Abstract
Drosophila spermatogenesis has become a paradigmatic system for the study of mechanisms that regulate adult stem cell maintenance, proliferation and differentiation. The dramatic cellular differentiation process from germline stem cell (GSC) to mature sperm is accompanied by dynamic changes in gene expression, which are regulated at transcriptional, post-transcriptional (including translational) and post-translational levels. Post-transcriptional regulation has been proposed as a unique feature of germ cells. However, recent studies have provided new insights into transcriptional regulation during Drosophila spermatogenesis. Both signaling pathways and epigenetic mechanisms act to orchestrate the transcriptional regulation of distinct genes at different germ cell differentiation stages. Many of the regulatory pathways that control male gamete differentiation in Drosophila are conserved in mammals. Therefore, studies using Drosophila spermatogenesis will provide insight into the molecular mechanisms that regulate mammalian germ cell differentiation pathways.
Keywords: germline stem cell, cyst stem cell, niche, spermatogonia, spermatocyte, spermatid, Drosophila testis, spermatogenesis
Overview of Drosophila Spermatogenesis
The Drosophila testis is a long tubular structure with a stem cell niche at the apical tip and a linear distribution of germ cells that are progressively differentiated toward the basal end of the tube. A cluster of post-mitotic cells, termed the hub, anchors both germline stem cells (GSCs) and cyst stem cells (CySCs) in the testis niche. The GSCs divide asymmetrically to self-renew and produce gonialblasts (GBs), which are displaced from the niche to start the cellular differentiation program. Each GB undergoes four rounds of mitosis as transit-amplifying spermatogonial cells that remain interconnected by incomplete cytokinesis. These 16 spermatogonia then undergo a pre-meiotic S phase before switching to an elongated G2 phase as spermatocytes. A robust transcription program is turned on in spermatocytes to actively express genes required for meiotic division and terminal differentiation.1
Along with GSC division, the CySCs, two of which encapsulate each GSC, also divide asymmetrically.2 While one daughter cell retains its stem cell identity, the other becomes a cyst cell, which never divides again. These two cyst cells continue to enclose the synchronously dividing and differentiating germ cells to form a distinct cyst until the individualization stage when the spermatids are separated, followed by release into the seminal vesicle as mature sperm.
Because of the physical association between cyst cells and germ cells, these two cell types act cooperatively throughout spermatogenesis. Here we will review transcriptional regulation of Drosophila spermatogenesis in a stepwise manner and in the context of the intimate soma-germline interaction.
Transcriptional Regulation in the Stem Cell Niche
Adult stem cells normally reside in a microenvironment called the niche. The Drosophila male GSC niche is one of the best characterized niches. In this niche, GSCs associate with two types of somatic cells: hub cells located at the tip of the testis and CySCs. The niche provides a polarized extrinsic environment where GSCs are maintained through cell-cell adhesion and niche-to-GSC signaling.3 At the cellular level, GSCs undergo stereotypical asymmetric cell division,4,5 and at the molecular level, GSCs probably maintain a unique chromatin structure and gene expression profile.6,7 Because both signaling pathways and epigenetic mechanisms change the transcriptional profile of cells, we will discuss both of them here. Although highly anticipated, a direct link between these two mechanisms has not been reported in the male GSC niche.
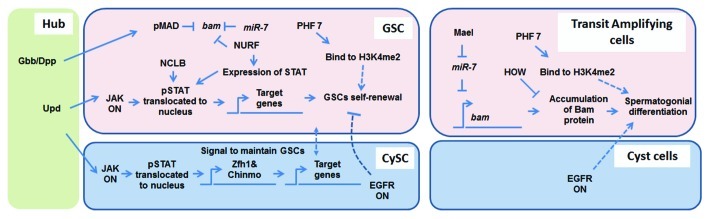
Two major signaling pathways play important roles in the male GSC niche: Bone Morphogenetic Protein (BMP) pathway and Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) pathway. In both pathways, ligands [Glass bottom boat (Gbb) and Decapentaplegic (Dpp) for BMP; Unpaired (Upd) for JAK-STAT] emanating from the niche act upon their corresponding receptors at stem cells [Saxophone (Sax), Thick veins (Tkv) and Punt for BMP; Domeless for JAK-STAT] to promote phosphorylation and translocation of their downstream transcription factors [Mothers against Dpp (Mad) for BMP; Stat92E for JAK-STAT].8-13 A recent study revealed that a regulator of BMP signaling called Magu is specifically expressed in hub cells and required for GSC maintenance.14 Activated transcription factors subsequently initiate a cascade of gene expression in GSCs and CySCs. Although direct target genes of both pMad and pStat92E have been identified in other cell types in Drosophila,15-18 their direct targets in GSCs or CySCs remain unidentified. However, it has been shown that GSCs unable to respond to the BMP pathway have ectopic transcription of a differentiation gene called bag of marbles (bam), which leads these GSCs to undergo premature differentiation and leave the niche.12,13 Therefore, it is possible that normal BMP activity represses bam transcription in male GSCs, just as it does in female GSCs.15 Recent studies demonstrate that the major role of JAK-STAT in GSCs is to increase GSC-hub adhesion,11 suggesting that cell-cell adhesion molecules, such as Drosophila E-cadherin homolog (DE-cadherin, DE-cad), are potential downstream targets of Stat92E. To search for Stat targets at a genome-wide level, microarray analysis was performed to identify genes whose expression dramatically changes in response to hyperactivated Stat.19 Interestingly, validation of the Stat-responsive genes revealed that most of them are expressed in CySCs instead of GSCs, suggesting that active Stat signaling in somatic cells predominates and is required for maintaining GSCs. Consistent with this finding, ectopic expression of the Stat92E target genes Zinc-finger homeodomain protein 1 (Zfh-1) or Chronologically inappropriate morphogenesis (Chinmo), both encoding transcription factors, in cyst cells is sufficient for GSC self-renewal outside of the niche.10,20 Zfh-1 has been implicated in guiding GSC self-renewal, probably by activating BMP signaling in CySCs,10,11 as well as providing crosstalk between the BMP and JAK-STAT pathways. Another particularly interesting Stat target gene in somatic cells is Suppressor of cytokine signaling 36E (Socs36E), which encodes an antagonist of the JAK-STAT pathway and acts to maintain a balanced ratio of CySCs and GSCs in the niche (Fig. 1 and Table 1).21 Because the microarray analysis was performed using the entire tissue (i.e., testes), it does not provide a cell type-specific transcriptional profile. Furthermore, some of the identified target genes are transcription factors that regulate other genes. Thus, many of the genes with changed expression may not be direct target genes. Further studies with purified cells, in combination with chromatin immunoprecipitation (ChIP) using specific antibodies against pMad or pStat92E, will reveal genes that are direct targets of BMP and JAK-STAT in the testis niche.
Figure 1. Summary of transcriptional regulation in stem cell niche and mitotic germ cells. Hub cells are in green, GSC and transit-amplifying cells are in pink, CySC and cyst cells are in blue. Solid lines denote direct regulation, dashed lines denote indirect regulation or lack of evidence for direct regulation. See text for detailed discussion.
Table 1. Factors required in transcriptional regulation of spermatogenesis.
| Cell Types | Stage | Gene product(s) | Function(s) | Reference(s) |
|---|---|---|---|---|
| Somatic Cells |
Hub cells |
Gbb/Dpp |
Ligand of BMP signaling pathway |
Refs. 12–13 |
| |
|
Upd |
Ligand of JAK-STAT signaling pathway |
Refs. 8–10 |
| |
|
Magu |
Regulates BMP signaling and GSC maintenance |
ref. 14 |
| |
Cyst Stem Cells |
Rac and Rho |
Small GTPases, downstream of Egf pathway. Rac promotes Egf signaling while Rho inhibits Egf signaling. Egf pathway inhibits GSC self-renewal and promotes GSC to GB transition. |
Refs. 38, 41 |
| |
|
pSTAT |
Downstream transcription factor of JAK-STAT pathway, sufficient for GSC self-renewal |
ref. 10 |
| |
|
Chinmo |
Transcription factor, a target gene of JAK-STAT signaling, sufficient for GSC self-renewal |
ref. 20 |
| |
|
Zfh1 |
Transcription factor, a target gene of JAK-STAT signaling, sufficient for GSC self-renewal |
ref. 10 |
| |
|
Socs36E |
An inhibitor and also a target gene of JAK-STAT signaling, maintains a balanced ratio of CySCs and GSCs in the niche |
ref. 21 |
| |
Cyst Cells |
Rac and Rho |
Small GTPases, downstream of Egf pathway. Egf pathway inhibits spermatogonial division and promotes spermatogonia-to-spermatocyte transition. |
Refs. 38, 41 |
| Germ Cells |
GSCs |
Nucleoporin98–86 |
Nuclear envelope components, regulate proper GSC-to-GB transition, act upstream of BMP, JAK-STAT and Egfr pathways |
ref. 40 |
| |
|
pMAD |
Downstream transcription factor of BMP pathway, important for GSC self-renewal |
Refs. 12–14 |
| |
|
pSTAT |
Downstream transcription factor of JAK-STAT pathway, required for GSC-Hub cell adhesion |
ref. 11 |
| |
|
Scny |
A deubiquitinating enzyme targeting mono-ubiquitinated H2B, represses differentiation gene expression in GSCs |
ref. 23 |
| |
|
NCLB |
A chromatin factor that has male-specific roles for GSC maintenance through regulating JAK-STAT pathway |
ref. 26 |
| |
|
NURF |
A chromatin remodeler that positively regulates JAK-STAT signaling |
ref. 22 |
| |
|
PHF7 |
An epigenetic reader that recognizes H3K4me2 and regulates GSC self-renewal |
ref. 48 |
| |
|
miR-7 |
A microRNA that binds to bam mRNA 3′UTR and downregulates bam expression |
ref. 34 |
| |
|
Msi |
A RNA binding protein required for GSC maintenance |
ref. 27 |
| |
|
HOW |
A RNA binding protein required for GSC maintenance |
ref. 28 |
| |
|
Imp |
A RNA binding protein that stabilizes upd mRNA and maintains GSC |
ref. 29 |
| |
Spermatogonia |
Bam |
A differentiation factor required for spermatogonial differentiation |
Refs. 30–32 |
| |
|
Mael |
A RNA binding protein that represses miR-7 and upregulates bam expression |
ref. 34 |
| |
|
PHF7 |
An epigenetic reader that recognizes H3K4me2 and regulates spermatogonia differentiation |
ref. 48 |
| |
|
HOW |
A RNA binding protein required for spermatogonial proliferation |
ref. 28 |
| |
|
Nucleoporin98–86 |
A nuclear envelope component, regulates proper spermatogonia-to-spermatocyte transition and act upstream of BMP, JAK-STAT and Egfr pathways |
ref. 40 |
| |
Spermatocyte |
tMAC (Aly,Achi/VisComr, Topi, Tomb,Mip40) |
Regulate tTAF proper nuclear localization and binding to target genes, transcription of meiotic cell cycle and spermatid differentiation genes |
Refs. 61–66, 71–73 |
| |
|
tTAFs) (Can, Sa, Mia, Nht, Rye) |
Transcriptional activation of spermatid differentiation genes, antagonizes PcG-mediated gene silencing, accumulation of Boule protein |
Refs. 69–70 |
| |
|
WUC |
Maturation of spermatocyte and meiotic divisions |
Refs. 64, 82 |
| |
|
THO-complex |
Maturation of spermatocyte and meiotic divisions |
ref. 83 |
| |
|
NURF |
Maturation of spermatocyte and meiotic divisions |
ref. 81 |
| Spermatid | Unknown | Post-meiotic transcription of cup and comet genes, required for individualization of sperm | Refs. 84, 90–93 |
In addition to signaling pathways, epigenetic mechanisms also play important roles in regulating GSC activity.7 Epigenetic regulation changes chromatin structure and gene expression without changing DNA sequences. Two major classes of chromatin regulators have been identified for their functional roles in the male GSC niche: ATP-dependent chromatin remodelers and histone modifying enzymes. For example, one chromatin remodeler, the Nucleosome Remodeling Factor (NURF) complex, has been shown to positively regulate JAK-STAT signaling.22 Because chromatin remodelers act in both transcriptional activation and repression, it is possible that NURF either promotes transcription of JAK-STAT activators or represses transcription of JAK-STAT inhibitors. A deubiquitinating enzyme of mono-ubiquitinated H2B encoded by scrawny (scny) serves as an example of the histone-modifying enzymes. Since Scny normally functions in gene silencing, it was postulated to maintain GSCs by repressing the transcription of differentiation genes.23 Both NURF24,25 and Scny23 also regulate female GSC function. By contrast, a novel chromatin factor encoded by no child left behind (nclb) specifically regulates male, but not female, GSC maintenance.26 Nclb is enriched at chromatin regions with active transcription. In nclb mutant GSCs, Stat92E has decreased expression or accumulation,26 suggesting that Nclb may act via signaling pathways to determine GSC fate in the niche (Fig. 1 and Table 1). However, no direct connection between epigenetic mechanisms and signaling pathways has been reported. This partly results from the difficulty in precisely mapping their direct target genes in different cell types from the niche. Finally, RNA-binding proteins, such as Musashi (Msi),27 Held-out-wings (HOW)28 and IGF-II mRNA binding protein (Imp),29 are all required for GSC maintenance, suggesting an important role of post-transcriptional regulation in the testis niche.
Transcriptional Regulation in the Transit-Amplifying Cells
After GSCs exit the niche, they enter a transit amplification stage consisting of mitotically dividing GBs and spermatogonial cells. The bam gene encodes a differentiation factor that is detected in 4- to 16-cell spermatogonia with a peak level in 8-cell spermatogonia,30 but not in GSCs.31,32 Ectopic expression of Bam in GSCs causes their premature differentiation or cell death.31,33 The HOW RNA-binding protein28 and microRNA-7 (miR-7) have both been implicated in binding to bam mRNA and downregulating bam expression34 post-transcriptionally. Another RNA binding protein, Maelstrom (Mael), is required in spermatogonia to repress miR-7 and upregulate bam expression so that GB can enter the normal transit-amplification stage and divide as spermatogonia (Fig. 1 and Table 1).34 The Bam protein subsequently accumulates to a threshold level that is required for spermatogonia to become spermatocytes.32 As this threshold is never reached in the absence of Bam, bam mutant testes are filled with continuously dividing spermatogonial cells.30,35 Although regulation of bam expression has been elucidated at both transcriptional and post-transcriptional levels, the exact mechanism Bam utilizes to regulate transit-amplifying cell differentiation is not yet clear. Another differentiation gene, benign gonial cell neoplasm (bgcn), has been shown to have mutant phenotypes similar to bam in both male and female germline.30 Although the mechanism that Bam utilizes to regulate male germ cell differentiation is unknown, studies in female germ cells demonstrate that Bam and Bgcn form a protein complex to antagonize factors for GSC self-renewal and promote differentiation gene expression in transit-amplifying cells.36 Because Bgcn is predicted to be an RNA-binding protein, further characterization of proteins and RNAs with which the Bam-Bgcn complex interacts will illuminate their functions in transit-amplifying cells.
In spermatogenesis, the switch from mitosis to meiosis is critical. Too early transition to meiosis may lead to fewer germ cells and decreased fertility, while failure in this transition may lead to germline tumors. The Epidermal growth factor (Egf) signaling pathway plays an important role in the regulation of this switch. The Egf receptor (Egfr) ligand Spitz is processed by Stet, a transmembrane protease, in germ cells.37 Activated Spitz then acts on Egfr expressed in somatic cells.38 Egf signaling acts through the guanine nucleotide exchange factor (GEF) Vav to activate Rac-type small GTPases, which are antagonized by the Rho-type small GTPases.39 Egfr signaling acts in cyst cells to restrict GSC self-renewal and spermatogonial proliferation, while promoting GSC-to-GB and spermatogonia-to-spermatocyte transitions.38 Recent studies also demonstrate that Egfr signaling decreases the frequency of GSC division in adult, but not larval, testes, suggesting a dynamic mode of Egfr regulation.40 In addition, mutations in a serine/threonine kinase signal transducer encoded by raf result in a phenotype similar to the Egfr mutant, suggesting that the receptor tyrosine kinase (RTK) pathway is, in general, required in cyst cells for proper transit-amplification.41 The direct target genes for the Egfr/Raf pathway have not been identified; however, because compromised Egf signaling leads to defects in germline-soma interaction and overproliferation of spermatogonial cells, it is possible that the target genes regulate proper encapsulation of germ cells by cyst cells.37,39 Furthermore, a recent study reported that a nuclear envelope component, Nucleoporin98–86, regulates proper GSC-to-GB and spermatogonia-to-spermatocyte transitions and that its function is upstream of the BMP, JAK-STAT and Egfr signaling pathways.42 These results highlight the importance of nuclear structure in regulating cellular differentiation during spermatogenesis.
In order to study the transcriptional profile and chromatin state in transit-amplifying cells, bam mutant testes were used because they are enriched with over-proliferative spermatogonial cells, as described previously. In addition, wild-type (wt) testes were used as a comparison because they contain germ cells at all stages. High-throughput mRNA sequencing (RNA-seq) studies reveal that both chromatin remodeling factors and histone-modifying enzymes have more abundant transcripts in bam testes compared with wt testes.43 Furthermore, ChIP followed by high-throughput sequencing (ChIP-seq) reveals a different chromatin structure in bam testes compared with other stem cell lineages, such as embryonic stem cells (ESCs).44 In ESCs, it has been shown that differentiation genes have both the repressive H3K27me3 and the active H3K4me3 modifications (i.e., “bivalent” chromatin signature), as well as stalled RNA polymerase II (Pol II, i.e., “poised” genes), at their promoter regions.45-47 By contrast, differentiation genes required for spermatocyte maturation and spermiogenesis are either enriched with H3K27me3 only, or deprived of H3K4me3 and H3K27me3, in bam testes, and they are not associated with stalled Pol II.44 This distinct chromatin structure in bam testes may prevent ectopic transcription of the differentiation genes in transit-amplifying cells. On the other hand, the chromatin structure in spermatogonia-enriched bam testes suggests that dramatic changes at the promoter regions of differentiation genes are needed to turn on their robust transcription in spermatocytes. In addition to these genome-wide studies, it was recently reported that an epigenetic reader-encoding Plant Homeodomain Finger 7 (PHF7) gene is specifically expressed in GSCs and transit-amplifying cells. PHF7 recognizes the active H3K4me2 histone modification and is required for GSC maintenance and proper spermatogonial differentiation.48 Further studies to identity the target genes of PHF7, which should be enriched with H3K4me2 or H3K4me3, will shed light on its roles in maintaining GSCs and regulating differentiation of transit-amplifying cells.
Recent studies also revealed that spermatogonia can dedifferentiate to reoccupy the niche and become GSC-like cells.33,49-52 Although dedifferentiated spermatogonia have cellular features that are distinct from bona fide GSCs,50 it is less clear whether they have a different transcriptional profile and chromatin structure. Nevertheless, the dedifferentiated spermatogonia can undergo asymmetric cell division just like GSCs,50 suggesting that they properly respond to signaling from the niche, possibly because of a permissive chromatin landscape in spermatogonia. By contrast, spermatocytes are unable to dedifferentiate to become GSCs,33 suggesting that germ cells commit to an irreversible program at the spermatocyte stage. Similar irreversible commitment may also apply to female meiotic germ cells because only 4- to 8-cell transit-amplifying cells have been reported to undergo dedifferentiation to become GSC-like cells in the ovary.53
Transcriptional Regulation in Meiotic Spermatocytes
The transition from spermatogonia to spermatocytes is accompanied by a series of transcriptional, epigenetic and morphological changes. After transit-amplification, germ cells undergo the last S phase followed by an extended G2 phase that initiates the spermatocyte stage. Spermatocytes grow 25 times in volume and turn on a robust transcription program to activate genes required for spermatocyte maturation, as well as genes needed for meiotic divisions and terminal differentiation.54 Most genes required for meiotic divisions, as well as terminal differentiation, are under translational repression until a later time when their encoded proteins are required.55
The G2/M transition in meiosis I requires Cyclin B, Boule (a RNA-binding protein) and Twine (Cdc25 homolog), all transcribed in spermatocytes.54,56,57 Boule translocates from the nucleus to the cytoplasm to trigger the G2/M transition in meiosis I by allowing translation of Twine.58 At this point in time, Cyclin B protein also escapes from translational repression and accumulates in the cytoplasm of spermatocytes.54 In both boule and twine mutant testes, spermatid differentiation occurs independently of meiotic cell cycle progression, suggesting that these two processes can be uncoupled.56,59 However, the discovery of two classes of genes expressed in early spermatocytes reveals a high degree of coordination between meiotic divisions and spermatid differentiation.60 Mutations in any of these genes arrest meiosis and block spermatid differentiation, leading to testes filled with immature spermatocytes. These genes are named “meiotic arrest” genes, which are classified into “aly-class” and “can-class” based on morphological differences in the chromosomal structure of the mutant spermatocytes54,60 and the distinct target genes they regulate.54,60-69 For example, transcription of meiotic cell cycle genes, such as Cyclin B, boule and twine, rely on aly-class, but not can-class, genes.54 However, Boule protein accumulation requires the can-class genes.70 Because meiotic arrest genes regulate transcription or translation of meiotic cell cycle genes, their functions ensure that the meiotic cell cycle does not proceed until terminal differentiation genes are robustly transcribed.54,60
The five known aly-class genes are always early (aly), cookie monster (comr), matotopetli (topi), tombola (tomb) and achintya/vismay (achi/vis). All of the aly-class genes, except achi/vis, are expressed exclusively in primary spermatocytes.62-65,71,72 Four of the five aly-class proteins have putative DNA binding domains, including Comr, which contains a winged helix; Topi, which contains multiple Zn-finger motifs; Tomb, which has a CXC domain; and Achi/Vis, products from a gene duplication, which have homeodomains. Thus, it is thought that these proteins regulate transcription of target genes by directly binding to DNA sequences, although their direct target genes have not been identified. Immunoaffinity purification studies have revealed that Aly and Tomb proteins are copurified with Mip40 (Myb interacting protein, 40kDa) to form the testis meiotic arrest complex tMAC, which also contains Topi, Comr and CAF1.66 A second form of tMAC contains Aly, Comr and Achi/Vis.71 The tMAC resembles the MIP/dREAM complex in mammals and the SynMuv complexes in C. elegans.54,61-66 While the mode of action of tMAC is not fully elucidated, it is thought to have an active, rather than repressive, transcriptional role. This is based on results demonstrating that expression of Achi/Vis fused with a strong transactivation domain, VP16, rescued the achi/vis mutant phenotype, while the fusion of Achi/Vis with a repression domain, EnR, failed to rescue this phenotype.73 Consistent with these findings, all tMAC subunits have been found to co-localize with euchromatin in primary spermatocytes.61,63,64,71
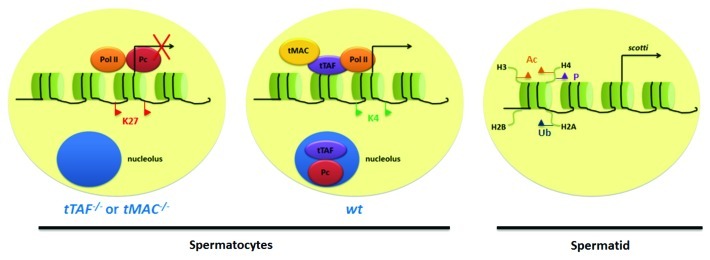
The can-class genes encode testis-specific homologs of the ubiquitously expressed subunits of the general transcription factor II D (TFIID). TFIID is one of the general transcription factors that constitute the RNA Pol II preinitiation complex composed of TATA-binding protein (TBP) and 13–14 TBP-associated factors (TAFs).74-76 TFIID coordinates the interaction between RNA Pol II and gene promoter regions. The characterized can-class genes include cannonball (can, TAF5L), meiosis I arrest (mia, TAF6L), no hitter (nht, TAF4L), ryan express (rye, TAF12L) and spermatocyte arrest (sa, TAF8L). Among these five TAF homologs, four, including Mia, Nht, Rye and Sa, share similar structural domains called histone folding motifs for protein-protein interaction, and Can is a WD40-repeat-containing protein.68 Indeed, Nht and Rye form a heterodimer in vitro.67 These testis-specific TAFs (tTAFs) are thought to form a testis-specific complex required for the transcriptional activation of terminal differentiation genes.67,68 Such predicted functions of tTAFs suggest that they localize at the euchromatin in spermatocyte nuclei. However, while a proportion of the total protein of each tTAF associates with chromosomes in spermatocytes, most tTAF protein is localized to a subcompartment within the nucleolus.70,77 Interestingly, Polycomb (Pc) and other components of the Polycomb Repressive Complex 1 (PRC1) are co-localized to the same nucleolar subcompartment with tTAFs in spermatocytes. Furthermore, localization of PRC1 components to the spermatocyte nucleolus is coincident with tTAF expression and dependent on wild-type tTAF function.70 These results suggest that tTAFs act as derepressors by sequestering PRC1 to the spermatocyte nucleolus to counteract Polycomb Group protein (PcG)-induced repression. However, removing PcG activity is not sufficient to turn on terminal differentiation genes in the absence of tTAFs,69 suggesting that the chromatin-associated tTAFs are required for activating terminal differentiation genes. Consistent with this observation, tTAFs were reported to turn on the transcription of more than 1,000 genes, and many of them are required for spermatid differentiation.54,69 Among the tTAF-dependent genes, three are shown to be tTAF direct target genes by ChIP assay: fuzzy onions (fzo), which encodes a protein required for mitochondrial fusion in early spermatids;78 mst87F, which encodes a component of the sperm tail79 and don juan (dj), which encodes a sperm-specific DNA-binding protein that also localizes to mitochondria.80 ChIP analysis at the promoter regions of these three tTAF direct target genes showed that levels of the repressive H3K27me3 mark and paused Pol II are high, while levels of the active H3K4me3 mark are low in can and aly mutant testes.69 These data suggest that tTAFs and tMAC might recruit the Trithorax group complex (TrxG), whose activities antagonize PcG, to methylate H3K4 at promoters of terminal differentiation genes (Fig. 2 and Table 1).70
Figure 2. Summary of transcriptional regulation in meiotic and post-meiotic germ cells. A schematic diagram outlines potential chromatin state in spermatocyte mutants for tMAC or tTAF (left, analyzed with can or aly mutant testes) compared with mature wt spermatocytes (middle, analyzed with wt testes). K27: H3K27me3, K4:H3K4me3. Adapted from Chen et. al.69 On the right, a schematic diagram outlines potential chromatin state in spermatids prior to the histone-to-protamine transition. The scotti gene is transcribed in elongating spermatids. Ac, acetylation; P, phosphorylation; Ub, ubiquitylation.
Although the mode of interaction between tMAC components (aly-class) and tTAFs (can-class) is not fully understood, it was reported that the function of aly is required for the binding of TAF8L to target gene promoters. Aly is also required for the proper nucleolar localization of several tTAFs and Pc in spermatocytes, suggesting that tMAC acts upstream of tTAFs.69 This is consistent with assays using northern blot, in situ hybridization and microarray analysis.54,68,69 In addition, while Mip40 is co-immunoprecipitated with tMAC components, loss of mip40 results in spermatocytes with condensed chromosomes, a phenotype similar to mutants of can-class genes,66 suggesting that Mip40 might mediate the interaction between tMAC and tTAFs. Both tMAC and tTAFs have their canonical counterparts that act generally in other tissues. Similarly, the canonical chromatin remodeler NURF has a germline-specific function in regulating meiotic divisions and spermatocyte differentiation,81 most likely through using an alternatively spliced isoform.
Recently, two other meiotic arrest genes were identified, which cannot be classified as either the aly-class or the can-class.82,83 Wake-up-call (Wuc) was identified by its physical interaction with Aly in a yeast-two-hybrid screen.64 In spermatocytes, the Wuc protein is highly expressed and associated with chromatin, similar to other tMAC components.82 However, unlike tMAC or tTAF mutants, loss of wuc does not abolish expression of either meiotic cell cycle genes or spermatid differentiation genes.82 Another study showed that disruption of a component of the THO complex, THOC5, led to the meiotic arrest phenotype.83 The THO complex is known to export mRNAs from nucleus to cytoplasm. However, no mRNA export defects were detectable in the thoc5 mutant. Moreover, neither meiotic cell cycle genes nor spermatid differentiation genes have decreased transcription in the thoc5 mutant.83 Finally, THOC5 is localized to a perinucleolar region, and loss-of-function in thoc5 leads to disrupted nucleolar structure and tTAF localization.83 Identification of wuc and thoc5 mutants and detailed characterization of their phenotypes demonstrate the existence of meiotic arrest genes other than aly-class and can-class. Understanding their molecular mechanisms will lead to new information about spermatocyte maturation.
Transcriptional Regulation in Post-Meiotic Cells
After germ cells exit the extended G2 phase, they undergo two meiotic divisions followed by spermatid differentiation. One of the major epigenetic events in post-meiotic germ cells is the displacement of histones by transition nuclear proteins (Tnps), followed by protamines (Prms).84 The replacement of histones with protamines allows for DNA condensation and packaging in the sperm nuclei. To prepare for this replacement, histones undergo a series of post-translational modifications that open up the chromatin, including hyperacetylation of H3/H4 tails,84,85 mono-ubiquitylation of H2A,84 and phosphorylation of H4S1 (Fig. 2).86
Replacement of histones with protamines causes chromatin condensation, which may block transcription in spermatids.84 Autoradiography studies of nucleic acid synthesis demonstrate a lack of transcription in post-meiotic cells.87,88 Thus, consensus has held that spermatid differentiation genes are mostly transcribed in spermatocytes, and remain under translational repression. For example, dj is transcribed in spermatocytes, but remains translationally repressed until the Dj protein is required for sperm tail formation in elongated spermatids.89 However, immunostaining showed that phosphorylated and active Pol II reappears in canoe-stage spermatids, indicating reinitiation of active transcription at this late stage during spermatogenesis.84 Using a sensitive single-cyst quantitative RT-PCR method, another study identified 24 genes, termed “comets and cups” based on their localization patterns, which showed active transcription in Drosophila spermatids.90 For example, scotti is a post-meiotically transcribed comet gene required for normal actin cone progression during spermatid individualization.91 Consistent with its function, scotti mutant males are sterile.90 More recently, microarray analysis92 and 5-bromouridine (BrU) incorporation assay93 were used to profile and visualize de novo transcription in spermatid bundles. These new results demonstrate that transcription is reactivated in 20–30% of testis genes prior to the histone-to-protamine change.90,92
Active transcription in spermatids has been described in mammals.94,95 These new discoveries of actively transcribed genes in Drosophila spermatids provide a striking similarity between fly and mammalian spermatogenesis. However, although genes transcribed in mammalian spermatids almost always encode components of the mature sperm, most genes transcribed in Drosophila spermatids (22 of the 24) do not encode sperm proteins.90 Further investigation of these genes is warranted to better understand their functions during spermatogenesis.
Conclusions and Perspectives
In conclusion, Drosophila spermatogenesis has provided an ideal model system to study transcriptional regulation of the cellular differentiation pathway in an endogenous stem cell linage. Recently, new technologies, including RNA-seq and ChIP-seq, have greatly improved our understanding of transcriptional regulation. In the future, the combination of highly sensitive genomic techniques with purified cell types at distinct stages of spermatogenesis will reveal transcriptional profiles with much higher spatiotemporal resolution. Furthermore, ongoing efforts to identify direct target genes downstream of key signaling pathways will illuminate regulatory networks that control transcriptional changes during spermatogenesis.
Acknowledgements
We apologize to people whose work cannot be discussed in this review due to space limitations. This work was supported by F31CA165781 from NCI, NIH to L.T. The work in the Chen lab has been supported by Research Grant No.05-FY09-88 from the March of Dimes Foundation, the R00HD055052 NIH Pathway to Independence Award, R01HD065816 and R21 HD065089 from NICHD, the 49th Mallinckrodt Scholar Award from the Edward Mallinckrodt Jr. Foundation, the American Federation of Aging Research, the David & Lucile Packard Foundation, and the Johns Hopkins University start-up funding for X.C.
Footnotes
Previously published online: www.landesbioscience.com/journals/spermatogenesis/article/21775
References
- 1.Fuller MT. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin Cell Dev Biol. 1998;9:433–44. doi: 10.1006/scdb.1998.0227. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, Tiyaboonchai A, Yamashita YM, Hunt AJ. Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development. 2011;138:831–7. doi: 10.1242/dev.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Losick VP, Morris LX, Fox DT, Spradling A. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21:159–71. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita YM. Cell adhesion in regulation of asymmetric stem cell division. Curr Opin Cell Biol. 2010;22:605–10. doi: 10.1016/j.ceb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamashita YM, Fuller MT, Jones DL. Signaling in stem cell niches: lessons from the Drosophila germline. J Cell Sci. 2005;118:665–72. doi: 10.1242/jcs.01680. [DOI] [PubMed] [Google Scholar]
- 6.de Cuevas M, Matunis EL. The stem cell niche: lessons from the Drosophila testis. Development. 2011;138:2861–9. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eun SH, Gan Q, Chen X. Epigenetic regulation of germ cell differentiation. Curr Opin Cell Biol. 2010;22:737–43. doi: 10.1016/j.ceb.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–5. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 9.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–9. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 10.Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–11. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawase E, Wong MD, Ding BC, Xie T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131:1365–75. doi: 10.1242/dev.01025. [DOI] [PubMed] [Google Scholar]
- 13.Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr Biol. 2003;13:2065–72. doi: 10.1016/j.cub.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Q, Wang Y, Vargas E, DiNardo S. magu is required for germline stem cell self-renewal through BMP signaling in the Drosophila testis. Dev Biol. 2011;357:202–10. doi: 10.1016/j.ydbio.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–91. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–64. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 17.Rushlow C, Colosimo PF, Lin MC, Xu M, Kirov N. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 2001;15:340–51. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 19.Terry NA, Tulina N, Matunis E, DiNardo S. Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol. 2006;294:246–57. doi: 10.1016/j.ydbio.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 20.Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, et al. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 2010;18:556–68. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–6. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherry CM, Matunis EL. Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell. 2010;6:557–67. doi: 10.1016/j.stem.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–51. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–9. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- 25.Ables ET, Drummond-Barbosa D. The steroid hormone ecdysone functions with intrinsic chromatin remodeling factors to control female germline stem cells in Drosophila. Cell Stem Cell. 2010;7:581–92. doi: 10.1016/j.stem.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casper AL, Baxter K, Van Doren M. no child left behind encodes a novel chromatin factor required for germline stem cell maintenance in males but not females. Development. 2011;138:3357–66. doi: 10.1242/dev.067942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A. 2006;103:8402–7. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monk AC, Siddall NA, Volk T, Fraser B, Quinn LM, McLaughlin EA, et al. HOW is required for stem cell maintenance in the Drosophila testis and for the onset of transit-amplifying divisions. Cell Stem Cell. 2010;6:348–60. doi: 10.1016/j.stem.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–10. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gönczy P, Matunis E, DiNardo S. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development. 1997;124:4361–71. doi: 10.1242/dev.124.21.4361. [DOI] [PubMed] [Google Scholar]
- 31.Schulz C, Kiger AA, Tazuke SI, Yamashita YM, Pantalena-Filho LC, Jones DL, et al. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics. 2004;167:707–23. doi: 10.1534/genetics.103.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Insco ML, Leon A, Tam CH, McKearin DM, Fuller MT. Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc Natl Acad Sci U S A. 2009;106:22311–6. doi: 10.1073/pnas.0912454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheng XR, Brawley CM, Matunis EL. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell. 2009;5:191–203. doi: 10.1016/j.stem.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pek JW, Lim AK, Kai T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev Cell. 2009;17:417–24. doi: 10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 35.McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4(12B):2242–51. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A. 2009;106:9304–9. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz C, Wood CG, Jones DL, Tazuke SI, Fuller MT. Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development. 2002;129:4523–34. doi: 10.1242/dev.129.19.4523. [DOI] [PubMed] [Google Scholar]
- 38.Kiger AA, White-Cooper H, Fuller MT. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750–4. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, Schulz C. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr Biol. 2007;17:1253–8. doi: 10.1016/j.cub.2007.06.048. [DOI] [PubMed] [Google Scholar]
- 40.Parrott BB, Hudson A, Brady R, Schulz C. Control of germline stem cell division frequency--a novel, developmentally regulated role for epidermal growth factor signaling. PLoS One. 2012;7:e36460. doi: 10.1371/journal.pone.0036460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran J, Brenner TJ, DiNardo S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature. 2000;407:754–7. doi: 10.1038/35037613. [DOI] [PubMed] [Google Scholar]
- 42.Parrott BB, Chiang Y, Hudson A, Sarkar A, Guichet A, Schulz C. Nucleoporin98-96 function is required for transit amplification divisions in the germ line of Drosophila melanogaster. PLoS One. 2011;6:e25087. doi: 10.1371/journal.pone.0025087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, et al. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010;20:763–83. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, et al. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11:R42. doi: 10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125:233–6. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 47.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang SY, Baxter EM, Van Doren M. Phf7 controls male sex determination in the Drosophila germline. Dev Cell. 2012;22:1041–51. doi: 10.1016/j.devcel.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–4. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 50.Cheng J, Türkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong C, Jones DL. Efficiency of spermatogonial dedifferentiation during aging. PLoS One. 2012;7:e33635. doi: 10.1371/journal.pone.0033635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallenfang MR, Nayak R, DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell. 2006;5:297–304. doi: 10.1111/j.1474-9726.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- 53.Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature. 2004;428:564–9. doi: 10.1038/nature02436. [DOI] [PubMed] [Google Scholar]
- 54.White-Cooper H, Schäfer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–34. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- 55.Schäfer M, Nayernia K, Engel W, Schäfer U. Translational control in spermatogenesis. Dev Biol. 1995;172:344–52. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- 56.Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–88. doi: 10.1016/0092-8674(92)90616-K. [DOI] [PubMed] [Google Scholar]
- 57.Courtot C, Fankhauser C, Simanis V, Lehner CF. The Drosophila cdc25 homolog twine is required for meiosis. Development. 1992;116:405–16. doi: 10.1242/dev.116.2.405. [DOI] [PubMed] [Google Scholar]
- 58.Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat Cell Biol. 1999;1:171–4. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 59.Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–5. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 60.Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–41. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- 61.White-Cooper H, Leroy D, MacQueen A, Fuller MT. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development. 2000;127:5463–73. doi: 10.1242/dev.127.24.5463. [DOI] [PubMed] [Google Scholar]
- 62.Ayyar S, Jiang J, Collu A, White-Cooper H, White RA. Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development. 2003;130:2841–52. doi: 10.1242/dev.00513. [DOI] [PubMed] [Google Scholar]
- 63.Jiang J, White-Cooper H. Transcriptional activation in Drosophila spermatogenesis involves the mutually dependent function of aly and a novel meiotic arrest gene cookie monster. Development. 2003;130:563–73. doi: 10.1242/dev.00246. [DOI] [PubMed] [Google Scholar]
- 64.Jiang J, Benson E, Bausek N, Doggett K, White-Cooper H. Tombola, a tesmin/TSO1-family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with always early. Development. 2007;134:1549–59. doi: 10.1242/dev.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perezgasga L, Jiang J, Bolival B, Jr., Hiller M, Benson E, Fuller MT, et al. Regulation of transcription of meiotic cell cycle and terminal differentiation genes by the testis-specific Zn-finger protein matotopetli. Development. 2004;131:1691–702. doi: 10.1242/dev.01032. [DOI] [PubMed] [Google Scholar]
- 66.Beall EL, Lewis PW, Bell M, Rocha M, Jones DL, Botchan MR. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 2007;21:904–19. doi: 10.1101/gad.1516607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, et al. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- 68.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–30. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen X, Lu C, Prado JR, Eun SH, Fuller MT. Sequential changes at differentiation gene promoters as they become active in a stem cell lineage. Development. 2011;138:2441–50. doi: 10.1242/dev.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–72. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, Mann RS. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development. 2003;130:2853–65. doi: 10.1242/dev.00510. [DOI] [PubMed] [Google Scholar]
- 72.White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 2010;139:11–21. doi: 10.1530/REP-09-0083. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Wang L, Wang Z. Transgenic analyses of TGIF family proteins in Drosophila imply their role in cell growth. J Genet Genomics. 2008;35:457–65. doi: 10.1016/S1673-8527(08)60063-6. [DOI] [PubMed] [Google Scholar]
- 74.Cler E, Papai G, Schultz P, Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci. 2009;66:2123–34. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matangkasombut O, Auty R, Buratowski S. Structure and function of the TFIID complex. Adv Protein Chem. 2004;67:67–92. doi: 10.1016/S0065-3233(04)67003-3. [DOI] [PubMed] [Google Scholar]
- 76.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–5. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 77.Metcalf CE, Wassarman DA. Nucleolar colocalization of TAF1 and testis-specific TAFs during Drosophila spermatogenesis. Dev Dyn. 2007;236:2836–43. doi: 10.1002/dvdy.21294. [DOI] [PubMed] [Google Scholar]
- 78.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–9. doi: 10.1016/S0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 79.Schäfer M, Börsch D, Hülster A, Schäfer U. Expression of a gene duplication encoding conserved sperm tail proteins is translationally regulated in Drosophila melanogaster. Mol Cell Biol. 1993;13:1708–18. doi: 10.1128/mcb.13.3.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Santel A, Blümer N, Kämpfer M, Renkawitz-Pohl R. Flagellar mitochondrial association of the male-specific Don Juan protein in Drosophila spermatozoa. J Cell Sci. 1998;111:3299–309. doi: 10.1242/jcs.111.22.3299. [DOI] [PubMed] [Google Scholar]
- 81.Kwon SY, Xiao H, Wu C, Badenhorst P. Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet. 2009;5:e1000574. doi: 10.1371/journal.pgen.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doggett K, Jiang J, Aleti G, White-Cooper H. Wake-up-call, a lin-52 paralogue, and Always early, a lin-9 homologue physically interact, but have opposing functions in regulating testis-specific gene expression. Dev Biol. 2011;355:381–93. doi: 10.1016/j.ydbio.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moon S, Cho B, Min SH, Lee D, Chung YD. The THO complex is required for nucleolar integrity in Drosophila spermatocytes. Development. 2011;138:3835–45. doi: 10.1242/dev.056945. [DOI] [PubMed] [Google Scholar]
- 84.Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120:1689–700. doi: 10.1242/jcs.004663. [DOI] [PubMed] [Google Scholar]
- 85.Awe S, Renkawitz-Pohl R. Histone H4 acetylation is essential to proceed from a histone- to a protamine-based chromatin structure in spermatid nuclei of Drosophila melanogaster. Syst Biol Reprod Med. 2010;56:44–61. doi: 10.3109/19396360903490790. [DOI] [PubMed] [Google Scholar]
- 86.Krishnamoorthy T, Chen X, Govin J, Cheung WL, Dorsey J, Schindler K, et al. Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis. Genes Dev. 2006;20:2580–92. doi: 10.1101/gad.1457006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gould-Somero MHL. The timing of RNA synthesis for spermiogenesis in organ cultures of Drosophila melanogaster testes. Development Genes and Evolution. 1974;174:133–48. doi: 10.1007/BF00573626. [DOI] [PubMed] [Google Scholar]
- 88.Olivieri G, Olivieri A. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat Res. 1965;2:366–80. doi: 10.1016/0027-5107(65)90072-2. [DOI] [PubMed] [Google Scholar]
- 89.Santel A, Winhauer T, Blümer N, Renkawitz-Pohl R. The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mech Dev. 1997;64:19–30. doi: 10.1016/S0925-4773(97)00031-2. [DOI] [PubMed] [Google Scholar]
- 90.Barreau C, Benson E, Gudmannsdottir E, Newton F, White-Cooper H. Post-meiotic transcription in Drosophila testes. Development. 2008;135:1897–902. doi: 10.1242/dev.021949. [DOI] [PubMed] [Google Scholar]
- 91.Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev Cell. 2010;19:160–73. doi: 10.1016/j.devcel.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vibranovski MD, Chalopin DS, Lopes HF, Long M, Karr TL. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics. 2010;186:431–3. doi: 10.1534/genetics.110.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reynard LN, Turner JM, Cocquet J, Mahadevaiah SK, Touré A, Höög C, et al. Expression analysis of the mouse multi-copy X-linked gene Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR. Biol Reprod. 2007;77:329–35. doi: 10.1095/biolreprod.107.061101. [DOI] [PubMed] [Google Scholar]
- 95.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–6. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]