WA-314 equally colonize the organs of co-infected mice, we applied the optimized colony hybridization experiment (see Methods) on bacteria extracted from both spleens and livers (Figure 2). Y. enterocolitica strain WA-314, on average, accounted for 80% of the total Yersinia loads in spleens and livers of the co-infected mice. Only in mouse number 13 strain 8081 surpassed strain WA-314. However, the total bacterial count in this mouse’s spleen was the lowest in this study (5.81 x 106), suggesting that inoculation was not correctly performed. From our own experience, there is one strongly deviating result within a group of ten mice. For these reasons, mouse number 13 was not taken into account for statistical analysis. Thus, the results of the co-infection experiments clearly indicated that strain WA-314 could out-compete strain 8081 for colonization of host organs.
Figure 2.
Bacterial strain identification from co-infected mice. Results are obtained with the colony hybridization experiments from spleens and livers of the 8 co-infected mice. Y. enterocolitica strain WA-314 is more abundant than Y. enterocolitica strain 8081 in 7 mice out of 8.
Whole genome comparison of two Y. enterocolitica 1B/O:8 strains
General features
We present here the first whole-genome high-quality sequence of Y. enterocolitica strain WA-314 and compare it to the published genome of Y. enterocolitica strain 8081 [12] (Table 1). Genome sequencing and assembly of the 129 contigs of strain WA-314 genome resulted in 4 large scaffolds and 8 contigs, with an average G + C content of 47.78%; one additional scaffold constituted the pYV plasmid (see Additional file 2). The genomes of the two Y. enterocolitica 1B strains share 3,736 orthologous coding sequences (CDSs). The Yersinia Genomic Islands 1 and 2 (YGI-1 and YGI-2) are conserved, as are the virulence-associated determinants ail, inv, the LPS operon, the flagella and urease clusters, the two General Secretion Pathways Yts1 and Yts2 and the chromosomal T3SS Ysa (Table 2).
Table 1.
Genome properties of the chromosomes ofY. enterocoliticastrains WA-314 and 8081
| Property | Y. enterocoliticaWA-314 | Y. enterocolitica8081[[12]] |
|---|---|---|
| Genome size |
~ 4,459,631 |
4,615,899 |
| G + C content |
47.78% |
47.27% |
| Number of CDSs |
> 4,045 |
4,037 |
| tRNA |
≥ 65 |
81 |
| Prophage regions | 7 | 4 |
Table 2.
Established and putative virulence determinants ofY. enterocolitica
| Genomic origin | Protein | Function | Sequence similarity |
|---|---|---|---|
|
Virulence-associated determinants | |||
|
Plasmid (pYV) | |||
|
yop |
Yops |
Anti-phagocytic action |
YopM: 75% (protein) |
|
ysc |
Ysc |
Deliver of Yops |
YscP: 72% (protein) |
|
yadA |
YadA |
Attachment, invasion |
97% (protein) |
|
Chromosome | |||
|
inv |
Invasin |
Attachment, invasion |
100% (protein) |
|
ail |
Ail |
Attachment, invasion |
96% (protein) |
|
myfEFABC |
Myf fibrillae |
Intestinal colonization |
97% (DNA) |
| HPI |
Yersiniabactin |
Dissemination in the host |
98% (DNA) |
|
ureABC |
Urease |
Bacterial survival in acidic environments |
100% (protein) |
|
flhDC |
Flagella |
Migration and adherence to host cells |
100% (protein) |
| Ysa (PZ) |
T3SS |
Secretion of Ysps proteins |
98% (DNA) |
| Yts1 (PZ) |
GSP |
Dissemination into deep tissues |
99% (DNA) |
| Yts2 |
GSP |
Unclear |
98% (DNA) |
|
ystA |
Enterotoxin |
Fluid loss and diarrhea |
100% (protein) |
|
Putative virulence determinants identified in this study | |||
|
Chromosome | |||
| YE0694 |
Putative adhesin |
Adherence |
75% (protein) |
| YE3700 |
Outer membrane protein/ Autotransporter |
Adherence and invasion |
23% (protein) |
| YE1111 | Fimbrial protein | Adherence | 40% (protein) |
Names and locus tags refer to the genome of Y. enterocolitica strain 8081 [12].
Mobile elements
The largest source of genetic variation between strains 8081 and WA-314 is represented by genomic islands and prophages (Figure 3 and Table 3). In particular, the YAPI (Yersinia Adhesion Pathogenicity Island) and the YGI-4 are absent from Y. enterocolitica strain WA-314. The YGI-4 of strain 8081 is a putative integrated plasmid variably present in other Y. enterocolitica 1B strains, whereas the virulence-associated YAPI is a 66-kb region located within the Plasticity Zone of Y. enterocolitica strain 8081 [12]. This pathogenicity island is integrated between an intact and a partial tRNA-Phe copy that is complete in WA-314. The YAPI carries encoded plasmid-related functions and virulence determinants such as a type IV secretion system, a putative hemolysin, a toxin-antitoxin system (CcdAB family) and an arsenic-resistance operon. Most differences between the genomes of strains WA-314 and 8081 are due to prophages or prophage remnants. We identified 7 prophage-like regions (see Additional files 3 and 4) in Y. enterocolitica strain WA-314, encoding numerous hypothetical proteins that may also contribute to the high virulence of this strain. Considering the prophage diversity, two of the WA-314 prophage regions, YWA-1 and YWA-2, appear to be highly similar to prophages in strain 8081, ΦYE185 and ΦYE200, and Φ98, respectively. Since these regions are not located in the same chromosomal context, they are likely strain-independent acquisitions. A 28-kb P4-like prophage in strain WA-314 (YWA-4) is located in a region corresponding to YGI-3, a 19-kb putative integrated plasmid in strain 8081. This WA-314-specific prophage encodes a PilV-like protein (locus tag: YWA314_12491), which may be a part of an ancient type IV pil operon, and contains a Shufflon N-terminal region highly similar to adhesion exoproteins in Y. enterocolitica subsp. palearctica and in Y. intermedia. It has been shown that type IV pili are used by enterobacteria during adhesion to and invasion of the human intestinal cells [16-18]. Whether the WA-314-specific PilV-like protein enhances bacterial intestinal colonization has to be addressed.
Figure 3.
Genomic DNA comparison. Circular representation of the genome comparison between Y. enterocolitica strains 8081 and WA-314, obtained with BRIG [41]. G + C content of the reference strain (8081) is also shown. The position of significant regions and virulence genes is marked in the outer ring.
Table 3.
Significant regions of difference betweenY. enterocoliticastrains 8081 and WA-314
| Region or gene cluster | Size in strain 8081 | Size in strain WA-314 | General description | Comment |
|---|---|---|---|---|
| YAPI (PZ) |
66 kb |
absent |
Pathogenicity island |
Absent from strain WA-314 |
| YGI-3 |
19 kb |
28 kb |
Putative integrated plasmid in strain 8081, putative prophage in strain WA-314 |
Strain-specific region |
| YGI-4 |
15 kb |
absent |
Putative integrated plasmid |
Absent from strain WA-314 |
| YE1922 |
0.2 kb |
absent |
Toxin/Antitoxin system (HigBA) |
Absent from strain WA-314 |
| YE1923 |
0.3 kb |
absent |
||
| YWA314_17584-YWA314_17599 |
absent |
4 kb |
Type II restriction-modification system |
Absent from strain 8081 |
| YWA314_20244-YWA314_20259 |
absent |
3 kb |
Putative group A colicin operon |
Absent from strain 8081 |
| Present in subsp. palearctica | ||||
| YWA314_07469 | absent | 1 kb | Xenobiotic-acyltransferase | Absent from strain 8081 |
| Present in subsp. palearctica |
Y. enterocolitica strain WA-314 specific genes
Genes specifically present in Y. enterocolitica strain WA-314 include an additional restriction-modification (RM) system, a four-gene putative colicin cluster and a xenobiotic-acyltransferase (XAT)-encoding gene. The RM cluster, present also in Y. frederiksenii, consists of 4 genes (locus tags: YWA314_17584-17599), encoding an EcoRII-like type II restriction endonuclease, a very-short-patch repair (Vsr) endonuclease, a cytosine methylase and a DNA-binding protein. Restriction modification systems defend bacteria against foreign DNA, by means of the endonuclease that recognizes non-methylated cytosines of incoming DNA and cleaves it at defined sites [19]. The host DNA is protected from cleavage as the internal cytosines are modified by the methylase activity; however, this methylation increases C to T mutations, which are in turn recognized and repaired by Vsr proteins [20]. Colicins are bacteriocins, proteins produced by bacteria which are lethal for closely related strains. Colicins are genetically organized in operons, with a variable genetic structure containing one or more genes encoding colicin, immunity, and lysis proteins [21]. The cluster found in Y. enterocolitica strain WA-314 genome is composed of four annotated genes (locus tags: YWA314_20244-20259), encoding two putative immunity proteins and two putative colicins. It is located near a phage anti-termination protein-encoding gene, a transposase and a pilus chaperone-encoding gene, reflecting a similar structure in other Yersinia species, such as Y. enterocolitica subsp. palearctica bioserotype 4/O:3 [13]. Our spot-on-lawn assay could not detect any colicin activity in Y. enterocolitica strain WA-314 against strain 8081 and the control strain E. coli K12, even after mitomycin C induction (data not shown). Thus, the colicin operon seems to be not active under the tested in vitro conditions. The XAT protein (locus tag: YWA314_07469) belongs to a family of resistance enzymes that catalyze the acetylation of a variety of hydroxyl-bearing acceptors such as chloramphenicol and streptogramin. It may be implicated in the inactivation of xenobiotics, leading to resistance.
The insecticidal toxin cluster, described in biotype 2–5 strains, was not found in the sequence of Y. enterocolitica strain WA-314 genome, in contrast to previously reported experimental data [22].
Newly identified potential virulence genes
Y. enterocolitica strain 8081 harbors a 635-aa outer membrane protein (locus tag: YE3700), which has only 23% of sequence similarity with its orthologous in strain WA-314, a 902-aa putative autotransporter (locus tag: YWA314_14949). These two proteins are situated in the same genomic region, indicating a common chromosomal origin and subsequent mutations during the evolution of the two strains. Interestingly, both genes have a low G + C content: 43.1% in strain 8081 and 39.2% in strain WA-314. Autotransporters are known virulence factors in Gram-negative bacteria, as they mediate bacterial aggregation and biofilm formation, as well as adhesion and invasion of epithelial cells. All classical autotransporters share a common organization: a signal peptide followed by an N-terminal passenger domain and a C-terminal translocator domain, with the passenger domain being involved in pathogenesis [23]. The protein encoded by strain 8081 has an autotransporter beta-domain at the C-terminus, but no known domains at the N-terminus. The protein of strain WA-314 contains a pertactin-like passenger domain at the central region and an autotransporter beta-domain at the C-terminus, a typical organization found in the homologous AidA-I protein in Escherichia coli[24]. Both proteins carry no signal peptide, according to in silico prediction algorithms (SignalP, version 4.0 [25]). However, as signal peptides have no high sequence homology among autotransporters [26], the SignalP program may not recognize the presence of signal peptides in the two analyzed amino acid sequences. The dissimilar sequences of the passenger domain of these two proteins may be responsible for the specific adhesion properties of Y. enterocolitica strains 8081 and WA-314. Further support for this possibility comes from another putative adhesin (locus tag: YE0694) in strain 8081, which is only 75% identical to the homologous protein in strain WA-314 (locus tag: YWA314_00878).
Fimbriae (or pili) are biological structures involved in adherence to host epithelial cells and important virulence factors for several diseases affecting the urinary, genital and gastrointestinal tracts [27]. A type 1 fimbrial operon was identified in Y. enterocolitica strain 8081 as region of difference among the Yersinia species [12]. The DNA sequence of this operon is 84% similar to the operon in strain WA-314. In addition, two fimbrial proteins in strain 8081 (locus tags: YE1111 and YE1114) show only 40% of sequence similarity with the orthologous proteins in strain WA-314 (locus tags: YWA314_11901 and YWA314_11891). These proteins are absent from Y. enterocolitica subsp. palearctica strains and, therefore, can be considered specific factors for highly virulent strains.
pYV plasmid-encoded genes
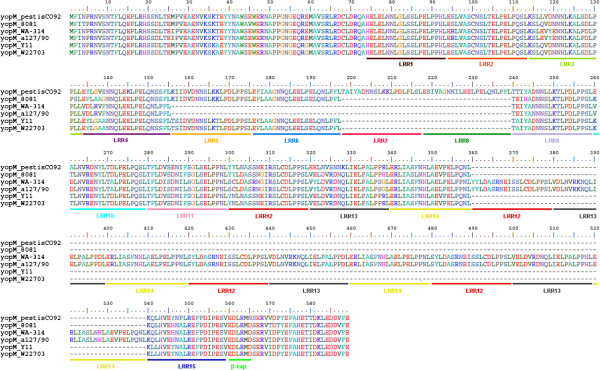
The nucleotide sequence of the pYVWA-314 was, as expected, nearly identical to the previously published sequence [15]. The pYVWA-314 sequence was also compared to the pYV plasmid sequence of strain 8081. Besides two repeat regions, the main differences were found in two T3SS protein-encoding genes, yscP and yopM. The amino acid sequence of the YscP protein, a component of the T3SS injectisome which determines the length of the needle, varies within Y. enterocolitica species in the specific number of three repeated motifs (of 14 aa, 25 aa and 46 aa respectively) [28]. Consistent with these data, strain 8081 harbors a 1359-bp gene, encoding a 452-aa YscP protein, while in strain WA-314 yscP is a 1617-bp gene encoding a protein of 538 residues, with 3 additional repeats (Figure 4). Interestingly, YscP proteins from low virulence strains Y. enterocolitica Y11 and W22703 (both of 515 residues) possess 2 extra repeats absent from highly virulent strains of serotype O:8. The T3SS effector YopM is a leucine-rich repeat (LRR) protein of unclear function. YopM sequences from different Yersinia strains contain duplications and deletions in the number of LRRs, with consequent variability in the YopM length [29]. YopM from Y. pestis CO92, for example, has 15 LRRs, while YopM from Y. enterocolitica strains Y11 and W22703 have 13 LRRs. The yopM gene in strain 8081 encodes a protein of 367 residues with 13 LRRs, while in strain WA-314 YopM is a 505-aa protein with 24 LRRs (Figure 5). In particular, the YopM sequence from pYVWA-314, which is 100% identical to the YopM sequence from the O:8 strain a127/90, has 3 additional copies of LRR12-LRR13-LRR14.
Figure 4.
Protein sequence alignment of YscP. The analyzed YscP protein sequences are taken from Y. enterocolitica strains 8081 (O:8), WA-314 (O:8), a127/90 (O:8), Y11 (O:3) and W22703 (O:9) (see Table 6 for accession numbers). The green, red and blue lines highlight the repeats of 14 aa, 25 aa and 46 aa, respectively.
Figure 5.
Alignment of the sequences of YopM proteins. Protein sequences are obtained from Y. enterocolitica strains 8081 (O:8), WA-314 (O:8), a127/90 (O:8), Y11 (O:3) and W22703 (O:9), and Y. pestis CO92, as reference strain (see Table 6 for accession numbers). LRR numbers follow the published notation [29].
Phylogenetic position of Y. enterocolitica strain WA-314
To gain insights into the evolution of Y. enterocolitica subspecies, we selected 9 strains representing the three Y. enterocolitica groups, classified according to the virulence grade: avirulent (biotype 1A, strains IP2222 and NF-O); low virulence (biotypes 3 and 4, strains Y11, Y5.27P, 105.5R(r), Y5307 and Y8265) and highly virulent (bioserotype 1B/O:8, strains 8081 and WA-314). The Y. pestis strain CO92 was selected as outgroup, known a priori to be an outlier to the ingroup sequences and chosen to root the tree.
The concatenated tree (Figure 6) sorts three monophyletic clusters for the three Y. enterocolitica groups, as expected. The intragroup genetic distances show that biotypes 3 and 4 are clustered more tightly than biotypes 1A and 1B; therefore genomes of low virulence strains have lower plasticity, when compared to avirulent and highly virulent strains, which present a higher degree of genetic variation. Concerning the evolution of Y. enterocolitica, this analysis indicates that the three groups evolved independently from a common ancestor, with biotype 1A strains being the first to diverge, followed by biotype 1B and 2–5 strains. According to our phylogeny reconstruction, non-pathogenic strains are evolutionarily closer to Y. pestis than the pathogenic Y. enterocolitica.
Figure 6.
Phylogenetic analysis of theYersinia enterocoliticaspecies. The constructed phylogram is based on DNA sequences and rooted using the genes of Y. pestis CO92. Nine different Y. enterocolitica strains were used: IP2222 and NF-O (avirulent); 8081 and WA-314 (highly virulent); Y11, Y5307, Y8265, Y5.27P and 105.5R(r) (low virulence). The distance is scaled in units of the expected fraction of changed nucleotides.
Discussion
Highly virulent Yersinia enterocolitica strains have been extensively used to clarify the virulence/fitness mechanisms of this heterogeneous gastrointestinal pathogen. The use of different bioserotype 1B/O:8 strains is normally not considered a variable factor in experimental animal infection procedures. In this study, however, two Y. enterocolitica 1B/O:8 strains, 8081 and WA-314, demonstrated different virulence behaviors in mice, both in single strain infection and in competition assays, with strain WA-314 showing a higher virulence/fitness level. A further characterization of these two strains was therefore necessary to uncover the genetic background behind the phenotypic differences.
Genome comparison between a high-quality sequence of Y. enterocolitica strain WA-314 and the published sequence of Y. enterocolitica strain 8081 [12] revealed, besides an overall similarity in gene composition, important differences in genomic islands and prophages. In pathogenic bacteria, genomic islands and prophages represent a large source of inter- and intra-species genetic variation [30]. Pathogenicity islands also play an essential role in spreading virulence genes through lateral gene transfer, an evolutionary process by which bacterial pathogens acquire new virulence factors en bloc. Our analysis revealed that the YGI-4 and the pathogenicity island YAPI, carrying a type IV secretion system known to be involved in Y. pseudotuberculosis virulence [31], are absent from strain WA-314. Moreover, both 8081 and WA-314 strains harbor prophages that have no or low similarity. We observed that the mobile genetic elements tend to occupy the same positions in both 1B/O:8 genome backbones. The P4-like prophage YWA-4, for example, is inserted in strain WA-314 in the same genomic region as the YGI-3 plasmid-like element in the chromosome of strain 8081. This speaks in favor of the presence of “hot-spots” for the integration of the acquired genetic material. Such hot spots, besides being integration sites, might represent genome regions with high gene expression potential, an important factor for the homing of laterally acquired genetic clusters. Taken together, these data emphasize the important role of horizontal gene transfer and mobile genetic elements in the evolution and genetic diversification among pathogenic Yersinia.
Besides mobile elements, the genomes of Y. enterocolitica strains 8081 and WA-314 differ in a number of gene clusters and single protein-encoding genes. Strain WA-314 specific acquisitions include a XAT-encoding gene, a RM system and a putative colicin cluster. Strain 8081, on the other hand, harbors a specific toxin/antitoxin system, similar to the HigBA family. Nucleotide polymorphisms in homologous genes also contribute to the genetic variation between strains 8081 and WA-314. The plasticity of Y. enterocolitica 1B/O:8 genomes mirror the versatile lifestyles of this heterogeneous bacterial species, found in human, animal and environmental sources, and results from the on-going process of adaptation. By living in contact with various microbial communities in different niches, Y. enterocolitica experiences frequent opportunities for exchanging genetic material.
One of the main challenges of comparative genomics is to identify genes involved in pathogenesis. Virulence factors are generally involved in adherence, invasion, colonization of the host, interference with host defense mechanisms and damage to the host [32]. A putative autotransporter, an adhesin and two fimbrial proteins show low sequence similarity in the genomes of strains 8081 and WA-314. Together with a type IV pilus, present only in the genome of strain WA-314, they may contribute to adhesion and colonization of host tissues. Such new potential virulence determinants may be able to explain the phenotypic differences observed in vivo between Y. enterocolitica strains 8081 and WA-314. The real involvement of these proteins in virulence, however, needs to be elucidated.
Established virulence-associated determinants of Y. enterocolitica have been extensively studied and reviewed [33]. One group is pYV plasmid-encoded, such as YadA, the T3SS secretion machinery Ysc and the T3SS effectors Yops, while the other group is encoded within the chromosome, for example Inv, Ail and the HPI. The genomes of Y. enterocolitica strains 8081 and WA-314 show no significant differences in the sequences of these classical virulence markers, except for the plasmid-encoded effector protein YopM and YscP, the “needle ruler” of the Yersinia injectisome. The function of the YopM protein is not completely understood. However, it has been shown to form a complex with two intracellular serine/threonine kinases, protein kinase C-like 2 (PRK2) and ribosomal S6 protein kinase 1 (RSK1) [34]. In Y. pseudotuberculosis, the interaction with RSK1 requires the region from LRR12 to C-terminus of YopM [35], whereas PRK2 binding involves the LRR6 to LRR15 region of YopM [36]. Both RSK1 and PRK2 interaction domains of YopM are critical for virulence, e.g. by inducing production of IL-10, as demonstrated by different YopM mutant proteins [36]. Interestingly, Y. enterocolitica strain 8081 encodes a YopM of 367 residues with 13 LRRs, while YopM of strain WA-314 has 505 residues and 24 LRRs. Therefore, further studies are required to elucidate whether the different number of LRRs in YopM proteins of strain 8081 and WA-314: i) generally results in the interaction of the corresponding YopM proteins with different targets in the host, ii) especially causes distinct interaction or binding affinity with PRK2 and RSK1 and iii) has different consequences on the virulence of Y. enterocolitica in the mouse infection model. YscP, a protein highly variable within Y. enterocolitica species, determines the needle length of the Yersinia spp. injectisome, with a linear correlation between the size of YscP and the needle length [28]. It has been shown that the Y. enterocolitica needle needed to have a minimal length to be fully functional [37]. Such a minimal needle length, which also correlated with the length of the YadA adhesin, provided optimal contact between the needle and the host cell membrane. Thus shorter YscP proteins or longer YadA proteins led to suboptimal Yop translocation [37]. Curiously, YscP of Y. enterocolitica strain 8081 contains 452-aa, while in strain WA-314 YscP is 538-aa long. As YadA length is unchanged between Y. enterocolitica strains WA-314 and 8081 (as predicted by gene sequence comparison), we propose that the longer YscP protein in strain WA-314 would allow higher Yop translocation efficiency than strain 8081 and, therefore, improved virulence activity. This would partly explain the different phenotypes of strains 8081 and WA-314 observed in the co-infection experiment.
Small reproducible differences between the in vitro growth rates of Y. enterocolitica strains 8081 and WA-314 have been documented, with strain WA-314 growing slightly faster than strain 8081 (data not shown). Thus the lower in vivo colonization ability of strain 8081 might be related to its growth behavior and to metabolic and regulatory factors, without regard to virulence determinants. However, no obvious differences in metabolic and nutrient acquisition systems have been found between strain 8081 and WA-314 genomes. In vitro growth conditions for Yersiniae are also extremely different from those in vivo: for example, Yersinia optimal growth temperature is 27°C, in contrast to the in vivo temperature of 37°C, and most virulence factors are only expressed at 37°C. Thus a correlation between the in vitro growth rates of Y. enterocolitica strains 8081 and WA-314 and their in vivo colonization properties is unlikely, but such a possibility cannot be completely excluded.
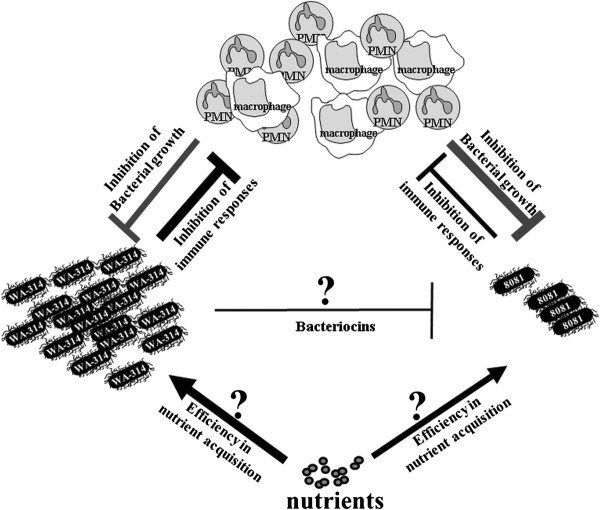
It is well known that pathogenesis induced by bacteria is dependent on bacterial proliferation in the host. Successful invasion of and growth in host niches requires adaptation of the pathogen to the host micro-environment. Micro-environmental factors influencing bacterial colonization of the host include nutrient composition and availability in the host niches, on one hand, and the host defense mechanisms, on the other hand. In the case of co-infection or co-existence of different pathogens in the same micro-environment, a further important factor is represented by the ability of each pathogen to compete against each other for host niches and nutrient sources. Pathogens are adapted to those micro-environmental factors. They carry adhesion molecules, which enable invasion of specific tissue and cell types, and metabolic systems, such as iron-binding proteins. They also express virulence factors, which counteract immune responses of the host, and molecules, like bacteriocins, which inhibit the growth of closely related bacterial strains, thus enabling competition against other pathogens. In our co-infection model (intra-peritoneal route) with two highly virulent Y. enterocolitica strains, strain WA-314 seemed to be better adapted than strain 8081 to the micro-environmental factors encountered in host tissues like spleen and liver (Figure 7). Thus the present data suggest that Y. enterocolitica strain WA-314 is a hyper-virulent strain. In fact, mice infected with strain 8081 showed lower bacterial loads than mice infected with strain WA-314 and strain 8081 could not colonize the host as efficiently as strain WA-314, even in the absence of a competitor pathogen. The colicin operon found in strain WA-314 showed no in vitro activity, but we cannot exclude that this colicin could be effectively expressed in vivo at the host’s environmental conditions, thus probably conferring additional advantage to strain WA-314 against strain 8081. Finally, the identified putative adhesion proteins, that have been shown here to be specifically acquired by strain WA-314, may also contribute to the higher capacity of this strain to invade the host.
Figure 7.
Representation of the micro-environmental factors influencing replication and bacterial pathogenesis ofY. enterocoliticastrains WA-314 and 8081. Strain WA-314 appears to be better adapted than 8081 strain, probably by counteracting more efficiently the bactericidal activities of the host immune cells. The growth of strain 8081 may be also inhibited by the direct effect of bacteriocins produced by strain WA-314 bacteria. The thickness of the arrows reflects the strength and efficiency of the depicted processes.
Conclusions
This study demonstrated that Y. enterocolitica highly virulent strains exhibit significant strain-to-strain genotypic and phenotypic differences, resulting in differences in their pathogenicity. Accordingly, virulence of pathogens can be increased, e.g. by acquiring new genes and/or improving the function of essential virulence proteins. Thus, understanding the genetic factors which allow bacterial hyper-virulence would enable the design of improved therapeutic strategies against such strains.
Methods
Ethics statement
All animal work was performed in strict accordance with the German regulations of the Society for Laboratory Animal Science (GV-SOLAS) and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA). The protocol was approved by the Regierung von Oberbayern, Sachgebiet 54 (Verbraucherschutz und Veterinärwesen): animal licensing committee permission no. 2531–6509. All efforts were made to minimize suffering.
Bacterial strains and growth conditions
This study was conducted with wild-type Y. enterocolitica strains 8081 [10] and WA-314 [11], obtained from the strain collection of our Institute. Both strains were mouse-passaged twice, to select the most virulent bacteria, and stored in 20% glycerol medium at −80°C. For mouse infection, exponential-phase bacteria were grown overnight in LB medium at 27°C, diluted 1:50 in fresh LB medium and grown for 80 min at 37°C to allow the expression of virulence factors. After pelleting and washing in Dulbecco’s Phosphate Buffered Saline (DPBS), bacteria were adjusted to the appropriate CFU/ml as infection dose.
Mouse infection and bacterial count
Female C57/BL6 mice were individually identified with ear tags and randomly assigned to three experimental groups containing eight mice per group: groups consisted of mice to be infected with either Y. enterocolitica strain 8081, strain WA-314 or both in combination (Table 4). Seven-week old mice were challenged by intra-peritoneal (i.p.) injection with 0.6 ml PBS, containing 1.7 x 104 bacteria for the single-strain-infection and 1.2 x 104 bacteria for the co-infection (6 x 103 bacteria of each strain were 1:1 mixed prior to injection). Bacterial numbers in the co-infection mix was confirmed by colony hybridization (see below). Mice were weighed every day and sacrificed 5 days post infection by inhalation of carbon dioxide. Spleens and livers were recovered and 1 ml of DPBS was added to each organ, which was homogenized with a tissue blender for 4 min. Bacterial counts were determined by plating serial dilutions of organ homogenates on LB agar followed by growth at 27°C for 20–24 hours. Differences in mice bacterial colonization were assessed by unpaired t tests and P values ≤ 0.05 were considered significant.
Table 4.
C57/BL6 mice and injected bacteria doses used in this study
| Mouse number | InjectedY. enterocoliticastrain | Infection dose |
|---|---|---|
| 1-2-3-4-5-16-17-18 |
8081 |
1.7 x 104 bacteria |
| 6-7-8-9-10-19-20-21 |
WA-314 |
1.7 x 104 bacteria |
| 11-12-13-14-15-22-23-24 | 8081 + WA-314 | 1.2 x 104 bacteria |
Digoxigenin-labeled probes
We designed two digoxigenin-dUTP-labeled probes to be used in the DNA-DNA colony hybridization experiment for distinguishing Y. enterocolitica strain 8081 from strain WA-314, by selecting strain-specific regions in the 8081 and WA-314 genomes. The 162-bp probe specific for detecting strain 8081 targets part of the putative hemolysin gene (locus tag: YE3454) encoded by the YAPI, whereas the 234-bp probe specific for strain WA-314 targets a region inside the colicin cluster (locus tag: YWA314_20259). Blast analysis revealed that our probe sequences did not align to any genomic regions in other bacteria, thus confirming species- and strain-specificity of these probes. Both probes were generated as PCR products with digoxigenin-11-dUTP incorporated as a labeling molecule, using the PCR DIG Probe Synthesis Kit (Roche, Mannheim, Germany). Primers and PCR conditions are given in Table 5.
Table 5.
Primers and PCR conditions for generation of the digoxigenin-labeled probes
| Primer | Sequence | Probe | PCR conditions |
|---|---|---|---|
| Hem_8081_FW |
5'-CAATATGACTACCGACCCGGTTAC-3' |
Hem_8081 |
- Denaturation at 95°C for 2 min |
| Hem_8081_RV |
5'-GGATACATCTGCTGGGCGATATAC-3' |
162 bp |
- 30 cycles: |
| denaturation at 95°C for 30 sec | |||
| Col_WA_FW |
5'-CGATCGTAGTAGTAAGGCAACTCC-3' |
Col_WA |
annealing at 60°C for 30 sec |
| Col_WA_RV | 5'-GACGGTATCATGCCCATAACTG-3' | 234 bp | elongation at 72°C for 40 sec |
| - Final elongation at 72°C for 7 min |
Preparation of membranes for colony hybridization
Plates from overnight growth were chilled at 4°C before colony lifts. Nylon membranes (Roche) were placed onto the LB agar surface for 5 min, transferred onto new LB agar plates and incubated at 27°C for 4 h to allow further bacterial growth. Membranes were removed from the LB agar plates and placed colony-side up on filter papers soaked with 10% w/v sodium dodecyl sulphate (SDS) for 10 min. This procedure was repeated with a denaturation solution (0.5 M NaOH and 1.5 M NaCl; pH 11.5) for 15 min, a neutralization solution (1.5 M NaCl and 1.0 M Tris–HCl; pH 7.4) for 15 min and 2 X SSC (stock solution of 20 X SSC: 3 M NaCl and 0.3 M Na3-citrate; pH 7.0) for 10 min. Membranes were then air-dried and baked at 80°C for 60 min to cross-link the transferred DNA. All membranes were stored at 4°C until hybridization.
Hybridization and detection protocol
The treated nylon membranes were placed in hybridization glass bottles and pre-hybridized at 50°C in a hybridization oven for 1 h in hybridization buffer (50% v/v formamide; 5 X SSC; 1% v/v blocking reagent diluted in 0.1 M maleic acid and 0.15 M NaCl, pH 7.5; 0.1% N-lauroylsarcosine; 0.02% v/v SDS), as previously described [38]. Labeled probes were denatured at 97°C for 5 min, placed on ice, mixed with pre-warmed hybridization buffer (2 μl probe/ml buffer) and transferred in sterile tubes with the appropriate membrane. Hybridization buffers containing the labeled probes were stored at -20°C and reused several times, after denaturation at 65°C. Hybridization was carried out at 50°C for 3 h. Nylon membranes were then stringently washed in two washing solutions with constant agitation: 2 X SSC-0.1% SDS for 2 x 5 min at room temperature; 0.5 X SSC-0.1% SDS for 2 x 15 min at 67°C. The detection step was performed with the DIG Nucleic Acid Detection Kit (Roche), according to the manufacturer’s instructions. Briefly, unspecific binding sites were blocked with 1% blocking reagent for 30 min, successively the labeled probes were bound to anti-digoxigenin Fab-fragments conjugated to alkaline phosphatase for 30 min and, finally, antibody-binding was visualized by membrane incubation in NBT/BCIP solution until the color reaction was completed (30–60 min). Membranes were photographed, scanned and wet stored in plastic bags at 4°C for any further stripping and re-hybridization.
Bacterial strain identification from mouse co-infection
From the plated serial dilutions used to determine the numbers of bacterial CFU recovered after mouse infection, plates with 200–600 single colonies were selected and examined by colony hybridization, as described above, in order to distinguish between colonies of Y. enterocolitica strains 8081 and WA-314. Membranes were either hybridized first with probe Hem_8081 or probe Col_WA, were stripped and re-hybridized with probe Col_WA or Hem_8081, respectively. The specificity of the probes was confirmed by analyzing plates containing either strain 8081 or strain WA-314.
Colicin activity assay
Antibacterial activity of the Y. enterocolitica WA-314 colicin cluster was tested by the spot-on-lawn method for screening of inhibitory activity against Y. enterocolitica strain 8081 and Escherichia coli K12 strain MG1655, known to be susceptible to colicins. Y. enterocolitica strain WA-314 was grown overnight at 27°C in LB medium. From this liquid culture, two spots of 5 μl were made on solid LB medium; induction of colicin production was conducted adding mitomycin C in the solid LB medium at a final concentration of 0.5 μg/ml. The spotted WA-314 bacteria were incubated for 20 h at 27°C or 37°C. Cells were killed by exposure to 700 μl chloroform vapor for 10 min and dried for 20 min by aeration. The surface of the solid medium was overlaid with 7 ml of 0.7% soft agar containing 10 μl of an overnight culture of the indicator organism; Y. enterocolitica strain 8081-overlaid plates were grown at 27°C or 37°C and plates overlaid with E. coli K12 were grown at 37°C.
Genome sequencing and comparison
A high-quality Y. enterocolitica strain WA-314 genome sequence was obtained in cooperation with BGI-Hongkong Co. (Hong Kong). High-throughput Illumina sequencing technology was used to construct 500-bp library with expected data of 500 Mb, and 6-kbp library with expected data of 250 Mb. Assembly of 15-bp short reads by the SOAPdenovo assembler resulted in 32x genome depth. Genome sequence was annotated by the RAST server [39] and tRNA identification was confirmed using tRNAscan-SE [40]. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number AKKR00000000. The version described in this paper is the first version, AKKR01000000. Comparison with the Y. enterocolitica strain 8081 genome sequence [GenBank: AM286415 and AM286416 (plasmid)] [12] was performed using BRIG [41], SEED [42] and the progressive Mauve [43] algorithm with default settings and a 1,500 bp cutoff as the minimum LCB length. Orthologous proteins were determined considering a minimum of 50% of sequence similarity between bi-directional hit proteins. Specific genes and gene clusters were aligned with ClustalW [44] and manual homology searches were performed by BLAST analysis [45]. To identify protein similarity with characterized proteins and known functional domains, we searched the NCBI conserved domain database (CDD) [46] or the Pfam protein database [47].
Phylogeny reconstruction
To infer Yersinia enterocolitica phylogeny, we selected 9 strains with available genome sequences and we chose Y. pestis strain CO92 as the outgroup; accession numbers are listed in Table 6. Phylogenetic trees were constructed using 5 housekeeping genes that were head-to-tail concatenated into a string of about 9200 bps for each strain: glnA (glutamine synthetase), gyrA and gyrB (DNA gyrase subunit A and B), groEL (60 kDa chaperonin) and recA (recombinase A). Protein sequences were aligned using Muscle [48] and the protein alignment was used as a model to create the DNA alignment with RevTrans [49]. Uninformative characters were removed using Gblocks [50] and phylogenies based on both DNA and protein alignments were reconstructed with Phylip [51] under a neighbor-joining model. A majority rule-consensus tree of 100 bootstrap replicates was also computed to evaluate node support.
Table 6.
Accession numbers of the genome sequences used in this study
| Organism | Accession number |
|---|---|
|
Y. enterocolitica WA-314, 1B/O:8 |
GenBank: AKKR01000000 |
|
Y. enterocolitica 8081, 1B/O:8 |
GenBank: AM286415, AM286416 (plasmid) |
|
Y. enterocolitica a127/90, 1B/O:8 |
NCBI RefSeq: NC_004564 (plasmid) |
|
Y. enterocolitica Y11, 4/O:3 |
GenBank: FR729477, FR745874 (plasmid) |
|
Y. enterocolitica Y8265, 4/O:3 |
GenBank: CACU01000001-CACU01000014 |
|
Y. enterocolitica Y5307, 4/O:3 |
GenBank: CACV01000001-CACV01000018 |
|
Y. enterocolitica 105.5R(r), 3/O:9 |
GenBank: CP002246 |
|
Y. enterocolitica W22703, 2/O:9 |
NCBI RefSeq: NC_002120 (plasmid) |
|
Y. enterocolitica Y5.27P, 3/O:5.27 |
GenBank: CACW01000001-CACW01000020 |
|
Y. enterocolitica NF-O, 1A/O:5 |
GenBank: CACY01000001-CACY01000097 |
|
Y. enterocolitica IP2222, 1A/O:36 |
GenBank: CACZ01000001-CACZ01000074 |
| Y. pestis CO92 | GenBank: AL590842 |
Competing interests
The authors declare that they have no competing interests.
Authors’contributions
DG designed and executed all experiments, carried out the genome comparison and bioinformatics analysis and drafted the manuscript. HB performed the major part of the mouse infection experiments, provided ideas for the study and participated in the composition of the manuscript. JH and AR supervised the study and helped to draft the manuscript. AR also conceived of the study and participated in its design. All authors read and approved the final manuscript.
Supplementary Material
Colony hybridization experiment. A: nylon membrane from the liver of mouse number 13 (13 L), probed with Hem_8081. B: nylon membrane from A stripped and re-probed with Col_WA. C: original LB-agar plate. D: modified and superimposed images from A and B. (PDF 146 kb)
Assembly in scaffolds and accession numbers of the contigs ofY. enterocoliticastrain WA-314.
Predicted prophages inY. enterocoliticastrain WA-314. Excel table with prophage names, respective contigs, locus tags and encoded products.
Genetic structure of prophages inY. enterocoliticastrain WA-314. Annotation of selected genes is shown. YWA-1: possible degenerate P2-like prophage, 34.4 kbs. YWA-2: putative bacteriophage, 33 kbs. YWA-3: putative P2-like prophage, 11.7 kbs. YWA-4: putative P4-like prophage, 28 kbs. YWA-5: Mu-like prophage, 43.3 kbs. YWA-6: P4-like prophage, 14.6 kbs. YWA-7: putative defective prophage, 12.5 kbs.
Contributor Information
Debora Garzetti, Email: garzetti@mvp.uni-muenchen.de.
Hicham Bouabe, Email: hicham.bouabe@babraham.ac.uk.
Juergen Heesemann, Email: heesemann@mvp.uni-muenchen.de.
Alexander Rakin, Email: rakin@mvp.uni-muenchen.de.
Acknowledgements
We would like to thank Dr. C. Harrison for critical reading of the manuscript and L. Schneider for helpful discussion.
This work was partially supported by the German Bundesministerium für Bildung und Forschung (BMBF) Network Grant FBI-Zoo.
References
- Bottone EJ. Yersinia enterocolitica: overview and epidemiologic correlates. Microb Infect. 1999;1:323–333. doi: 10.1016/S1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- Bottone EJ. Yersinia enterocolitica: The Charisma Continues. Clin Microbiol Rev. 1997;10(2):257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. The Virulence Plasmid of Yersinia, an Antihost Genome. Microbiol Mol Biol Rev. 1998;62(4):1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters G, Kandolo K, Janssens K. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol. 1987;9:14–21. [PubMed] [Google Scholar]
- Heesemann J, Laufs R. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J Bacteriol. 1983;155:761–767. doi: 10.1128/jb.155.2.761-767.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Rakin A, Heesemann J. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int J Med Microbiol. 2004;294:83–94. doi: 10.1016/j.ijmm.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Kay BA, Wachsmuth K, Gemski P, Feeley JC, Quan TJ, Brenner DJ. Virulence and phenotypic characterization of Yersinia enterocolitica isolated from humans in the United States. J Clin Microbiol. 1983;17:128–138. doi: 10.1128/jcm.17.1.128-138.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Qouqa IA, El Jarou MA, Samaha AS, Al Afifi AS, Al Jarousha AM. Yersinia enterocolitica infection among children aged less than 12 years: a case–control study. Int J Infect Dis. 2011;15(1):e48–e53. doi: 10.1016/j.ijid.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Rosner BM, Stark K, Werber D. Epidemiology of reported Yersinia enterocolitica infections in Germany, 2001–2008. BMC Public Health. 2010;10:337. doi: 10.1186/1471-2458-10-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Moseley SL, SF. Characterization of Plasmids and Plasmid-Associated Determinants of Yersinia enterocolitica Pathogenesis. Infect Immun. 1981;31(2):775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PB, Varga CF, Keet EE. New strain of Yersinia enterocolitica pathogenic for rodents. Appl Microbiol. 1973;26(6):1016–1018. doi: 10.1128/am.26.6.1016-1018.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, Howard S, Wren BW, Holden MTG, Crossman L, Challis GL, Churcher C, Mungall K, Brooks K, Chillingworth T, Feltwell T, Abdellah Z, Hauser H, Jagels K, Maddison M, Moule S, Sanders M, Whitehead S, Quail MA, Dougan G, Parkhill J, Prentice MB. The Complete Genome Sequence and Comparative Genome Analysis of the High Pathogenicity Yersinia enterocolitica Strain 8081. PLoS Genet. 2006;2(12):e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzilla J, Antonenka U, Höper D, Heesemann J, Rakin A. Yersinia enterocolitica palearctica serobiotype O:3/4 - a successful group of emerging zoonotic pathogens. BMC Genomics. 2011;6(12):348. doi: 10.1186/1471-2164-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Jing H, Ren Y, Zhou Z, Wang S, Kan B, Xu J, Wang L. Complete Genome Sequence of a Yersinia enterocolitica "Old World" (3/O:9) Strain and Comparison with the "New World" (1B/O:8) Strain. J Clin Microbiol. 2011;49(4):1251–1259. doi: 10.1128/JCM.01921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhettinger P, Schütz M, Raddatz G, Keller H, Autenrieth IB, Linke D. The sequence of the pYV virulence plasmid from Yersinia enterocolitica strain WA-314 biogroup 1B serotype O:8. Plasmid. 2011;65:20–24. doi: 10.1016/j.plasmid.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Bieber D, Ramer SW, Wu CY, Toru MWJT, Fernandez R, Schoolnik GK. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Tsui ISM, Yip CMC, Fung AWY, Wong DKH, Dai X, Yang Y, Hackett J, Morris C. Salmonella enterica serovar typhi uses type IVB pili to enter human intestinal epithelial cells. Infect Immun. 2000;68(6):3067–3073. doi: 10.1128/IAI.68.6.3067-3073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning PA. The tcp gene cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/S0378-1119(97)00036-X. [DOI] [PubMed] [Google Scholar]
- Pingouda A, Fuxreiterb M, Pingouda V, Wendea W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Lieb M. Cooperation and competition in mismatch repair: very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Molec Microbiol. 2002;44(6):1421–1428. doi: 10.1046/j.1365-2958.2002.02989.x. [DOI] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. Colicin Biology. Microbiol Mol Biol Rev. 2007;71(1):158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanier B, Starke M, Higel F, Scherer S, Fuchs TM. Yersinia enterocolitica Infection and tcaA-Dependent Killing of Caenorhabditis elegans. Appl Environ Microbiol. 2010;76(18):6277–3285. doi: 10.1128/AEM.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol. 2011;301(1):461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Wells TM, Totsika M, Schembri MA. Autotransporters of Escherichia coli: a sequencebased characterization. Microbiology. 2010;156:2459–2469. doi: 10.1099/mic.0.039024-0. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria – structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Sorg I, Degiacomi M, Journet L, Dal Peraro M, Cornelis GR. The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome. Molec Microbiol. 2009;71(3):692–701. doi: 10.1111/j.1365-2958.2008.06556.x. [DOI] [PubMed] [Google Scholar]
- Vieux EF, Barrick D. Deletion of internal structured repeats increases the stability of a leucine-rich repeat protein, YopM. Biophys Chem. 2011;159:152–161. doi: 10.1016/j.bpc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Hacker J. Whole genome plasticity in pathogenic bacteria. Curr Opin Microbiol. 2001;4:550–557. doi: 10.1016/S1369-5274(00)00250-2. [DOI] [PubMed] [Google Scholar]
- Collyn F, Billault A, Mullet C, Simonet M, Marceau M. YAPI, a new Yersinia pseudotuberculosis pathogenicity island. Infect Immun. 2004;72(8):4784–4790. doi: 10.1128/IAI.72.8.4784-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Hughes J, Moxon ER. Using the Genome to Understand Pathogenicity. Methods Mol Biol. 2004;266:261–287. doi: 10.1385/1-59259-763-7:261. [DOI] [PubMed] [Google Scholar]
- Fàbrega A, Vila J. Yersinia enterocolitica: Pathogenesis, virulence and antimicrobial resistance. Enferm Infecc Microbiol Clin. 2012;30(1):24–32. doi: 10.1016/j.eimc.2011.07.017. [DOI] [PubMed] [Google Scholar]
- McDonald C, Vacratsis PO, Bliska JB, Dixon JE. The Yersinia Virulence Factor YopM Forms a Novel Protein Complex with Two Cellular Kinases. J Biol Chem. 2003;278(20):18514–18523. doi: 10.1074/jbc.M301226200. [DOI] [PubMed] [Google Scholar]
- McCoy MW, Marré ML, Lesser CF, Mecsas J. The C-Terminal Tail of Yersinia pseudotuberculosis YopM Is Critical for Interacting with RSK1 and for Virulence. Infect Immun. 2010;78(6):2584–2598. doi: 10.1128/IAI.00141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JB, Mena P, Bliska JB. Delineation of Regions of the Yersinia YopM Protein Required for Interaction with the RSK1 and PRK2 Host Kinases and Their Requirement for Interleukin-10 Production and Virulence. Infect Immun. 2010;78(8):3529–3539. doi: 10.1128/IAI.00269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR. Bacterial Injectisomes: Needle Length Does Matter. Science. 2005;307(5713):1278. doi: 10.1126/science.1107679. [DOI] [PubMed] [Google Scholar]
- Durisin MD, Ibrahim A, Griffiths MW. Detection of pathogenic Yersinia enterocolitica using a digoxigenin labelled probe targeting the yst gene. J Appl Microbiol. 1998;84:285–292. doi: 10.1046/j.1365-2672.1998.00344.x. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. ProgressiveMauve: Multiple Genome Alignment with Gene Gain. Loss and Rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31(13):3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylip: Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colony hybridization experiment. A: nylon membrane from the liver of mouse number 13 (13 L), probed with Hem_8081. B: nylon membrane from A stripped and re-probed with Col_WA. C: original LB-agar plate. D: modified and superimposed images from A and B. (PDF 146 kb)
Assembly in scaffolds and accession numbers of the contigs ofY. enterocoliticastrain WA-314.
Predicted prophages inY. enterocoliticastrain WA-314. Excel table with prophage names, respective contigs, locus tags and encoded products.
Genetic structure of prophages inY. enterocoliticastrain WA-314. Annotation of selected genes is shown. YWA-1: possible degenerate P2-like prophage, 34.4 kbs. YWA-2: putative bacteriophage, 33 kbs. YWA-3: putative P2-like prophage, 11.7 kbs. YWA-4: putative P4-like prophage, 28 kbs. YWA-5: Mu-like prophage, 43.3 kbs. YWA-6: P4-like prophage, 14.6 kbs. YWA-7: putative defective prophage, 12.5 kbs.