Abstract
Determining the mechanism of action of bacterial growth inhibitors can be a formidable challenge in the progression of small molecules into antibacterial therapies. To help address this bottleneck, we have developed a robust transposon mutagenesis system using a suite of outward facing promoters in order to generate a comprehensive range of expression genotypes in Staphylococcus aureus from which to select defined compound-resistant transposon insertion mutants. Resistance stemming from either gene or operon over/under-expression, in addition to deletion, provides insight into multiple factors that contribute to a compound’s observed activity, including means of cell envelope penetration and susceptibility to efflux. By profiling the entire resistome, the suitability of an antibacterial target itself is also evaluated, sometimes with unanticipated results. We herein show that for the staphylococcal signal peptidase (SpsB) inhibitors, modulating expression of lipoteichoic acid synthase (LtaS) confers up to a 100-fold increase in the minimal inhibitory concentration. As similarly efficient transposition systems are or will become established in other bacteria and cell types, we discuss the utility, limitations and future promise of Tnp mutagenesis for determining both a compound’s mechanism of action and in the evaluation of novel targets.
Keywords: transposon mutagenesis, antibacterials, chemical genomics, Staphylococcus aureus, drug discovery, lipoteichoic acid, signal peptidase, arylomycin
Text
Transposons (Tnp) are mobile genetic elements capable of self excision and reinsertion within the genome at DNA sequence sites that vary depending on the particular transposon, from site specific to nearly random.1 As such, Tnp systems are powerful tools for altering gene expression on a genome wide scale, and Tnp mutagenesis is the enabling methodology behind numerous microbial forward genetic strategies.2,3 Tnp libraries have been extensively utilized to define essential gene sets through genetic footprinting, as well as to identify genes that confer fitness under challenge by negative selection as in signature tagged mutagenesis, transposon site hybridization and drug-dependent haploinsufficiency screens.4-7 Genes that are important for viability or growth are inferred by lack of transposon insertions or selective depletion from a given population, respectively, and these approaches have made valuable contributions to our understanding of microbial physiology and virulence.
One limitation with Tnp based approaches for identifying the molecular target of a small molecule growth inhibitor is that Tnp mediated resistance generally only arises through target independent mechanisms. Since the majority of antibiotics interact with essential gene products, target related Tnp insertions typically confer deleterious growth phenotypes that preclude representation. Consequently, Tnp mediated resistance to an inhibitor is confined to mechanisms involving disruption of nonessential genes. Deletion of a membrane transporter, of a transcription factor repressing the target gene, or of a branch pathway that consumes the substrate of the enzyme being targeted can all confer resistance but do not report on the molecular target itself.
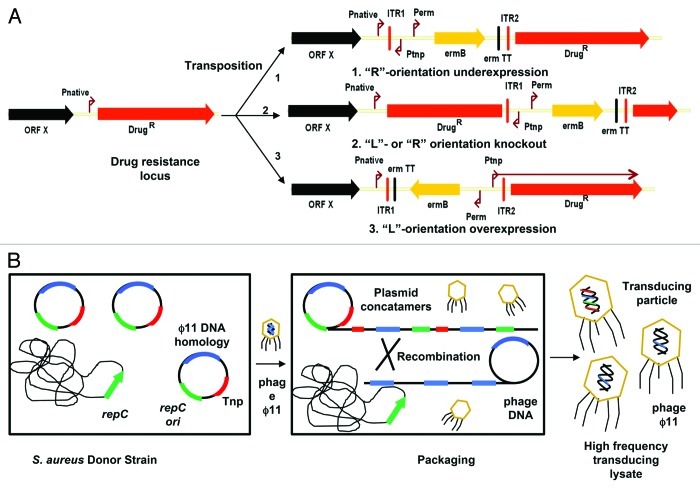
In order to directly implicate the target in addition to capturing these null allele resistant genotypes, we fitted a suite of HMAR mariner Tnp cassettes with a panel of outward facing promoters encompassing a range of promoter strengths for use in the gram positive pathogen Staphylococcus aureus.8 We envisioned that readthrough transcription from the Tnp cassette across the junction into neighboring genes could potentially confer resistance by overexpression of the target (Fig. 1A). Transcriptional coupling proximal to Tnp insertion sites is well documented in other genera, including Bacillus subtilis,9 Salmonella enterica,10-12 Vibrio cholerae13 and Escherichia coli.14 We opted to use a three member panel of outward directed promoters, with each promoter having different intrinsic strength and subject to unique temporal regulation. The opposite side of each Tnp cassette was fitted with a transcriptional terminator, which upon insertion in front of downstream target genes could prematurely terminate transcripts originating at the native promoter. Depending on the orientation of the Tnp cassette, the amount of transcript could therefore be increased or decreased by insertion of either the promoter or the transcriptional terminator, respectively, into the coding DNA strand. When combined with expression attenuation due to insertion site proximity, a wide range of downstream gene expression levels can thus be achieved through cis- acting polarity.
Figure 1. Modulation of drug resistance genes (DrugR) by Tnp insertion and strategy for bacteriophage mediated delivery. (A) A Tnp cassette with an outward facing promoter can reduce (pathway 1) or induce (pathway 3) expression of neighboring genes depending on insertion site location and orientation. Insertion of a transcriptional terminator (“R”-orientation) upstream of an open reading frame divorces the native promoter from the coding region by blocking transcription, whereas the opposite orientation (“L”-orientation) increases transcription by placing a promoter in front of the target gene. Insertion within the coding region destroys gene function, irrespective of orientation (pathway 2). Pnative- native promoter; ITR1/ITR2- inverted terminal repeat; Perm- erythromycin promoter; Ptnp- promoter on Tnp cassette; ermB- ribosomal methylase conferring erythromycin resistance; erm TT- transcriptional terminator of ermB gene. (B) An S. aureus donor strain is used to conditionally replicate a high copy number plasmid containing the Tnp cassette (red). A chromosomally encoded replication protein (repC) drives replication in trans by recognizing an origin (ori) retained on the donor plasmid (green). A short region of DNA (~1 kb) on the plasmid (blue) homologous to bacteriophage ϕ11 stimulates plasmid packaging into capsids presumably through recombination between phage and plasmid concatameric intermediates. High frequency transducing lysates containing Tnp concatamers are then transduced into S. aureus and dual resistance to inhibitor and erythromycin selected in situ.
To deliver the mini Tnp cassettes into recipient bacteria with high efficiency, we utilized bacteriophage to package and transduce plasmid DNA harboring Tnp cassettes (Fig. 1B). Plasmids with rolling circle-type replicons and as little as ~1 kb of bacteriophage DNA are transduced at remarkably high frequency via phage induced concatameric replication and bacteriophage-plasmid homologous recombination.15 Donor strains providing plasmid replication protein in trans were used as hosts for Tnp plasmids, which become packaged as non-replicating concatamers upon infection with generalized transducing phage. The packaging efficiency (1 in 3 progeny virus particles contain Tnp harboring DNA8) approaches that of specialized bacteriophage transduction Tnp delivery systems,16,17 while offering the advantages of working with small plasmids during the assembly of different Tnp promoter constructs. In order to realize highly efficient, non-biased and stable transposition, recipient S. aureus strains harbored a temperature sensitive plasmid constitutively expressing the HMAR mariner transposase. The mariner transposase inserts into substrate DNA between TA base dinucleotides with minimal regional bias,18 making it an ideal choice for generating insert site diversity in the AT-rich S. aureus genome. The unstable plasmid replicon ensured the transposase would be lost under non-selective growth conditions, preventing further transposition post selection. To prevent phage replication and cell lysis of recipient strains, we either inserted the cI-like repressor gene (ORF5 of bacteriophage ϕ11) into the chromosome to block phage replication (as with S. aureus RN4220) or used strains that were already resistant due to resident prophages (as in methicillin resistant S. aureus COL). The high titer transducing lysate coupled with an optimized transposition protocol routinely achieved 1 transposant per ~104 recipient CFU in S. aureus RN4220,8 allowing high quality Tnp mutant libraries to be generated and screened in situ for dual resistance to the Tnp selection marker (erythromycin) and the growth inhibitor under study. As bacteriophage induced high frequency transduction of rolling circle type plasmids is a generalized mechanism common to many bacteria,19 this Tnp delivery approach may be of broader utility.
With a highly efficient Tnp system in hand, we then tested a panel of control antibiotics with diverse mechanisms of action (MOA) to ascertain whether all types of gene expression related resistance [underexpression, overexpression, and null] could be uncovered in a single experiment (Fig. 2).8 In a typical experiment, a ~2 × 106 member Tnp library (providing 2 to 3–fold bi-directional insertion site coverage at each genomic TA dinucleotide position) was suspended in top agar and plated over selective media in a single Petri dish to isolate transposants that had acquired drug resistance. Multiple colonies were then sequenced to determine Tnp insertion site and orientation bias. By analyzing the Tnp insertion pattern and genomic context, resistance associated gene/operon candidates were implicated with high confidence for the majority of cases.8 For instance, subsets of Tnp mutants clustering upstream in a single overexpression orientation suggested that upregulation of a downstream target gene imparts resistance (as seen with overexpression of the triclosan target fabI), while random nondirectional insertion within a protein coding region was consistent with resistance due to gene deletion (as seen with a subset of indolmycin resistant mutants clustering within guaA).
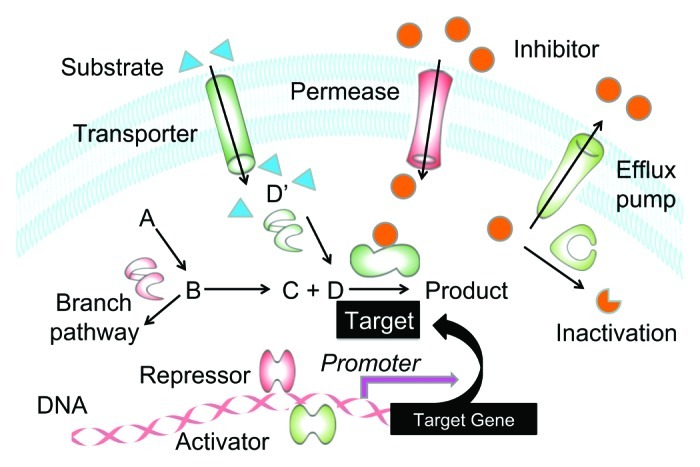
Figure 2. Inhibitor resistance mechanisms amenable to detection by Tnp mutagenesis using a suite of outward facing promoters. Resistance to a particular compound can be imparted by direct overexpression of target by Tnp promoter insertion (pink arrow), or indirectly by either off target gene induction (green shaded) or underexpression/deletion (red shaded). Upregulation of transcriptional activators controlling target gene expression, of transporters that import extracellular nutrients utilized by the targeted pathway, of periphery enzymes funneling substrates into the target pathway, of efflux pumps that remove inhibitor and of inhibitor modifying/degrading enzymes can all lead to phenotypic resistance. Likewise, downregulation or deletion of a permease utilized by an inhibitor to enter the cell, of a target gene transcriptional repressor or of a branch pathway can all suppress growth inhibition by decreasing intracellular compound concentration, by increasing amounts of target protein, and by redirecting pathway flux. For examples of Tnp mediated resistance mechanisms to specific antibodies, see Wang, et al.8
In this approach, the overexpression class of Tnp mutants is arguably the most important in regard to target identification. Transposants upregulating target genes clustered anywhere from immediately 5′ to as far as 7 kb upstream.8 A common theme among the Tnp target overexpressing transposants was a strong bias to keep the total increased transcript level bounded within a relatively narrow range; sufficient to impart resistance under the antibiotic challenge conditions but not enough to impact fitness. High-level expression of essential genes often impairs growth as pathway flux is altered. Rate limiting steps can be changed, accumulated intermediates are often competitive inhibitors of downstream pathway enzymes, and regulatory feedback mechanisms are ablated. Consistent with this, most Tnp mutants that arose through overexpression of the target were only modestly shifted [2- to 4-fold shift in minimum inhibitory concentration (MIC)]. While a minor MIC shift in liquid broth, substantial growth advantages of antibiotic resistant mutants often occurs at concentrations well below the final MIC.20,21 This makes diffusion agar (where compound diffuses into an agar overlay impregnated with bacteria) an ideal choice for differentiating low level Tnp mediated resistance. Sampling a range of expressions is critical in order to ensure adequate inhibitor resistance without compromising fitness through overexpression. By introducing promoters with different intrinsic strength, in contrast to using a regulated promoter with varied concentrations of inducer, competitive outgrowth and selection could be performed within the same agar plate without the need for interplate comparisons. Selection conditions are thus optimal for differentiating transposants with minor fitness advantages. In some cases, drug resistant transposants were selectable despite having 2-fold MIC increase when measured by standard liquid broth assay.8
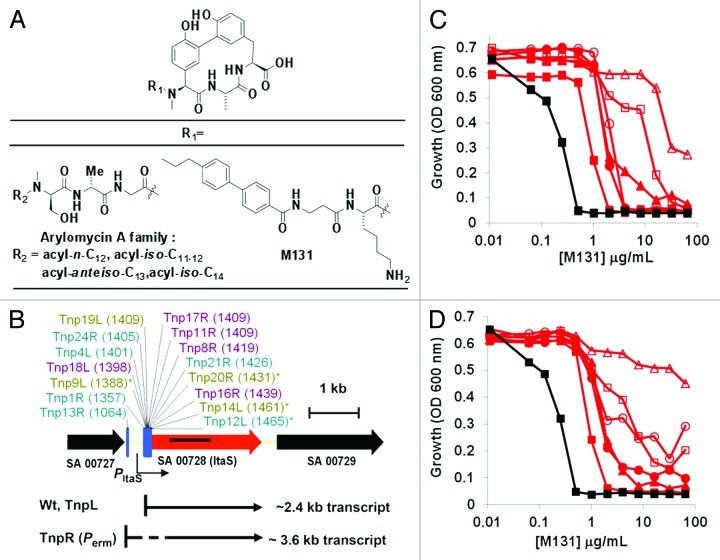
Tnp libraries are useful for revealing off target resistance mechanisms as well, which in many ways are as valuable as the on target information. Resistance mechanisms not related to the compound, but rather to the biology of the target being engaged, cannot be addressed by simply making better molecules. The diverse range of genotypes produced by Tnp mutagenesis probes for potential compensatory mechanisms, and can reveal critical functional genetic interactions not previously anticipated during standard target evaluation. For instance, bacterial signal peptidases are membrane bound endopeptidases that cleave the N-terminal leader sequence from nascently transported pre-proteins destined for extracellular roles.22,23 Being located in the relatively drug accessible environment of the outer leaflet of the cytoplasmic membrane, having a nonconventional Ser-Lys catalytic dyad active site that is unique among serine proteases, and being essential to bacterial viability, the type I signal peptidase SpsB has been extensively investigated as a new antibacterial target in S. aureus.24-27 Genetic potentiation of SpsB by antisense interference restores the activity of β-lactams against methicillin resistant S. aureus (MRSA),28 also making SpsB an attractive target for novel β-lactam combination therapies. Promising molecular scaffolds that inhibit SpsB have been identified, including the β-lactam (5S,6S)-penem core29 and most notably the natural product arylomycins.30,31 Starting with the lead bi-aryl bridged lipopeptide arylomycin A (Fig. 3A), we developed a more potent series represented by analog M131 that demonstrated good whole cell activity against the model MRSA strain COL (MIC = 0.5–1 μg/mL).32 We applied the Tnp system to explore non SpsB related routes to M131 (the experimental protocol for Tnp mutagenesis,8 northern blotting,8 immunoblotting36 and drug susceptibility testing28 have all been previously described). A 15 member subset of highly resistant Tnp mutants with insertions that mapped directly 5′ to the lipoteichoic acid synthase (ltaS) gene was identified (Fig. 3B). Lipoteichoic acid (LTA) is an important cell envelope component that is critical for proper cell division and morphogenesis in S. aureus.33-35 LTA consists of a glycerolphosphate (GroP) polymer connected to a membrane embedded diacyl glycerol gentiobiose unit. The LtaS enzyme synthesizes the GroP polymer using diacyl glycerol gentiobiose as acceptor and phosphatidyl glycerol as the GroP donor on the extracellular side of the membrane.36 Resistance levels to M131 increased markedly for some S. aureus COL transposants, with more than a 100-fold MIC increase in comparison to the wildtype (Fig. 3C and D). Surprisingly, no Tnp orientation bias was observed as either rightward (R, underexpression) or leftward (L, retaining expression) facing inserts imparted resistance. No insertions were isolated within the ltaS open reading frame, consistent with an essential role for LtaS/LTA in S. aureus viability and general fitness.36
Figure 3. Tnp mediated resistance to the arylomycin derivative M131. (A) Structure of the natural product arylomycin A family and of the analog M131.32 (B) The M131 resistant transposants were selected in S. aureus COL with a final post diffusion concentration of 2 μg/mL of M131 as described.8 All 15 resistant transposants harbored insertions immediately 5′ to the lipoteichoic acid synthase gene ltaS. Inserts are labeled with transposant isolate number (Tnp#), orientation [Tnp promoter (L) or transcriptional terminator (R) on coding strand], which promoter [Pcap (teal), Ppen (purple) or Ptuf (olive)], and base pair insertion position relative to extreme left of sequence (base pair 1).8 Transposant labels marked with an asterisk (*) denote concatameric insertion events. The putative position of the native ltaS promoter (arrow) and the location of the ltaS probe (black bar) used for northern blotting in Figure 4A is indicated. Gene locus numbers correspond to those used in the reference S. aureus NCTC 8325 genome. (C) Dose-response growth curve of S. aureus COL M131 resistant transposants that underexpress ltaS (“R”-orientation). Strains were innoculated into TSB (~5x105 CFU/mL) and grown overnight at 37°C. M131 sensitive wildtype- black squares; M131 resistant transposants- red [solid circle (Tnp1R), open circle (Tnp8R), solid square (Tnp13R), open square (Tnp16R), solid triangle (Tnp17R), open triangle (Tnp20R)]. (D) Dose-response growth curve of S. aureus COL M131 resistant transposants with disregulated ltaS expression (“L”-orientation). Experimental details are as in Figure 3C. M131 sensitive wildtype- black squares; M131 resistant transposants- red [open circle (Tnp4L), open triangle (Tnp9L), solid square (Tnp12L), open square (Tnp14L), solid circle (Tnp18L), solid triangle (Tnp19L)].
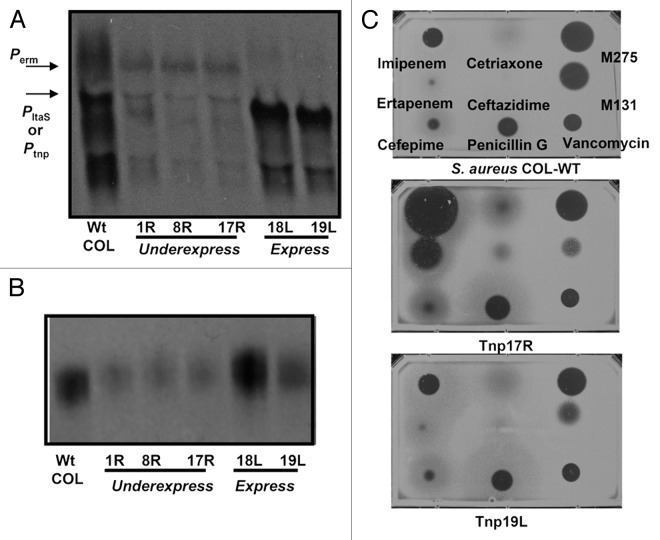
To confirm the anticipated polarity, we analyzed total RNA transcripts for ltaS message by northern blotting (Fig. 4A). Transcripts encoding ltaS of the predicted size and relative quantity were identified; robust ltaS expression was only observed when the Tnp promoter was on the coding strand (“L”-orientation) while just a trace of transcript when on the opposite strand (“R”-orientation). Lengths were consistent with transcription initiating from either the Tnp promoter or from transcriptional terminator read through by the erythromycin promoter, respectively. The levels of LTA glycolipid were directly compared by immunoblotting (Fig. 4B). Consistent with the levels of ltaS mRNA, only the “L”-orientation Tnp mutants retained LTA amounts comparable to wildtype. We then confirmed reduced LTA expression in the “R”-orientation Tnp mutants by measuring β-lactam susceptibility in an agar diffusion assay (Fig. 4C). Since decreasing LTA levels increases β-lactam susceptibility,37,38 only the “R”-orientation Tnp subset should have enlarged zones of clearing in a halo assay. Indeed, all of the underexpressing ltaS Tnp insertion mutants (“R”-orientation) were specifically more susceptible to β-lactam antibiotics (particularly imipenem and ertapenem). Only the most distal insert Tnp13R, which is also the least resistant to M131 (Fig. 3C), did not display notably enlarged zones of clearing and likely reflects some degree of residual native ltaS promoter activity. The ltaS expressing set (“L”-orientation), in comparison, phenotypically mirrored wildtype for all test antibiotics and were not shifted except for being more resistant to M131.
Figure 4. Northern and LTA immunoblot analysis of M131 resistant Tnp mutants. (A) Northern blot analysis of ltaS transcripts in selected M131 resistant transposants using 5 μg of total mRNA. Transposant number and Tnp orientation are noted. Transcripts lengths and the location of the ltaS specific probe is depicted in Figure 3B. Original image used with permission from Nature Chemical Biology. (B) LTA immunoblot using anti polyglycerophosphate specific monoclonal antibody mAB55 as described.36 (C) Halo drug susceptibility assay of a representative member from each Tnp subset using agar seeded with S. aureus COL wt (top panel), the ltaS disregulated strain Tnp17R (middle panel) or the ltaS expressing strain Tnp19L. Zones of clearing indicate sensitivity to spotted drug. The control compound M275 is a putative wall teichoic acid inhibitor (unpublished).
A regulated IPTG-inducible ltaS strain in S. aureus COL was constructed (Fig. 5) to independently confirm the inverse relationship between ltaS gene dose and SpsB inhibitor resistance suggested by the underexpressing “R”-orientation Tnp subset (Fig. 5). Since the 5′ intergenic region upstream of ltaS is intact in ANG2506, an M131 resistant phenotype in this strain would rule out the possibility that M131 resistance actually arises through disruption of a small unannotated open reading located immediately upstream of ltaS (Fig. 3B). LTA immunoblotting of cell associated extracts from the ltaS inducible strain ANG2506 revealed tight regulation of LTA glycolipid biosynthesis when comparing noninducing [tryptic soy broth (TSB) only] to inducing (TSB plus 1 mM IPTG) growth conditions (Fig. 6A). However, while IPTG addition restored LTA production to near wildtype levels, minimal LtaS protein was detected by western blotting using an LtaS specific antibody under either non-inducing or inducing conditions (Fig. 6B). The S. aureus COL strain apparently tolerates a large depletion in ltaS expression before the amount of LTA decreases. We thus chose to examine M131 resistance under ltaS inducing conditions (TSB plus 1 mM IPTG), as the growth rate was similar to wildtype, the β-lactam potentiation was minimal (data not shown) and yet the amount of LtaS was reduced. The M131 MIC in ANG2506 increased 8-fold (Fig. 6C), confirming that underexpression of ltaS imparts resistance to M131 as predicted by the Tnp results.
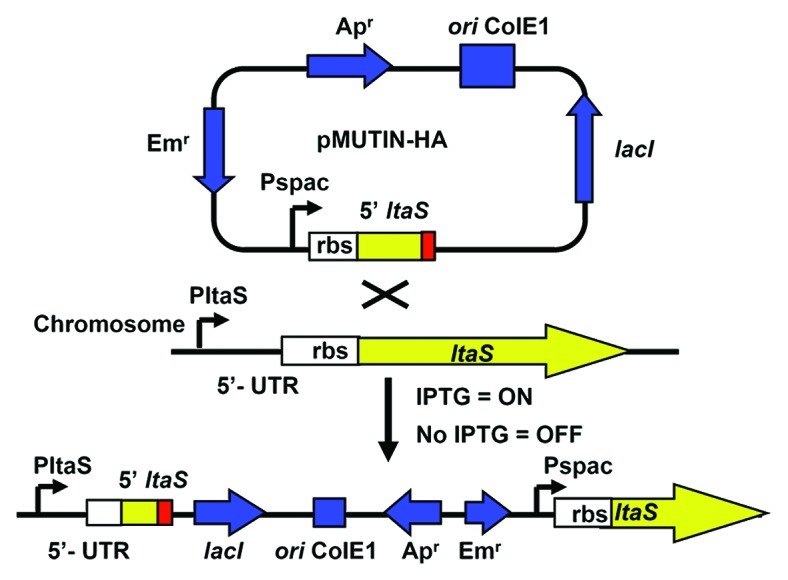
Figure 5. Construction and analysis of IPTG-inducible ltaS in S. aureus COL. The plasmid pMUTIN-HA containing an N-terminal fragment of ltaS (truncated reading frame ending at box) in front of the IPTG inducible promoter Pspac was first integrated into S. aureus strain RN4220.36 The regulated ltaS construct was then transduced into S. aureus COL using bacteriophage 85 to give strain ANG2506. Note the intergenic region located 5′ to ltaS remains intact after integration of plasmid.
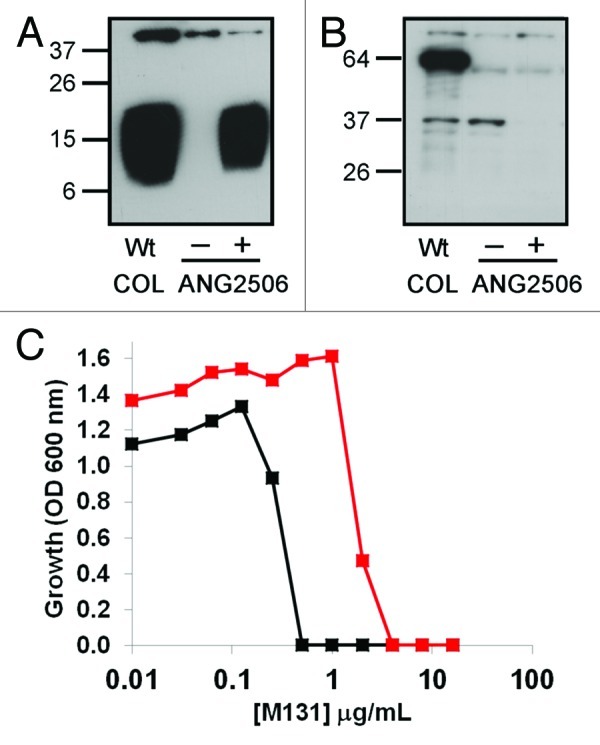
Figure 6. Characterization of the IPTG inducible ltaS strain ANG2506. (A) LTA immunoblot analysis using cell associated fraction of ANG2506 grown under noninducing (TSB only) and inducing (plus 1 mM IPTG) conditions at 37°C. (B) western blot analysis of cell associated LtaS protein from ANG2506 grown in the presence (+) and absence (─) of 1 mM IPTG. The ~64 kDa band corresponds to the soluble cleaved LtaS fragment eLtaS previously observed in S. aureus strains.39,52 Under these conditions, the membrane bound full length LtaS protein is not detectable for all samples. (C) Dose-response growth curve of wildtype S. aureus COL ANG2506 (seeded at ~5x105 CFU/mL) in TSB supplemented with 1 mM IPTG and various concentrations of M131 at 37°C; COL wildtype- black; ANG2506- red.
Intriguingly, both underexpression (in the Tnp “R”-orientation subset and ANG2506) and disregulation (in the Tnp “L”-orientation subset) of ltaS in S. aureus COL confer substantial resistance to the SpsB specific inhibitor M131. A similar pattern of resistance was seen with a second SpsB inhibitor from a distinct chemical class (data not shown), suggesting the LtaS-SpsB inhibitor resistance connection is not related to the intrinsic molecular properties of M131 (i.e., LTA dependent cell penetration/exclusion). Mature LtaS is cleaved in staphylococci by SpsB at a non canonical amino acid recognition site,39,40 which in turn may act as a regulatory mechanism by inactivating LtaS.39 It is conceivable that LtaS activity is somehow transmitted or sensed by the cell, inducing an SpsB inhibitor tolerant phenotype. Antagonism of SpsB inhibition through LTA/ltaS expression modulation thus provides an effective target-independent route for pan-resistance to SpsB inhibitors in S. aureus COL, and perhaps in other S. aureus strains. Indeed, significant strain-to-strain MIC variability and high MIC90 values have been reported for arylomycin type inhibitors.41-43 Mechanistically, the basis of LtaS suppression of SpsB inhibition is unclear. However, the use of genome wide Tnp mutagenesis enhanced with polarity modulating cassettes in evaluating antibacterial targets by revealing otherwise unforeseen resistance mechanisms is apparent.
The most prominent class of inhibitors for which we failed to get direct target information through Tnp mediated overexpression was for ribosome inhibitors.8 Being a multi subunit nucleic acid-protein particle, upregulation of a single ribosomal gene would not necessarily be expected to impart resistance. In comparison, downregulation of a single gene component of a ribosome or of other complexes generally does impart hypersensitization to cognate inhibitors.44 To extend the usefulness of Tnp mutagenesis in identifying inhibitor targets belonging to multi-subunit complexes, generating a gradient of underexpression genotypes from which to find transposants with reduced fitness is an attractive strategy. Tnp cassettes harboring transcriptional terminators of varying strength would complement the promoter overexpression Tnp system described above, providing more detailed MOA information when applied in tandem. Attenuation of transcriptional termination through Tnp insertion has potential advantages over simply using Tnp cassettes with weak promoters, as (1) the native promoter is left intact, (2) temporal promoter activity and regulation is retained and (3) a small number of terminator sequence variants is needed since the fractional decrease in transcript abundance is intrinsic to the terminator itself and independent of the native promoter strength. The second challenge to implementing such a system is to identify Tnp insertion sites that become underrepresented when counterselected with inhibitor. High density oligonucleotide tiled microarrays45-47 and more recently next generation sequencing technologies48,49 are proving to be highly efficient in mapping Tnp junction sites among large transposant pools with base pair resolution. When disrupted by Tnp insertion, genes that confer sensitivity to aminoglycosides in Pseudomonas aeruginosa50 and to bile acid in Salmonella typhymurium51 have been identified using this approach. Developing methods for generating and probing Tnp libraries biased toward representing underexpression genotypes is likewise a promising avenue for antibacterial target elucidation.
Technological advances in molecular biology and bacterial genetics have made it possible to build genome wide ordered libraries with predefined genetic content, whether it be plasmid based overexpression, regulated antisense knockdown or gene knockout. While arrayed libraries have been widely adopted in systematic MOA antibacterial studies and in some ways have even supplanted traditional Tnp based approaches, the simplicity and power of Tnp mutagenesis should not be overlooked. The ability of any genetic approach to elucidate inhibitor resistance mechanisms is in many ways a function of the number of distinct genotypes generated and then screened for a given phenotype. The bacteriophage based delivery system we developed in S. aureus generates ~2x106 nonclonal transposants equipped with outward facing promoters that can be interrogated for resistance on a single agar plate. The cost of creating, curating and screening a pre-arrayed library of this size would be prohibitive for routine analysis. Further, Tnp mutagenesis systems can easily be moved to different strain backgrounds if need be. This is important as unoptimized lead molecules identified in cell growth assays often exhibit strain dependent MIC values that can vary substantially, even showing strain specific resistance mechanisms (M131, case in point). Tnp mutagenesis, particularly when fitted with polarity attenuating modules and combined with newer technology to map insertion sites, thus remains and will continue to be an attractive first line tool of choice for antibacterial MOA and target evaluation studies.
Acknowledgments
We would like to thank Alex Therien and Michel Gallant for insights into SpsB inhibitors and MOA discussions. A.G acknowledges funding from the European Research Council under ERC-StG grant LiSta-LTA 260371.
Glossary
Abbreviations:
- Tnp
transposon
- MOA
mechanism of action
- MIC
minimal inhibitory concentration
- LTA
lipoteichoic acid
- MRSA
methicillin resistant S. aureus
- TSB
tryptic soy broth
- GroP
glycerol phosphate
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/21647
References
- 1.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, D.C.: ASM Press, 2002. [Google Scholar]
- 2.Choi KH, Kim KJ. Applications of transposon-based gene delivery system in bacteria. J Microbiol Biotechnol. 2009;19:217–28. doi: 10.4014/jmb.0811.669. [DOI] [PubMed] [Google Scholar]
- 3.Hayes F. Transposon-based strategies for microbial functional genomics and proteomics. Annu Rev Genet. 2003;37:3–29. doi: 10.1146/annurev.genet.37.110801.142807. [DOI] [PubMed] [Google Scholar]
- 4.Judson N, Mekalanos JJ. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 2000;8:521–6. doi: 10.1016/S0966-842X(00)01865-5. [DOI] [PubMed] [Google Scholar]
- 5.Lehoux DE, Sanschagrin F, Levesque RC. Discovering essential and infection-related genes. Curr Opin Microbiol. 2001;4:515–9. doi: 10.1016/S1369-5274(00)00244-7. [DOI] [PubMed] [Google Scholar]
- 6.Reznikoff WS, Winterberg KM. Transposon-based strategies for the identification of essential bacterial genes. Methods Mol Biol. 2008;416:13–26. doi: 10.1007/978-1-59745-321-9_2. [DOI] [PubMed] [Google Scholar]
- 7.Oh J, Fung E, Schlecht U, Davis RW, Giaever G, St Onge RP, et al. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog. 2010;6:e1001140. doi: 10.1371/journal.ppat.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Claveau D, Vaillancourt JP, Roemer T, Meredith TC. High-frequency transposition for determining antibacterial mode of action. Nat Chem Biol. 2011;7:720–9. doi: 10.1038/nchembio.643. [DOI] [PubMed] [Google Scholar]
- 9.Bordi C, Butcher BG, Shi Q, Hachmann AB, Peters JE, Helmann JD. In vitro mutagenesis of Bacillus subtilis by using a modified Tn7 transposon with an outward-facing inducible promoter. Appl Environ Microbiol. 2008;74:3419–25. doi: 10.1128/AEM.00476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C, Wozniak C, Karlinsey JE, Hughes KT. Genomic screening for regulatory genes using the T-POP transposon. Methods Enzymol. 2007;421:159–67. doi: 10.1016/S0076-6879(06)21014-0. [DOI] [PubMed] [Google Scholar]
- 11.Rappleye CA, Roth JRA. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J Bacteriol. 1997;179:5827–34. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salipante SJ, Barlow M, Hall BG. GeneHunter, a transposon tool for identification and isolation of cryptic antibiotic resistance genes. Antimicrob Agents Chemother. 2003;47:3840–5. doi: 10.1128/AAC.47.12.3840-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Judson N, Mekalanos JJ. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat Biotechnol. 2000;18:740–5. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 14.Chow WY, Berg DE. Tn5tac1, a derivative of transposon Tn5 that generates conditional mutations. Proc Natl Acad Sci U S A. 1988;85:6468–72. doi: 10.1073/pnas.85.17.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick RP, Edelman I, Lofdahl S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J Mol Biol. 1986;192:209–20. doi: 10.1016/0022-2836(86)90360-8. [DOI] [PubMed] [Google Scholar]
- 16.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, et al. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10961–6. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–80. doi: 10.1016/0076-6879(91)04009-D. [DOI] [PubMed] [Google Scholar]
- 18.Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–9. [PMC free article] [PubMed] [Google Scholar]
- 19.Viret JF, Bravo A, Alonso JC. Recombination-dependent concatemeric plasmid replication. Microbiol Rev. 1991;55:675–83. doi: 10.1128/mr.55.4.675-683.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu A, Fong A, Becket E, Yuan J, Tamae C, Medrano L, et al. Selective advantage of resistant strains at trace levels of antibiotics: a simple and ultrasensitive color test for detection of antibiotics and genotoxic agents. Antimicrob Agents Chemother. 2011;55:1204–10. doi: 10.1128/AAC.01182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, et al. Type I signal peptidases of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:279–97. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Tuteja R. Type I signal peptidase: an overview. Arch Biochem Biophys. 2005;441:107–11. doi: 10.1016/j.abb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Black MT, Bruton G. Inhibitors of bacterial signal peptidases. Curr Pharm Des. 1998;4:133–54. [PubMed] [Google Scholar]
- 25.Paetzel M, Dalbey RE, Strynadka NC. The structure and mechanism of bacterial type I signal peptidases. A novel antibiotic target. Pharmacol Ther. 2000;87:27–49. doi: 10.1016/S0163-7258(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 26.Rao S, Bockstael K, Nath S, Engelborghs Y, Anné J, Geukens N. Enzymatic investigation of the Staphylococcus aureus type I signal peptidase SpsB - implications for the search for novel antibiotics. FEBS J. 2009;276:3222–34. doi: 10.1111/j.1742-4658.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 27.Smitha Rao CV, Anné J. Bacterial type I signal peptidases as antibiotic targets. Future Microbiol. 2011;6:1279–96. doi: 10.2217/fmb.11.109. [DOI] [PubMed] [Google Scholar]
- 28.Lee SH, Jarantow LW, Wang H, Sillaots S, Cheng H, Meredith TC, et al. Antagonism of chemical genetic interaction networks resensitize MRSA to β-lactam antibiotics. Chem Biol. 2011;18:1379–89. doi: 10.1016/j.chembiol.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Harris DA, Powers ME, Romesberg FE. Synthesis and biological evaluation of penem inhibitors of bacterial signal peptidase. Bioorg Med Chem Lett. 2009;19:3787–90. doi: 10.1016/j.bmcl.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 30.Schimana J, Gebhardt K, Höltzel A, Schmid DG, Süssmuth R, Müller J, et al. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp. Tü 6075. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot (Tokyo) 2002;55:565–70. doi: 10.7164/antibiotics.55.565. [DOI] [PubMed] [Google Scholar]
- 31.Kulanthaivel P, Kreuzman AJ, Strege MA, Belvo MD, Smitka TA, Clemens M, et al. Novel lipoglycopeptides as inhibitors of bacterial signal peptidase I. J Biol Chem. 2004;279:36250–8. doi: 10.1074/jbc.M405884200. [DOI] [PubMed] [Google Scholar]
- 32.Therien AG, Huber JL, Wilson KE, Beaulieu P, Caron A, Claveau D, et al. Broadening the spectrum of β-lactam antibiotics through inhibition of signal peptidase type I. Antimicrob Agents Chemother. 2012;56:4662–70. doi: 10.1128/AAC.00726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichmann NT, Gründling A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett. 2011;319:97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol. 2010;300:148–54. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Fischer W. Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus. Med Microbiol Immunol. 1994;183:61–76. doi: 10.1007/BF00277157. [DOI] [PubMed] [Google Scholar]
- 36.Gründling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8478–83. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohta K, Komatsuzawa H, Sugai M, Suginaka H. Triton X-100-induced lipoteichoic acid release is correlated with the methicillin resistance in Staphylococcus aureus. FEMS Microbiol Lett. 2000;182:77–9. doi: 10.1111/j.1574-6968.2000.tb08877.x. [DOI] [PubMed] [Google Scholar]
- 38.Stapleton PD, Shah S, Ehlert K, Hara Y, Taylor PW. The beta-lactam-resistance modifier (-)-epicatechin gallate alters the architecture of the cell wall of Staphylococcus aureus. Microbiology. 2007;153:2093–103. doi: 10.1099/mic.0.2007/007807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wörmann ME, Reichmann NT, Malone CL, Horswill AR, Gründling A. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J Bacteriol. 2011;193:5279–91. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powers ME, Smith PA, Roberts TC, Fowler BJ, King CC, Trauger SA, et al. Type I signal peptidase and protein secretion in Staphylococcus epidermidis. J Bacteriol. 2011;193:340–8. doi: 10.1128/JB.01052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts TC, Schallenberger MA, Liu J, Smith PA, Romesberg FE. Initial efforts toward the optimization of arylomycins for antibiotic activity. J Med Chem. 2011;54:4954–63. doi: 10.1021/jm1016126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith PA, Roberts TC, Romesberg FE. Broad-spectrum antibiotic activity of the arylomycin natural products is masked by natural target mutations. Chem Biol. 2010;17:1223–31. doi: 10.1016/j.chembiol.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith PA, Powers ME, Roberts TC, Romesberg FE. In vitro activities of arylomycin natural-product antibiotics against Staphylococcus epidermidis and other coagulase-negative staphylococci. Antimicrob Agents Chemother. 2011;55:1130–4. doi: 10.1128/AAC.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Ondeyka JG, Zink DL, Basilio A, Vicente F, Salazar O, et al. Discovery of okilactomycin and congeners from Streptomyces scabrisporus by antisense differential sensitivity assay targeting ribosomal protein S4. J Antibiot (Tokyo) 2009;62:55–61. doi: 10.1038/ja.2008.8. [DOI] [PubMed] [Google Scholar]
- 45.Murry JP, Sassetti CM, Lane JM, Xie Z, Rubin EJ. Transposon site hybridization in Mycobacterium tuberculosis. Methods Mol Biol. 2008;416:45–59. doi: 10.1007/978-1-59745-321-9_4. [DOI] [PubMed] [Google Scholar]
- 46.Badarinarayana V, Estep PW, 3rd, Shendure J, Edwards J, Tavazoie S, Lam F, et al. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat Biotechnol. 2001;19:1060–5. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- 47.Winterberg KM, Reznikoff WS. Screening transposon mutant libraries using full-genome oligonucleotide microarrays. Methods Enzymol. 2007;421:110–25. doi: 10.1016/S0076-6879(06)21011-5. [DOI] [PubMed] [Google Scholar]
- 48.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–72. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Febrer M, McLay K, Caccamo M, Twomey KB, Ryan RP. Advances in bacterial transcriptome and transposon insertion-site profiling using second-generation sequencing. Trends Biotechnol. 2011;29:586–94. doi: 10.1016/j.tibtech.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2:e00315–10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–16. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu D, Wörmann ME, Zhang X, Schneewind O, Gründling A, Freemont PS. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc Natl Acad Sci U S A. 2009;106:1584–9. doi: 10.1073/pnas.0809020106. [DOI] [PMC free article] [PubMed] [Google Scholar]