Abstract
Meiosis entails sorting and separating both homologous and sister chromatids. The mechanisms for connecting sister chromatids and homologs during meiosis are highly conserved and include specialized forms of the cohesin complex and a tightly regulated homolog synapsis/recombination pathway designed to yield regular crossovers between homologous chromatids. Drosophila male meiosis is of special interest because it dispenses with large segments of the standard meiotic script, particularly recombination, synapsis and the associated structures. Instead, Drosophila relies on a unique protein complex composed of at least two novel proteins, SNM and MNM, to provide stable connections between homologs during meiosis I. Sister chromatid cohesion in Drosophila is mediated by cohesins, ring-shaped complexes that entrap sister chromatids. However, unlike other eukaryotes Drosophila does not rely on the highly conserved Rec8 cohesin in meiosis, but instead utilizes two novel cohesion proteins, ORD and SOLO, which interact with the SMC1/3 cohesin components in providing meiotic cohesion.
Keywords: meiosis, homolog pairing, homolog conjunction, conjunction complex, sister chromatid cohesion, sister centromere mono-orientation, pairing site, cohesin, chromosome territories, Drosophila males
Introduction
Meiosis generates haploid reproductive cells (gametes) from diploid precursors, an essential step in sexual reproduction. The unique challenge of meiosis is to segregate both homologous chromosomes and sister chromatids accurately so that each gamete receives exactly one copy of each chromosome. DNA is replicated only once, at the onset of meiosis, and from then on there are four copies of each chromosome (two homologous chromosomes with two sister chromatids each) that must be sorted and segregated. This is accomplished via two successive divisions, meiosis I and meiosis II, referred to as the “reductional” and “equational” divisions, respectively, in which first homologs, then sister chromatids are segregated.1,2 Errors in meiotic segregation lead to aneuploid offspring, which are often inviable or poorly viable due to gene dosage imbalance. Although aneuploidy due to meiotic missegregation is uncommon in flies and most other eukaryotes, it is remarkably common in human reproduction, accounting for approximately 1/3 of spontaneous miscarriages, and is the most common cause of developmental disability and mental retardation.3
For the most part, meiosis is highly conserved. As a result, it is possible to define a “standard meiotic script” that is followed with reasonable fidelity by creatures as diverse as baker’s yeast, mice, maize and female Drosophila. We will provide a brief outline of this standard script below and refer the interested reader to several excellent reviews. This will set the stage for a much more detailed review of meiosis in male Drosophila, which exhibits some major departures from the standard script.
Spindles and chromosome segregation
We will begin with a very brief summary of events that are common to mitosis and meiosis, then highlight the uniquely meiotic ones. Chromosome segregation in both mitosis and meiosis is mediated by a bipolar microtubule spindle apparatus in which the minus ends are anchored at the poles and the plus ends extend outward. The chromosomes attach to the spindle by binding to the plus ends of kinetochore microtubules through protein complexes called kinetochores that are assembled at centromeres. Each chromosome or chromatid pair normally has two functional kinetochores, and when all goes well, each kinetochore binds to a bundle of microtubules originating from the same pole while the kinetochore of its sister or homolog binds to a microtubule bundle from the opposite pole, an arrangement known as “bipolar orientation” or “bi-orientation.” Since spindle microtubules undergo treadmilling, kinetochores of bi-oriented chromosome pairs experience poleward forces that generate tension across the centromere regions. Bipolar orientation is achieved during prometaphase by a trial-and-error process in which improper kinetochore-microtubule attachments that fail to generate tension across the centromere region are eliminated and tension-generating attachments are preserved. Once all chromosome pairs have achieved stable bipolar orientation and congressed to the spindle equator at metaphase, the connecting proteins that restrain the chromosomes (discussed in detail below) are removed, freeing the chromosomes to segregate.4
Cohesion
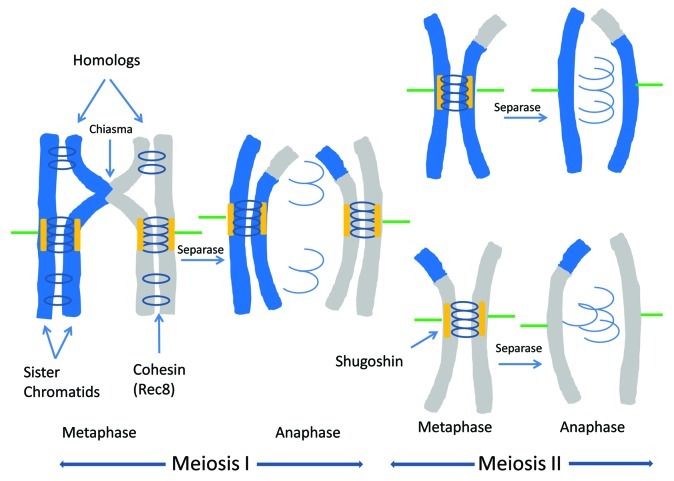
A prerequisite for achieving bipolar orientation is that the orienting chromosomes must be connected stably enough to resist the poleward forces experienced at the kinetochores during prometaphase and metaphase. In mitosis and meiosis II, it is sister chromatids that must bi-orient; the connections between sister chromatids, referred to as “cohesion,” are mediated by cohesin, a conserved ring-shaped protein complex that entraps pairs of sister chromatids. Cohesin is loaded all along chromosomes prior to or during S phase and establishes cohesion during DNA replication.2,5 The core cohesin components are two long coiled-coil proteins, SMC1 and SMC3, which interact with each other and with a “kleisin” subunit (SCC1/Rad21 in mitosis) to form a tripartite ring, and SCC3/SA/Stromalin, which attaches to the kleisin component and mediates various interactions. The main removal pathway at anaphase involves cleavage of the kleisin subunit by the protease enzyme Separase. Meiotic cohesins appear to have the same basic subunit composition but one or more of the core subunits may be replaced by a meiosis-specific paralog. Most such variants are restricted to fairly narrow taxonomic ranges and their specific functions are not well-characterized but the meiosis-specific kleisin Rec8 replaces SCC1/Rad21 in most meiotic cohesins in nearly all eukaryotes.2,6-8 We will consider the special properties of Rec8 below.
Bi-orientation of homologs and chiasmata
Homologous chromosomes must bi-orient at meiosis I (Fig. 1). Although each homolog consists of a pair of sister chromatids held together by cohesion, that cohesion alone is of no use because it does not connect homologs.2 Instead, the homolog connectors for most eukaryotes are “chiasmata” in which crossovers between homologous chromatids are stabilized by cohesion between the crossover chromatids and their sisters distal to the crossover sites (Fig. 1).9 These connections prevent homologs from dissociating prior to anaphase I, a major cause of nondisjunction and aneuploidy. Moreover, during prometaphase I and metaphase I, chiasmata provide the resistance that enables homolog pairs to achieve bipolar orientation on the meiosis I spindle. Activation of Separase at anaphase I causes dissolution of chiasmata (Fig. 1), thus releasing homologs to segregate.2 Formation of chiasmata is a complex process that encompasses three major steps: homolog pairing and alignment10; synapsis (formation of synaptonemal complexes (SC)s, an elaborate protein network that serves as the interface between paired homologs)11; and meiotic recombination, a specialized version of the double strand break repair pathway biased toward use of homologous chromatids as repair templates.12 These processes have been the focus of most meiosis research but, with the exception of homolog pairing, will not be further discussed because they play no role in Drosophila male meiosis.
Figure 1. Meiotic chromosome segregation: how most eukaryotes do it. Homologous chromosomes segregate during meiosis I; sister chromatids segregate during meiosis II. In both divisions, chromosomes achieve bipolar orientation during prometaphase (not shown) and align at the spindle equator during metaphase. Sister chromatids are held together by cohesin complexes containing the meiosis-specific Rec8 subunit. In meiosis I, homologous chromosomes are linked by chiasmata at sites where homologous chromatids have undergone a crossover. Sister kinetochores attach to microtubules emanating from the same spindle pole. At anaphase I, Separase cleaves Rec8 cohesin only on chromosome arms, releasing chiasmata; Rec8 at centromeres is protected by Shugoshin. In meiosis II, the residual centromeric cohesion facilitates the attachment of sister kinetochores to microtubules emanating from opposite poles of the spindle. Sister chromatid separation at anaphase II is triggered by cleavage of the remaining Rec8 by Separase.
Centromere orientation
As indicated above, bi-orientation requires a pair of properly organized kinetochores capable of binding to microtubule bundles originating from opposite poles. In mitosis and meiosis II, sister kinetochores adopt a “back-to-back” orientation and are often visibly separate and stretched toward the poles during prometaphase I. During meiosis I, a key prerequisite for bipolar orientation of homologs is “mono-orientation” (or “co-orientation”) of sister centromeres, in which sister centromeres form a functionally single kinetochore and orient to the same pole2,13 (Fig. 1). If not for this behavior, there would be four functionally independent centromeres/kinetochores in each bivalent but only two poles, a recipe for segregational chaos. In ultrastructural studies of meiosis I centromeres from a variety of eukaryotes, the kinetochore forming regions of the sister centromeres appear fused during prophase I but become resolved by anaphase I.14-16 This has led to the idea that sister centromeres may be clamped tightly together during meiosis I to prevent them from orienting to opposite poles. In budding yeast, a specialized complex called Monopolin localizes to centromeres during meiosis I and is required for mono-orientation.17-19 In fission yeast, Rec8 cohesin, assisted by Moa1, appears to function directly as the clamp.20-22 Neither the Monopolin complex genes nor Moa1 appear to be conserved outside of fungi, and little progress has been made in elucidating the mechanism of mono-orientation in higher eukaryotes.
Regulating release of chromosome connections
An additional challenge faced by meiotic cells is to regulate the release of connections to ensure that homologs segregate at meiosis I and sister chromatids segregate at meiosis II.2,5 Separase is activated both at anaphase I, when it cleaves arm cohesins and releases chiasmata, and at anaphase II, when it cleaves centromeric cohesins. Premature loss of centromere cohesion at anaphase I is prevented by a conserved centromere protein called Shugoshin (Fig. 1).23-26 Shugoshin recruits the phosphatase PP2A which functions to prevent Rec8 (and perhaps other subunits) of centromere cohesin from being phosphorylated and targeted for cleavage during meiosis I.27 SCC1/RAD21 cannot be so protected, so Rec8 cohesin is essential to maintain sister centromere cohesion after anaphase I. Curiously, Drosophila lacks Rec8 but does have Shugoshin; indeed the fly homolog MEI-S332 is the founding member of the Shugoshin family.23
Overview of Meiosis in Male Drosophila
The following is a brief overview of cytological aspects of male meiosis. Readers wishing more detail should consult one of the excellent descriptive reviews of meiosis and spermatogenesis in Drosophila.28-31 Meiosis I takes place near the apical (closed) end of the testis in “cysts” of 16 primary spermatocytes interconnected by ring canals, remnants of incomplete cytokinesis during the preceding spermatogonial mitoses. Figure 2 is a schematic representation of the main stages of meiosis I. Meiotic S phase initiates very soon after the completion of the last mitosis and is completed within about three hours.31 The ensuing G2 phase lasts approximately 80–90 h and serves as a growth phase; cells expand approximately 25-fold29 and RNAs and proteins needed in later stages of spermatogenesis accumulate. As described below, it is also during this period that homologous chromosomes achieve high levels of pairing, one of the hallmarks of prophase I. For that reason, we will refer to this period as prophase I, in contrast with most recent authors. However, since G2/prophase I chromosomes in Drosophila are decondensed and lack the structural features normally associated with meiotic chromosomes, the conventional prophase I stages are inapplicable and the nomenclature introduced by Cenci et al.31 which subdivides G2 into seven stages, S1-S6, with S2 further subdivided into S2a and S2b, on the basis of nuclear size, shape and positioning, and chromosome distribution, has become standard.
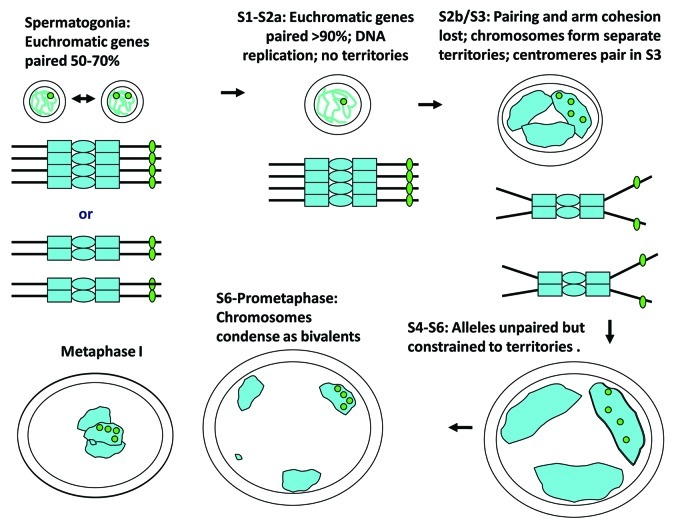
Figure 2. Meiosis I in Drosophila males. Meiosis occurs in cysts of 16 primary spermatocytes (only one shown). DNA replication is completed within 3 h of the last gonial mitosis and is followed by a lengthy growth period during which the three major chromosome pairs form separate territories by the start of stage S3 and remain separate throughout the rest of prophase I. Late in stage S6, the chromosomes condense to form four compact bivalents that achieve bipolar orientation (not shown) during prometaphase I and congress to the spindle equator at metaphase I. Euchromatic alleles are tightly paired during S1 and S2a but come unpaired, along with sister chromatids, at S2b/S3, coincident with territory establishment, and remain unpaired throughout the rest of meiosis I. The green dots represent fluorescent foci of GFP-LacI that accumulate at a specific chromosomal site where a 256mer array of lacO repeats is inserted. Pairing and cohesion status can be ascertained by counting spots, as shown. On chromosome diagrams, ovals represent centromeres and rectangles represent centromere-flanking heterochromatin.
Prophase I
In S1, the cells are morphologically indistinguishable from gonia (except that there are 16 germ cells per cyst instead of 2, 4 or 8) and the chromosomes are intermingled and largely fill the nucleus. But by S2a the cells and nuclei are visibly larger and the nucleus becomes clearly tri-lobular. S2b nuclei exhibit three distinct and separate chromatin masses that correspond to the three major bivalents (based on FISH and other molecular analyses, as described below). As the nuclei continue expanding, the bivalents continue to separate and come to occupy distinct, albeit formless, “territories” closely juxtaposed to the nuclear envelope. Throughout prophase I a single nucleolus is present; by S2b it is clearly associated with the sex chromosome territory (where the nucleolar organizers (NORs) are located) and serves as a convenient marker for it.29,31 The fourth chromosome pair, which is much smaller than the others, is sometimes separate from the other chromosome pairs (often in the middle of the nucleus) but other times co-mingled with one of the other territories, usually the X-Y territory.31,32 By stage S5 “mature spermatocytes” reach maximum size and the territories are not only separate but quite far apart. Although much of the nucleus appears devoid of DNA in fluorescently stained preparations, the nuclear lumen is actually full of highly decondensed and transcriptionally active DNA derived from the unwound “Y chromosome loops”33. For the most part, the chromosomes are decondensed throughout prophase I, rendering them largely inaccessible to classical cytological approaches. Homologs are presumed to be “conjoined” in some way since they share a common territory throughout prophase I but classical assays provide no insight into where or how they are conjoined (nor do molecular assays, as we will see). The major exceptions are the 4th chromosomes which, when they are not obscured by another territory, usually appear to be much more condensed and to have closely apposed homologs.32,34 (A terminological note: we use “conjunction”35 to refer to homolog connections for which molecular-level pairing is either known to be absent or for which no information is available, and reserve “pairing” for close associations of allelic sequences as detected by FISH or GFP spot assays.)
The meiotic division stages
The subsequent meiosis I and meiosis II division stages are subdivided into M1-M11 (see ref. 31 for details). At the onset of the meiotic divisions in late stage S6 and extending into M1, the nucleolus disintegrates and the chromatin masses rapidly condense into compact, roughly spherical chromosomes initially located peripherally in the nucleus. This is a landmark event as it renders the chromosomes accessible for cytological analysis. Indeed in classical (non-fluorescent) studies, this is the first stage at which chromosomes can even be clearly seen. Unfortunately, the chromosomes are so compact (“blobs,” in the common vernacular) that, at least for the major autosomes, individual homologs and chromatids usually cannot be distinguished. Nevertheless, the conjunction status of the bivalents can be readily assessed on the basis of size and number of blobs. In addition, in optimal squash preparations, the X and Y appear fully separate but remain connected by one or two string-like fibers.35 Once condensation is complete and the spindle has formed and penetrated the nucleus, prometaphase I begins, and the chromosomes begin migrating from peripheral locations toward the center. After a brief period of rapid chromosome movement driven by kinetochore-spindle interactions, the chromosomes congress to form a compact metaphase I cluster in the center of the spindle. Prometaphase I and metaphase I chromosomes remain highly compact but often have an elongated, stretched morphology that allows the homologs to be at least partly discriminated.28,35,36 The 4th homologs become clearly separated at prometaphase I but the distance between them is never more than about 1 um, indicating that they remain conjoined.32 Homologs split at the onset of anaphase I and migrate to the poles. Homolog splitting is accompanied by decompaction (distinct from chromatin decondensation which is delayed until telophase I) in which the “blob” morphology is replaced by a more conventional axial morphology in which chromosome arms can be clearly visualized. Throughout anaphase I and meiosis II, sister chromatid arms are split apart (due to early loss of arm cohesion, as described below) and chromatids are visibly connected at their centromere regions. Consequently these stages are optimal for analyzing cohesion phenotypes. At telophase I, nuclei re-form and the chromosomes briefly decondense. Meiosis II largely recapitulates the steps of the first division (at least cytologically), albeit with smaller (univalent) chromosomes, smaller cells and a much briefer prophase stage. Each post-meiotic cyst contains 64 spermatids with nuclei of equal size and DNA content. Mutations that disrupt meiotic segregation are readily identified at this stage on the basis of intra-cyst variation in nuclear size or DNA content. Starting from chromosome condensation in late stage S6, the division stages last about 4 ½ hours total with meiosis I and meiosis II being roughly equal in length.31
Homolog Pairing and Conjunction
Pairing in early prophase I
As mentioned above, Drosophila males dispense with both synapsis and recombination entirely. Prophase I chromosomes not only lack SCs but also the prominent axes decorated with cohesins and other lateral element proteins seen in nearly all other eukaryotes.34,37,38 Meiotic crossover frequencies are essentially zero throughout the genome, which in turn means that there are no chiasmata to connect bivalents.39 Another major departure from the standard script (this one shared with their sisters) is that the homologs enter meiosis already paired, obviating the need for a homology search. Mitotic pairing was first described over a century ago and was recently verified in spermatogonia by a fluorescent spot assay involving GFP-LacI proteins recruited to genomically inserted lacO arrays40-42 (Fig. 2). The resulting GFP foci appear as spots, the numbers of which can be used to monitor cohesion and pairing. In males homozygous for single lacO insertions, 50–70% of spermatogonia exhibit only one GFP spot, indicating that the allelic insertion sites (of which there are four at G2) are all paired; the remainder show two spots, indicating that homologous sites are unpaired but sister sites paired (presumably due to cohesion).42 Similar pairing frequencies have been reported for several types of mitotic cells in Drosophila.43,44 Entry into meiosis is accompanied by upregulation of pairing so that more than 90% of young spermatocytes (stages S1–S2a) display only one spot, the remainder two. However, there is no evidence for a mitosis-meiosis transitional loss of pairing, so no evidence for a de novo homology search.42 In female germ cells, pairing is also continuous through the mitosis-meiosis transition and pairing frequencies are upregulated in early female meiosis.45,46 The molecular basis for mitotic pairing remains unclear although several recent genetic studies in vivo and in tissue culture cells have identified genes that both promote and inhibit pairing.47-50 Interestingly, none of the genes involved in the male meiotic pairing and cohesion pathways (including the cohesins) discussed in this review are among the pairing promoters.
The fluorescent spot assay reveals that homologs are nearly always paired throughout S1 and S2a42 (Fig. 2). This is true for all autosomal lacO insertion sites that have been tested thus far (13) and is probably the general state for euchromatic regions. For technical reasons the assay has not yet been extended to heterochromatin. (X-Y pairing is a special case that will be discussed below.) A different fluorescent spot assay, based on probes (both antibodies and GFP fusion proteins) for the centromere-specific histone protein CID (Centromere IDentifier) has allowed the cohesion/pairing status of centromeres to be monitored.51,52 During S1 and S2a, the number of CID spots is quite variable, ranging from 1–8, although most nuclei exhibit 1–3 spots42,34. This likely reflects two different forces: cohesion between sister centromeres, which seems to be nearly absolute (else nuclei with more than 8 spots would be seen), and non-specific centromere clustering akin to chromocenter formation. Homologous pairing of centromeres cannot be absolute since cells with 5–8 spots are not uncommon, but because of clustering it is impossible to rule out some homologous pairing. Data for heterochromatic sites, obtained from FISH studies utilizing repeat sequence probes with unique or semi-unique chromosome distributions, are limited but interesting.32 Two target sites near the centromeres of chromosomes 2 and 3 exhibited around 50% pairing in stage S1, but three other non-centric heterochromatic sites, one each on the three autosomes, exhibited euchromatin-like pairing frequencies around 90%.
The mid-G2 transition
Three dramatic changes in pairing status and chromosome organization take place at the “mid-G2” transition in late S2b (Fig. 2). The first, already mentioned above, is the emergence of separate homologous chromosome territories.31 The second is the sudden and permanent loss of pairing and cohesion at all euchromatic sites. The single GFP-LacI spots dissociate into four spots and from then through the remainder of prophase I the four allelic spots diffuse independently of one another, although they usually remain within a common territory. When the chromosomes condense, the allelic spots ratchet in close to each other but never fuse.42 The third is homologous centromere pairing, a robust but transient event, largely restricted to stage S3, which usually manifests as four CID spots per nucleus, one spot per territory. The significance of centromere pairing for subsequent events is not known. Except for the 4th chromosome centromeres, which remain paired throughout prophase I, homologous centromere pairing is completely lost at the end of S3 and centromeres remain unpaired for the remainder of meiosis I42,34. However, unlike the earlier unpairing of the euchromatin, unpairing of centromeres is not accompanied by loss of centromere cohesion. Consequently, most prophase I cells after stage S3 exhibit seven spots (or six when the 4th chromosome pair is commingled with another territory), never more than eight.34 The loss of pairing extends to heterochromatic sites as well; six target sites in the centric heterochromatin of the 2nd and 3rd chromosomes, including sites both near and distant from centromeres, were assayed by FISH and all found to be largely unpaired from S3 through the end of meiosis I. However, as expected, a 7th target site on chromosome 4 exhibited very strong pairing throughout prophase I.32
It is worth noting the parallel between the pairing/unpairing cycle in Drosophila euchromatin revealed by the GFP-LacI spot assay and early prophase events in the standard version of meiosis. In most eukaryotes, allelic sites throughout the genome become closely aligned and exhibit high pairing frequencies during early prophase I, coincident with synapsis, and the disassembly of SC at mid-prophase I is accompanied by a general loss of intimate pairing between homologs. Except for the lack of SC, this description applies equally well to male Drosophila.
Site-specific pairing of the sex chromosomes
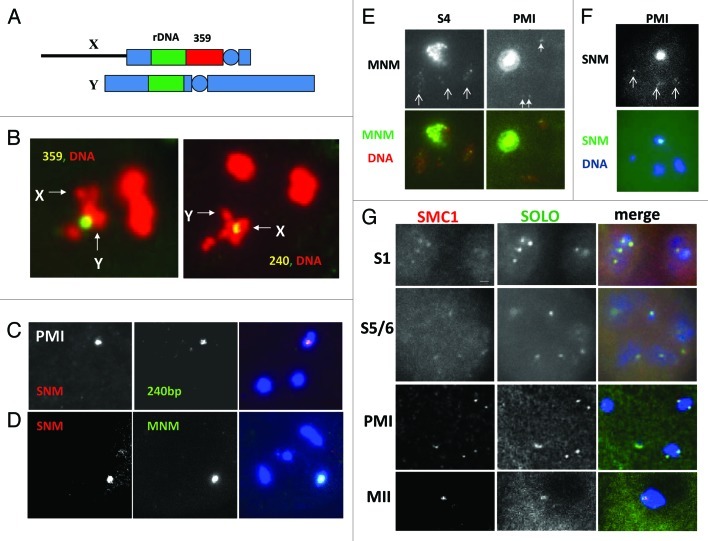
The sex chromosome pair faces a special challenge because the X and Y chromosomes are essentially devoid of homology except for the nucleolus organizers (NORs). Located in the central region of the heterochromatin of XL and near the base of YS, the NORs house the tandemly repeated rRNA (rRNA) genes (Fig. 3A). The basic repeat, which contains the genes for the 18S, 5.8S and 28S rRNAs as well as an upstream “intergenic spacer” (IGS) is about 12 Kb in length.53 Since there are usually about 150–250 copies per X and Y, the NORs provide some 2–3 Mb of homology. Three lines of evidence indicate that the NORs constitute the X-Y pairing/conjunction region. First, X heterochromatic deletions that remove all of the rDNA abolish X-Y conjunction and lead to random X-Y disjunction at anaphase I.54-57 Second, transgenes containing either single complete rRNA genes or partial genes that include components of the IGS can restore X-Y conjunction and segregation when inserted on heterochromatically deficient X chromosomes. Further studies led to the identification of a 240 bp repeat, present in 6–10 copies in each IGS region, as the main pairing site. Transgenes that contain arrays of at least six 240 bp repeats have pairing activity whereas transgenes that carry much larger rDNA fragments but without IGS regions are completely inert.58-62 Thus X-Y pairing seems to be mediated largely by a repeated 240 bp sequence that is present at copy numbers of around 1200–2000 per X and Y chromosome and that is absent from the autosomes. Third, X-Y pairing can be visualized in late stages of meiosis I by both conventional and fluorescent cytological methods. The universal observation is that pairing is restricted to discrete regions in the vicinity of the NORs35,63 but it is unclear from classical low-resolution studies whether pairing occurs in the rDNA or at nearby sites. However, FISH experiments using rDNA-derived probes clearly show that the rDNA regions are paired even though flanking heterochromatic repeats are not (Fig. 3B). Unfortunately it has not yet been possible to ascertain the pairing status of the X and Y prior to DNA condensation. rDNA sequences, including the 240 bp repeats, are dispersed throughout the nucleolus prior to S6 and although they serve as excellent FISH targets, there are invariably multiple signals and the X and Y-derived signals cannot be distinguished.64
Figure 3. Homolog pairing and localization of SNM, MNM and SOLO in wild-type spermatocytes. (A) Diagram of the X-Y pair showing restricted X-Y homology at shared rDNA loci. 359 is the 359 bp satellite sequence, a highly repeated element located between the centromere and the rDNA of the X chromosome and unique to the X. Circles represent centromeres, rectangles represent heterochromatin and line represents X euchromatin (not to scale). (B) FISH using fluorescently labeled probes specific for the 359 bp satellite (left panel) or the 240 bp repeats in the spacers of the rDNA repeats (right panel). DNA was counterstained with DAPI. Arrows point to X and Y chromosomes. 359 signal is clearly displaced from paired region in left panel, but 240 bp signal overlaps the paired region in the right panel. Note there is only one 240 bp signal signifying that the X and Y rDNA loci are paired. (C). Co-immuno-FISH analysis using 240 bp probe and anti-SNM antibody. DNA was counterstained with DAPI. After chromosome condensation and nucleolar dissolution, the 240 bp repeat and anti-SNM signals co-localize on the condensed X-Y pair until anaphase I. (D) MNM-GFP and SNM colocalize on the X-Y bivalent at prometaphase I. MNM-GFP detected by native fluorescence, SNM by an anti-SNM antibody and a FITC-conjugated secondary antibody. DNA was counterstained with DAPI. (E) MNM-GFP localizes to all four chromosome pairs throughout meiosis I (S4 and PMI shown). White arrows point to autosomal foci. X-Y signal is overexposed to allow autosomal signals to show. (F) SNM-Venus localizes to all four chromosome pairs throughout meiosis I (PMI shown). White arrows point to autosomal foci. DNA stained with DAPI. X-Y signal is overexposed in upper panel to allow autosomal signals to show. (G) Colocalization of Venus-SOLO and SMC1 during meiosis I. SMC1 was detected with an anti-SMC1 antibody and a FITC-conjugated secondary antibody, and Venus-SOLO was detected by native fluorescence. DNA was counterstained with DAPI. The spots of both proteins co-localize on centromeres throughout meiosis (based on colocalization with the centromere specific protein CID (not shown)).
Do autosomes also use specific pairing sites?
These findings stimulated speculation that specific sites might also be important for autosomal pairing/conjunction. After all, in the absence of recombination, why bother with genome-wide homology pairing? What purpose would it serve? The answer seems to be that euchromatic homology is the major determinant of chromosome segregation patterns, at least for the two large autosomes. The evidence for this comes from cytogenetic studies in which various types of chromosome rearrangements were tested for ability to perturb segregation patterns (reviewed in refs. 65 and 66). There were two main findings. First, manipulating the amount or linkage arrangement of heterochromatin on chromosome 2 or 3 made no difference whatsoever in how the chromosomes segregated.67-69 In one representative study that gives the flavor of this genre, the centromeres along with large blocks of flanking heterochromatin were swapped between chromosomes 2 and 3. In heterozygous males carrying the two swapped chromosomes and normal 2nd and 3rd chromosomes, the chromosomes segregated solely on the basis of euchromatic homology and appeared indifferent to the origin of their centromeres.67 These findings were reinforced by the FISH study involving repetitive heterochromatic probes, described above, that failed to find any stable pairing sites in the heterochromatin of chromosomes 2 or 3.32 Second, euchromatic homology directs conjunction and segregation patterns in a democratic manner, meaning that the likelihood of two chromosomes conjoining and segregating from one another is proportional to the amount of euchromatic homology they share. For example, Y chromosomes carrying inserted euchromatic segments from chromosome 2 will conjoin with a normal 2 (resulting in fusion of the X-Y and 2nd chromosome territories) at a frequency roughly proportional to the size of the segment. Moreover, in translocation heterozygotes, where one of the 2nd chromosomes is deficient for the transposed segment, the Y segregates preferentially away from the normal 2nd chromosome to a degree proportional to the size of the segment.69 Taken together, the genetic and cytological evidence indicate that the major autosomes utilize genome-wide homology pairing, not site-specific pairing to establish homology and determine segregation patterns. Moreover, these findings clearly indicate that the homology pairing mechanism is sufficiently robust that very limited regions of homology can be detected and utilized to direct segregation even when separated by interchromosomal rearrangements. Similarly, meiotic pairing in female meiosis is very robust and largely indifferent to translocations and inversions.45,46
It remains unclear how the autosomal homologs remain connected after the loss of intimate euchromatic and centromere pairing after S3. The search for stable homolog connection sites in the heterochromatic regions of the major autosomes has thus far come up empty.32 Nevertheless, upon condensation it is clear not only that homologs are conjoined but that the information needed for homolog alignment either has been preserved or regenerated. Allelic GFP-LacI spots ratchet in next to each other, and, in optimal cytological preparations, the arms of the major autosomes can be seen to be clearly aligned.35,36,42 Although we do not understand the mechanism by which homologs remain associated in late prophase I, some of the responsible proteins have been identified.
Homolog conjunction genes
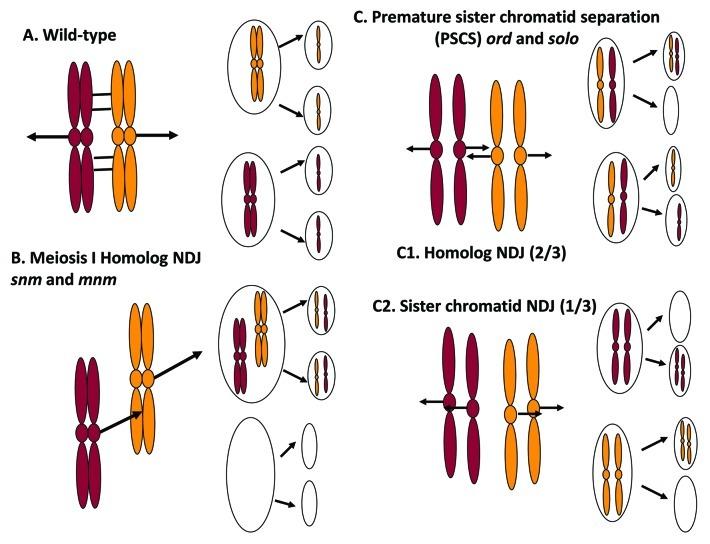
Alleles of three genes with central roles in the homolog pairing/conjunction pathway, teflon (tef), mod(mdg4) in meiosis (mnm) and stromalin in meiosis (snm) were identified in a multi-investigator genetic screen of the “Zuker” collection of mutagenized autosomes70-72,64. The null phenotype for all three genes is similar: premature homolog separation and high frequencies of homolog nondisjunction (NDJ, failure to disjoin to opposite poles) at male meiosis I. Figure 4 depicts the various types of NDJ that are caused by the mutations discussed extensively here. This phenotype is most obvious after the onset of chromosome condensation. In snm and mnm mutants, instead of condensing as four bivalents, the chromosomes condense as up to eight univalents and become dispersed along the metaphase I spindle64 (Fig. 5). In tef mutants, unpaired autosomes are frequently observed after condensation but the sex bivalent is always intact.72 Anaphase I chromosome segregation appears random in snm and mnm mutants and genetic cross tests confirm this impression. NDJ frequencies for the X-Y pair are near 50% for strong alleles, consistent with random assortment, and autosomal NDJ frequencies are also very high and probably random.64 In tef mutants, 4th chromosome segregation is random and 2nd and 3rd chromosome NDJ also very high, but X-Y segregation is entirely normal.72 Thus, tef is specific for autosomal conjunction and segregation while snm and mnm are required for conjunction and segregation of all four pairs. For mutants in all three genes, the disomic NDJ products are entirely of the “homolog NDJ” type, meaning that they inherit two homologous chromatids rather than two sister chromatids, (e.g., XY sperm but no XX sperm in snm and mnm mutants). Premeiotic and postmeiotic germ cell development appear normal, as do other aspects of meiosis including spindle formation and structure, chromosome condensation, kinetochore function, cytokinesis and all aspects of meiosis II. Thus tef, snm and mnm are specific for homolog conjunction and segregation. They are also apparently specific for male meiosis; no somatic or female meiotic phenotypes have been observed in any of the mutants, although Tef transcripts have been detected in embryos and in various somatic tissues.73 However, at least for snm and mnm, there is an earlier (pre-condensation) meiosis I phenotype: loss of territorial integrity in stages S5–S6 (Fig. 5). Although chromosome territories form normally, they gradually lose definition, becoming more diffuse and often seeming to bleed into each other. Accompanying this, GFP-LacI tagged alleles diffuse further from one another than in wild-type and are sometimes present in different territories. Thus it is clear that homolog connections are impaired in snm and mnm mutants long before the chromosomes condense. However, snm and mnm mutations do not perturb pairing either in spermatogonia or young spermatocytes in the GFP-LacI/lacO assay, indicating that they are not required for mitotic pairing or for the upregulation of genome-wide pairing that occurs at the onset of meiosis.64
Figure 4. Chromosome segregation patterns in Drosophila wild-type and meiotic mutants. (A) Wild-type bivalents are held together by the conjunction complex (cross-bars) which enables them to achieve bipolar orientation and segregate to opposite poles during meiosis I. Sister chromatids orient to the same pole (mono-orient) at meiosis I, then to opposite poles at meiosis II. (B) Homolog nondisjunction at meiosis I in snm and mnm mutants. Failure to maintain conjunction leads to premature homolog separation and random segregation at meiosis I. However, sister chromatids always mono-orient and segregate to the same pole at meiosis I. Meiosis II is normal and sister chromatids segregate to opposite poles. (C) Premature sister chromatid separation leads to random chromatid segregation in ord and solo mutants. Sister centromeres dissociate prematurely and orient randomly at meiosis I. The SNM-MNM conjunction complex is still present (not shown) and maintains bivalent integrity. In addition to the two types of balanced segregations pictured, unbalanced meiosis I segregations (3:1 or 4:0) can also occur but are mostly suppressed by the conjunction complex. (C1) Sister chromatids segregate to opposite poles at meiosis I (“equational” segregation) 2/3 of the time. Each secondary spermatocyte receives one chromatid from each homolog, which then segregate randomly. ¼ of the resulting spermatids will carry two homologous chromatids, resulting in homolog NDJ (same outcome as B but different mechanism). (C2) Sister chromatids segregate to the same pole (reductional segregation) at meiosis I 1/3 of time. Each secondary spermatocyte inherits a pair of sister chromatids (as in wild-type) but they are disconnected and segregate randomly at meiosis II. Diagram shows only the 2:0 segregations at meiosis II but 1:1 segregations are equally frequent. ¼ of the resulting spermatids will carry two sister chromatids, yielding sister chromatid NDJ.
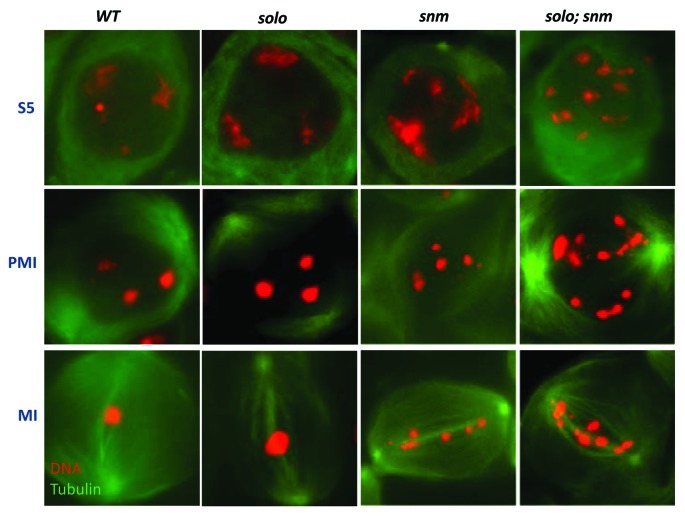
Figure 5. Meiosis I chromosomes from wild-type, snm, solo and solo; snm spermatocytes at S5, prometaphase I (PMI) and metaphase I (MI). DNA was stained with DAPI and tubulin with an anti-tubulin antibody and detected with a FITC-conjugated secondary antibody. Note the poorly formed territories at S5 and the prematurely separated univalents at PMI and MI in snm spermatocytes. The bivalents appear normal in solo spermatocytes in this preparation but other squash methods reveal abnormal, loosely packed bivalents often with protruding single chromatids. solo; snm double mutants form multiple mini-territories at S5 which condense into single chromatids at PMI and MI.
Molecular analysis of SNM and MNM
These findings suggest that snm, mnm and tef function primarily to maintain homolog connections after the loss of intimate pairing in mid-prophase, a role that can be thought of as analogous to that played by chiasmata in conventional meiosis. Analysis of the products of snm and mnm using both antibodies and fluorescently tagged constructs confirms this idea. SNM and MNM are chromosomal proteins that co-localize to all four chromosome pairs throughout meiosis I until anaphase I when they disappear suddenly and permanently. The X-Y localization pattern is most revealing. SNM and MNM colocalize to the nucleolus, where the rDNA is sequestered, during prophase I. After condensation, they colocalize with each other and a 240 bp repeat FISH probe to a single prominent focus on the X-Y pair that coincides with the paired rDNA repeats64 (Fig. 3C and D). Deletion of the X chromosome rDNA abolishes localization to the X but not to the Y, and X localization is restored by transgenes carrying 240 bp repeat arrays. SNM and MNM also localize to transgenes containing arrays of 240 bp repeats inserted on mini-X chromosomes where they promote conjunction and segregation of pairs of such minichromosomes from each other.74 This shows that 240 bp repeats function to recruit SNM and MNM to sex chromosomes and that they must be present on both homologs to mediate conjunction and segregation. Localization of SNM and MNM to condensed sex chromosomes during late meiosis I is mutually co-dependent; neither protein localizes in the absence of its partner. However, during prophase I, dependence is one-way; SNM localizes to nucleoli even in mnm mutants but MNM-GFP does not localize to nucleoli in snm mutants.64 Unfortunately the significance of nucleolar localization of SNM and MNM is unclear as we have no way of ascertaining whether the X and Y 240 bp repeat arrays are paired while in the nucleolus. Nevertheless, these findings suggest that SNM and MNM function together as a complex.
The autosomal localization pattern is more complex but consistent with MNM and SNM functioning together in a complex. Both MNM-GFP and SNM-Venus localize to multiple faint foci on all three autosomes throughout prophase I (Fig. 3E and F). The proteins are clearly present on chromosomes during S1 (but not earlier) even though they are dispensable for allelic pairing. After the onset of condensation, the foci coalesce into a small number of larger foci (sometimes just two) which persist until anaphase I when they disappear along with the X-Y focus (Fig. 3E).64 Interestingly, MNM-GFP localization to autosomes both during prophase I and in post-condensation stages is abolished by mutations in either snm or tef.64 These results indicate that the pathway for recruiting MNM and SNM to autosomes is different from that for the X-Y pair. X-Y localization is dependent on the 240 bp repeats but independent of TEF, whereas recruitment to the autosomes, which lack 240 bp repeats, is dependent on TEF. The autosomal sites of MNM and SNM localization have not yet been identified. These data are consistent with the idea that MNM and SNM (and perhaps TEF) are components of a specialized “homolog conjunction complex” that serves as a substitute for chiasmata, maintaining connections between homologs throughout prophase I and enabling conjoined homologs to achieve bipolar orientation on the metaphase I spindle.
How do TEF, MNM and SNM mediate homolog conjunction?
The structures of TEF, MNM and SNM provide potential clues to the roles in homolog conjunction. TEF is predicted to contain three zinc finger motifs, two at one end and one at the other, leading to the suggestion that it might use its zinc fingers to bind directly to both homologs (perhaps by dimerizing at one end and binding to chromosome conjunction sites at the other, much like SC transverse filament proteins), and thereby serve as a protein bridge.73 Unfortunately, it has not been possible to test this model as attempts to visualize TEF protein have thus far been unsuccessful. MNM is one of 31 predicted alternative splice products from the mod(mdg4) locus.64,75,76 Several others are chromosomal proteins expressed in somatic cells; the best characterized, Mod(mdg4)67.2, is a chromatin insulator protein that associates with Su(Hw) at gypsy insulator sites and regulates enhancer-promoter communication.77 Like other Mod(mdg4) proteins, MNM has a BTB domain at its N-terminus and a “FLYWCH domain” at its C-terminus.76 The BTB domain is a large dimerization/multimerization domain found in many classes of proteins that sometimes functions in formation of large protein complexes and may contribute to formation of the prominent chromosomal foci in spermatocytes.78 As in most Mod(mdg4) proteins, the FLYWCH domain includes a non-canonical (unusually spaced) C2H2 motif.76 A similar motif functions in DNA binding in the C. elegans protein PEB-1 but the corresponding motif of Mod(mdg4)67.2 is involved in binding to Su(Hw).79,80 Intriguingly, a substitution mutation in the MNM C2H2 motif (replacing the first histidine with a tyrosine) is a null allele in which the protein product is present in the nucleus but fails to localize to chromosomes.81 Thus the C2H2 motif is likely the primary determinant of chromosome localization, but whether it functions by binding to DNA, to another protein or something else remains to be determined. SNM is also intriguing. It is a paralog of the cohesin protein SA/Stromalin, the protein that is not part of the cohesin ring.64 This raises the possibility that homolog conjunction has something in common with cohesion, a possibility that is under active investigation.
The taxonomic distribution of SNM is a potentially interesting issue. Achiasmate male meiosis appears to be universal within the genus Drosophila and also extends to some “higher Dipterans” (but not mosquitoes). We have found SNM homologs in all sequenced Drosophila genomes but not in the Anopheles genome.64 We predict it will be present in genomes of dipterans with achiasmate male meiosis but not in those with chiasmate meiosis.
Homolog Segregation
Two recent reports have described mutations that impair segregation of homologs at anaphase I without disturbing homolog conjunction. Two of the affected genes, Cap-D3 and Cap-H2, encode non-SMC components of the condensin II complex, a conserved complex with roles in chromatin condensation and chromosome resolution and other processes during both mitosis and meiosis.82-87 The third is dtopors, which encodes a conserved ubiquitin/SUMO ligase that has been shown to interact with a variety of chromosomal proteins.88-90
Condensin II
All strong condensin II alleles were completely sterile86 and exhibited severely defective chromosome condensation. The DNA was smeared uniformly throughout the nucleus during prophase I and condensed bivalents were completely absent during S6-prometaphase I. Remarkably, however, the chromosomes eventually condensed and congressed to form reasonably normal metaphase I and anaphase I figures, except that 30–40% of the anaphase I cells exhibited chromatin bridges, some between homologs and others between non-homologs. Intriguingly, teflon mutants partially suppressed both the homologous and heterologous bridges, suggesting that condensin II may function in opposition to homolog conjunction in some way. One possibility is that condensin II is required to release the inter-homolog connections that the conjunction complex creates or preserves. A role as an anti-pairing factor is consistent with studies that show condensin II to be responsible for the suppression of polyteny in nurse cells in stage 6–10 egg chambers.49 A related suggestion is that inter-homolog connections might take the form of DNA entanglements, perhaps generated during the early prophase intimate pairing period.42 These tangles might be preserved (by, e.g., SNM and MNM) until anaphase I and then removed by the combined action of condensin and topoisomerase II. This suggestion has not been pursued but perhaps should be, especially in light of the failure to find any stable autosomal pairing sites. A role of condensin II in removing DNA tangles is certainly consistent with its established enzyme activities and its interaction with topoisomerase II.85
dTopors
Unlike the condensin II mutants, all of the dtopors alleles, even null alleles, were fertile, albeit weakly so, and NDJ frequencies were high for all chromosomes, approaching 50% for the sex and 4th chromosome pairs, consistent with random assortment.90 Interestingly, chromatin bridges were observed in nearly 100% of anaphase I cells and while such bridges might be expected to cause mostly chromosome loss, disomic NDJ products (e.g., XY) were recovered at frequencies comparable to those in snm and mnm mutants that undergo homolog pairing failure and NDJ without bridges. Moreover, no anaphase II bridges were reported and all the disomic exceptions resulted from homolog rather than sister chromatid NDJ (e.g., XY but not XX sperm), just like in snm and mnm mutants. The condensation phenotype was also similar to snm and mnm mutants: chromosome territories that were normal in mid-prophase but appeared somewhat less condensed than in wild-type during S5, S6 and M1. However, homolog conjunction at late prophase I and prometaphase I appeared to be intact. Moreover, mnm mutants did not suppress anaphase bridging, indicating that the bridges in dtopors mutants, unlike those in condensin II mutants, are not dependent on homolog conjunction. There were two other prominent phenotypes in dTopors primary spermatocytes, neither of which are seen in mnm, snm or condensin mutants: “nuclear blebbing,” associated with malformation of the nuclear envelope and lamina and mislocalization of nuclear lamins, and premature separation of centrosomes at prometaphase I (instead of telophase I as in wild-type), leading to tetrapolar spindles in roughly half of meiosis I cells. The phenotype is thus very complex. Any of the four main cytological phenotypes—nuclear envelope/lamina malformation, tetrapolar spindles, incomplete chromatin condensation and anaphase I bridging—could lead to homolog missegregation and it will be important to determine how the diverse phenotypes are related. However, it is not easy to reconcile any of them with the observed missegregation pattern, a pattern that is usually diagnostic of premature homolog separation.
Meiotic Cohesion in Drosophila
Cohesion in wild-type meiosis
A fairly comprehensive, albeit low-resolution, picture of cohesion dynamics in wild-type meiosis has emerged in the last decade. As described above, cohesion was shown, by the GFP-LacI/lacO assay, to be prevalent throughout the euchromatin of the major autosomes during early prophase but lost suddenly and permanently in late S2b/S3 (Fig. 2). A site in the X euchromatin was shown to behave the same way.42 However, cohesion at centromeres is preserved throughout meiosis until anaphase II, based on the CID spot assay42,34. What about heterochromatin more broadly? The FISH study involving repeat sequence probes described above provided information about cohesion at 10 widely distributed heterochromatic sites.32 The results were more complex than expected. Five non-centromeric sites, including all three tested sites on the Y chromosome and two on chromosome 2, behaved like euchromatin, showing little to no cohesion at either prometaphase I or metaphase I. Three other non-centromeric sites, two blocks of the 1.686 Kb satellite sequence on 2L and 3L, and one block of AATAT repeats on chromosome 4, behaved like centromeres, exhibiting strong cohesion throughout meiosis I and II. Finally, two centromere-proximal sites, Dodeca on chromosome 3 and Rsp on chromosome 2, exhibited high cohesion frequencies both early (S1–S3) and late (prometaphase I and prometaphase II) but only moderate cohesion frequencies (as low as 50%) in late prophase I (S4–S6). Thus, unlike in the euchromatin where all sites behave synchronously and are either fully cohesive (until S2b) or fully uncohesive (after S2b) or at centromeres, which maintain cohesion throughout meiosis, cohesion behavior in the heterochromatin varies among sites. The results suggest that the heterochromatin may be a patchwork of sites with different cohesion properties.32
Cohesin in Drosophila
The Drosophila genome encodes the four canonical cohesin subunits, SMC1, SMC3, Rad21 and SA, and the proteins have been shown to localize to chromosomes in the expected pattern in mitotic cells, broadly distributed during interphase, then concentrated on centromeres from prophase until metaphase.91-93 The genome also encodes homologs of all of the major known cohesin regulatory factors: SCC2/Nipped-B and SCC4 (which comprise the adherin complex required for cohesin loading on DNA); Pds5 and Sororin/Dalmation (a mostly conserved complex that colocalizes with cohesin and is needed for cohesion establishment and maintenance); San and Deco (two acetyl transferases, the latter of which is a homolog of yeast Eco1, required for SMC3 acetylation and cohesion establishment); and Wapl (which also complexes with Pds5 and promotes cohesin removal in prophase).5,94-99 All of these genes are essential, with lethality occurring for most late in larval development, as is typical for mitotic genes, although rad21 mutants are embryonic lethals. For most of them (including rad21, SA, SCC2/Nipped-B, Pds5, Sororin/Dalmation, san and deco), mutation or RNAi depletion has been shown to disrupt cohesion, often severely, either in vivo or in tissue culture cells or both. The most convincing evidence for cohesin-mediated cohesion in Drosophila has come from studies of Rad21. Rad21 has been eliminated in embryos by three different methods: RNAi depletion,92 mutation of the native rad21 gene100,101 and TEV protease-mediated cleavage of a Rad21 engineered to contain a TEV cleavage site in a rad21 null background.101 In all three studies, cohesion was drastically impaired and in the latter case, the timing of cohesion loss was determined by the timing of TEV protease expression. Further support came from the discovery that the Drosophila Separase homolog is also essential and that mutant embryos exhibit failure to release cohesion at metaphase.102
Meiotic cohesins
Although mitotic cohesion in Drosophila appears to adhere fairly closely to the standard script, the meiotic story line is quite a bit murkier. Cohesin is clearly present and important but its composition has been difficult to pin down and its roles remain only partially defined. Antibodies against SMC1 and SMC3 have enabled investigators to study cohesion distribution in situ. In oocytes, both proteins localize strongly to centromeres and to chromosome axes where they colocalize with the SC transverse filament protein C(3)G.103,104 Moreover, recent analyses of germ-line clones of an SMC1 deletion and of a germ-line specific SMC3 RNAi line have clearly established an essential role for both proteins in SC formation and homologous centromere pairing in early prophase I.103,105 In spermatocytes only SMC1 has been characterized thus far. The only prominent site of SMC1 localization in male germ cells is to centromeres (based on colocalization with CID).34,64 Centromere localization is evident from the onset of meiosis and is sustained until anaphase II. Genetic data detailed below show that SMC1 localization to centromeres correlates perfectly with the presence or absence of centromere cohesion (at least after stage S4). Perhaps surprisingly, the rDNA does not appear to accumulate much SMC1 and SMC1 does not visibly colocalize with SNM and MNM.64 Also surprising is the apparent lack of SMC1 on chromosome arms during S1 and S2 when arm cohesion is quite strong. However, it is certainly possible that SMC1 localizes to sites other than centromeres but not at high enough levels to detect with currently available antibodies.
The situation with respect to other cohesin subunits is less clear. Although antibodies against Rad21 and SA are available and work well on mitotic chromosomes, there have been no reports thus far of localization to meiotic chromosomes in either sex. The fly genome encodes meiosis-specific paralogs of both Rad21 and SA, but no apparent Rec8 ortholog. As discussed above, SNM, a SA paralog, is male-specific and central to the homolog conjunction pathway but has little or no role in cohesion.64 C(2)M is a kleisin homolog required for normal synapsis and recombination in female meiosis. It colocalizes with SMC1 on chromosome axes so may well be a cohesin component. However, C(2)M is female-specific (at least in function), expressed only during prophase I (apparently absent during S phase) and not essential for sister chromatid cohesion.106,107 Moreover, despite the presence of putative Separase cleavage sites in C(2)M, mutation of those sites did not impair C(2)M function or perturb meiotic segregation. Separase is clearly active in female meiosis, based on cleavage of a known substrate (Three Rows, a regulatory subunit of Separase) but its target in cohesin, if any, has yet to be identified.107
Roles in female meiosis for two of the cohesin co-factors have been described, but not yet thoroughly analyzed. Nipped-B/SCC2 localizes to SCs but not to centromeres in prophase I and is required for maintenance of SCs in mid-late pachytene.104 Pds5 is required for timely repair of meiotic double strand breaks but appears to be dispensable for SC formation and maintenance.108,109 San appears to be completely dispensable in the female germline despite being required for mitosis in embryos and somatic cells.110 None of the cohesin co-factors has been analyzed in male meiosis thus far.
MEI-S332 as a cohesin protector
Although a role of Separase in meiotic homolog segregation has yet to be directly demonstrated, indirect evidence strongly suggests that it has one. MEI-S332 is the founding member of the Shugoshin family and was shown many years ago to be required for centromere cohesion during meiosis II in both sexes.23,27 mei-S332 mutants lose centromere cohesion shortly after the onset of anaphase I and undergo high frequencies of sister chromatid NDJ during meiosis II. The MEI-S332 protein localizes to meiotic centromeres late in prophase I and remains there through anaphase II. It also localizes to mitotic centromeres but is not essential for mitotic cohesion.111,112 Shugoshins in other eukaryotes have been shown to prevent cleavage of Rec8 during anaphase I27 (Fig. 1). Until recently it has been unclear what, if anything, MEI-S332 protects in Drosophila. When MEI-S332 was depleted from S2 cells by RNAi, no MEI-S332 could be detected on centromeres yet Rad21 localized to centromeres at normal levels.113 However, it was recently shown that mei-S332 mutations lead to premature loss of SMC1 from male meiotic centromeres.34 Centromeric SMC1 foci disappear at anaphase I instead of anaphase II, in accord with the timing of cohesion loss in mei-S332 vs. wild-type. Thus, like other Shugoshins, MEI-S332 protects centromeric cohesin during anaphase I. Although the mechanism by which it does so in Drosophila remains to be determined, data from mitotic cells suggest that, like Shugoshins in other organisms, MEI-S332 may collaborate with the phosphatase PP2A.27 In S2 tissue culture cells, MEI-S332 was found to co-immunoprecipitate with several subunits of PP2A and its centromere localization was disrupted by RNAi depletion of the PP2A subunit Widerborst.114 In any case, the requirement for a Shugoshin to prevent cohesin removal strongly suggests that Separase is activated at anaphase I. This is intriguing in light of the absence of chiasmata and of arm cohesion.34 It will be of considerable interest to determine whether Separase is required for homolog segregation.
Regulation of MEI-S332
Several studies have addressed the regulation of MEI-S332 loading and removal at centromeres. Two kinases have been found to play particularly prominent roles, Aurora B in loading of MEI-S332 and POLO in its removal. Aurora B localizes, as a component of the Chromosome Passenger Complex (CPC), to inner centromere regions during prometaphase where it plays a major role in correcting erroneous kinetochore-microtubule attachments, thereby promoting bipolar orientation.115 A hypomorphic mutation, IncenpQA26, in the gene for INCENP, a CPC component that recruits and activates Aurora B, was found to significantly reduce accumulation of MEI-S332 on male meiotic centromeres.116 Subsequent analysis revealed that centromere accumulation of MEI-S332 depends both on a direct interaction with INCENP and phosphorylation of MEI-S332 by Aurora B. Moreover, a mutant MEI-S332 protein that could not be phosphorylated by Aurora B proved to be recruited inefficiently to centromeres in tissue culture cells.116 This mechanism may be conserved; components of the Passenger Complex are required for Shugoshin centromere localization in fission yeast, mice and humans. Moreover, mouse and human Shugoshin must be phosphorylated by Aurora B to recruit PP2A to centromeres.27
polo mutations, on the other hand, cause retention of MEI-S332 beyond anaphase (anaphase II in meiosis).117 MEI-S332 has two consensus POLO binding sites and is a strong POLO phosphorylation target. Mutation of one of the POLO binding sites strongly enhances MEI-S332 retention on mitotic centromeres, indicating that binding and phosphorylation by POLO are required for delocalization of MEI-S332. Analysis of polo mutants also revealed that delocalization of MEI-S332 is separate from its functional inactivation.117 In polo mutants, sister chromatids separate successfully, albeit with elevated NDJ, despite the retention of MEI-S332, indicating that MEI-S332 is no longer functional. Presumably it is inactivated sometime after anaphase I, the time when its protective role is really needed. The mechanism of this inactivation is unknown.
Perhaps surprisingly, binding of MEI-S332 to centromeres is unaffected by absence of cohesion or cohesin. MEI-S332 localizes normally to centromeres in embryos in which Rad21 has been depleted by RNAi and cohesion has been disrupted.118 In male meiosis, MEI-S332 localizes normally to the prematurely separated sister centromeres in ord mutants (discussed in depth below) that lack detectable centromeric SMC1.34,119
Core meiotic cohesion genes
Much of the information about meiotic cohesion in most eukaryotes has come from analysis of rec8 mutations. Thus the absence of a rec8 gene in Drosophila has hampered progress in defining the composition and functions of meiotic cohesins. However, in apparent compensation, nature has endowed Drosophila with two genes (maybe more, see below), orientation disruptor (ord) and sisters on the loose (solo), the products of which behave remarkably like Rec8 despite not sharing any sequence homology. ord and solo mutations cause high NDJ frequencies for all chromosomes in both sexes120-122,34,36. Strong alleles give total NDJ frequencies in the vicinity of 50% and generate substantial numbers of both “homolog” (e.g., XY) and “sister chromatid” (XX) NDJ sperm (Fig. 4C). The cytological phenotypes of ord and solo mutants are also similar. In chromosome squash studies, premature cohesion loss is particularly obvious in later stages of meiosis, from anaphase I on; most nuclei exhibit fully separated chromatids and metaphase II figures are largely absent. Not surprisingly, anaphase II segregation appears chaotic in both ord and solo mutants. Meiosis I phenotypes are considerably more subtle. Territory structure during prophase I and bivalent stability during S6-prometaphase I are not visibly perturbed in ord or solo mutants.34,36 However, bivalent morphology is decidedly abnormal. Bivalents often appear to be loosely packed with separated centromere domains and protrusions of single chromatid arms.34,36,121,122
Centromere cohesion has been assayed directly in solo and ord mutants both by FISH and by anti-CID staining. The most comprehensive study was an analysis of anti-CID spots in solo mutants at all stages of meiosis.34 Cells in stages S1-S4 exhibited a maximum of 8 (usually 6 or 7) CID spots in solo mutants as in wild-type, indicating that cohesion was intact. However, more than 8 CID spots (up to 15) were seen in 95% of nuclei at S5 and S6 and in 100% of nuclei at PMI. FISH studies in both solo and ord mutants yielded similar results and showed that cohesion is compromised at distal heterochromatic sites as well as pericentromeric sites by prometaphase I.32,34,123
solo mutants exhibit a novel “random 2:2” segregation pattern
solo and ord mutants abolish centromere cohesion prior to prometaphase I when centromeres must orient on the meiosis I spindle. In the absence of cohesion, sister centromeres would be expected to form independent kinetochores and to orient randomly. The expected outcome would be random chromatid segregation at both anaphase I and anaphase II. This should yield a 2:1 ratio of homolog to sister NDJ (e.g., XY to XX + YY sperm), a prediction in accord with a large volume of cross data involving strong alleles of both ord and solo.34,36,120-122 However, three observations are inconsistent with this hypothesis. First, in both solo and ord mutants, recovery of XXY and XXYY sperm (indistinguishable from each other for practical purposes) is far lower (by a factor of about 9) than would be expected from the random segregation model.34,120-122 Second, in solo mutants approximately 90% of anaphase I segregations appear balanced (roughly equal DNA masses at each pole), a far higher frequency than random segregation would predict.34 Third, FISH analysis using probes specific for the X and Y chromosomes revealed that the sex chromatids nearly always segregated XY:XY or XX:YY (at a 2:1 ratio); only 4% unbalanced segregations (XXY:Y, XYY:X or XXYY:O) were observed compared with an expected 53%.34 Evidently, the intact bivalents somehow constrain the chromatids to segregate two to each pole even though sister and homologous chromatids orient randomly with respect to one another. This explanation was confirmed by analyzing the effect of removing SNM/MNM on the solo phenotype. solo; snm double mutants exhibited completely detached chromatids throughout the meiosis I division stages and yielded a much higher frequency of unbalanced anaphase I segregations (43%) in the FISH assay.34 Evidently, in solo (and, we suspect, ord) single mutants, SNM and MNM serve to bundle the four chromatids into a semi-functional bivalent in which the four chromatids are constrained to orient two to each pole even though partner choice is random. The basis for this numerical constraint has not yet been determined.
Subcellular localization of SOLO and ORD
Unfortunately the predicted sequences of ORD and SOLO proteins have not been very revealing. Except for the N-terminal 137 amino acids of SOLO which are identical with the N-terminus of VASA (because the two genes share their first three exons) and which contain RGG motifs of unknown function, neither sequence matches any known proteins or protein domains.34,124 There are SOLO homologs in all Drosophila genomes but none have been found outside the Drosophilids. Surprisingly, SOLO is evolving quite rapidly within the genus; why a cohesion protein should evolve rapidly is a mystery.
The intracellular localization patterns, based on GFP or Venus tagged fusion proteins (partially confirmed for ORD by antibody staining, but for early stages only) are intriguing. The pattern for SOLO is the simpler of the two: Venus-SOLO localizes as distinct foci to centromeres (as judged by colocalization both with CID and SMC1) in mitotic spermatogonia and throughout meiosis until anaphase II when it disappears permanently34 (Fig. 3G). This pattern is seen whether expression is driven heterologously with Nanos-VP16 or with the native promoter of SOLO. At some stages, the SOLO foci appear distinctly broader than those of CID, suggesting SOLO may localize to heterochromatic domains outside centromeres as well. This impression is consistent with the observation that cohesion at prometaphase I at four heterochromatic loci, two of which are quite distant from centromeres, is abolished by solo mutations.32,34
ORD has a much more dynamic localization pattern. During S1-S3 ORD-GFP localizes predominantly to interchromosomal spaces in the nucleus and is largely excluded from the chromosomes. However, a subfraction of ORD-GFP associated with DAPI-bright spots in both spermatogonia and early prophase I spermatocytes; these were shown by colocalization with anti-CID to be centromeres.123,125 Beginning in stage S4 ORD gradually relocates to the chromosome territories where it becomes broadly distributed on the chromosomes while retaining the centromere-enriched subfraction. It then redistributes again, shortly before or during chromosome condensation, becoming restricted to centromere regions (again confirmed by colocalization with anti-CID), where it remains until disappearing at anaphase II. Interestingly, in prometaphase I and metaphase I spermatocytes, the CID and ORD foci overlap but do not fully coincide, suggesting that ORD, like SOLO, may localize to non-centromeric heterochromatic domains as well as to centromeres.123,125
The significance of the complex localization pattern of ORD is unclear. During S1-S3, when ORD is predominantly nonchromosomal, it colocalizes with two proteins—EAST, a nuclear skeleton protein, and dRING, a Polycomb Group protein and histone H2A ubiquitinase which interacts directly with ORD by yeast two-hybrid (and which continues to colocalize with ORD on chromosomes during S4-S6 but not on centromeres)—but neither of these proteins has been shown to have any role in cohesion. east mutations perturb the number and size of chromosome masses and their alignment at metaphase I in male meiosis but the basis for this phenotype remains to be determined.123,126-128 During S4-S6, when ORD is predominantly chromosomal but broadly distributed, arm cohesion has already been lost. Instead, ORD has been postulated to play a role in chromosome condensation, a suggestion supported by the “loosely-packed bivalent” phenotype in ord mutants.123
An intriguing aspect of SOLO and ORD localization is that both are present on pre-meiotic centromeres even though neither seems to be required for proper mitotic segregation.34,125 Although their roles in gonial cells are completely unknown, the fact that both proteins are present on centromeres in most 8-cell cysts as well as early G2 16-cell cysts strongly suggests that they are present during meiotic S phase, which occurs right after the end of the last gonial mitosis. This puts them in position to play a role in cohesion establishment.
SOLO, ORD and cohesin
Since the centromeric foci of SOLO (and, presumably, ORD) colocalize both spatially and temporally with SMC1, it seems likely that their roles in cohesion involve an interaction with cohesin. Two observations confirm this interpretation. One, mentioned above, is that the centomeric foci of ORD, SOLO and SMC1 are prematurely removed, at anaphase I instead of anaphase II, in mei-S332 mutants in which cohesion is lost at anaphase I.34,125 The second observation is that centromeric SMC1 foci are abolished at all stages of meiosis, including S1, by strong alleles of either solo or ord.34 Absence of SMC1 during meiotic S phase would seem to preclude establishment of cohesion, at least of the cohesin-dependent variety. This observation suggests that the cohesion and segregation phenotypes in strong solo and ord mutants might actually reflect a failure to establish cohesion during S phase rather than a failure to maintain it. These findings raise the question of whether SOLO and ORD are also required for maintenance of cohesion. Their centromere localization throughout meiosis and their shared dependence on MEI-S332 for persistence on centromeres after anaphase I strongly suggest such a role. There is supporting genetic evidence for such a role for ord—weak alleles have been shown to lose cohesion later in meiosis than strong alleles121,122—but not yet for solo, all extant alleles of which are null.34
Regulation of cohesion by the Passenger Complex
The hypomorphic allele of Incenp mentioned above was found to cause 16% sex chromosome NDJ in addition to disrupting MEI-S332 localization.116 While the occurrence of NDJ was no surprise given the importance of MEI-S332 in maintenance of cohesion, the pattern of NDJ was. Instead of mostly sister chromatid NDJ, as seen in mei-S332 mutants, IncenpQA26 males generated both XY and XX sperm at an approximately 2:1 ratio. Moreover, loosely packed bivalents, like those described in ord mutants, were common.111,116,121 Thus a hypomorphic Incenp allele causes phenotypes very similar to ord and solo mutations, suggesting a role for the CPC in regulating cohesion independent of its role in MEI-S332 maintenance. The finding that male-sterile mutations in the gene for Australin, which replaces the Borealin subunit of CPC specifically in the male meiotic divisions and is required for CPC centromere localization, disrupt both sister chromatid cohesion during prometaphase I and chromosome segregation, strongly supports such a role for the CPC.129 It will be of considerable interest to learn whether Aurora B and INCENP regulate centromere localization of SMC1, SOLO and ORD. Another possibility is a specific role in sister centromere mono-orientation; Aurora B has recently been shown to be involved in the mono-orientation pathways in both budding and fission yeast.130,131
How are Cohesion and Pairing Mediated in Early Prophase I?
Notably, neither solo nor ord appears to be required for cohesion in early prophase I. No defects in centromeric or heterochromatic cohesion have been reported prior to S5. Moreover, solo, snm and mnm are completely dispensable for both arm cohesion and euchromatic pairing, based on the GFP-LacI/lacO spot assay.34 This raises the question of how early prophase cohesion is mediated. Chromatid entanglement is an obvious possibility. However, if entanglement were the basis for arm cohesion, one might expect to observe gradual resolution of sister alleles during early prophase rather than a sudden and complete loss of arm cohesion at S2b/S3. It would also be difficult to explain the simultaneous loss of homolog and sister pairing. Could genome-wide homolog pairing in early prophase also be nothing more than entanglement? This possibility cannot be ruled out, but at least two alternatives seem plausible: there might be yet-undiscovered proteins that pair both sisters and homologs during early prophase I, or pairing might be in some way the default state of chromatin, not requiring any specific positive regulators, just absence of negative regulators. In the former case, the failure thus far to discover such proteins might be due to functional redundancy. In the latter case, it would be the unpairing of both sisters and homologs at S2b/S3 that would be actively regulated, not their prior pairing. It is not clear how this hypothesis would account for the increase in pairing frequency at the mitosis/meiosis transition. In any case, it is clear that non-cohesin-dependent early prophase cohesion is not sufficient to orient centromeres or prevent premature separation of sister chromatids. With respect to segregation patterns, it is the cohesin-based pathway that seems to matter.
Unresolved Questions
The past decade has been a time of significant progress in understanding the often idiosyncratic methods Drosophila males use to segregate their chromosomes. We now have a fairly comprehensive description of the timeline of cohesion and pairing patterns across the genome. We also have a growing list of core conjunction and cohesion proteins and much information about where and when these proteins do their jobs. But the list is almost certainly not complete, and we don’t understand in adequate molecular depth how the proteins we have already identified do their jobs. Many important questions remain unanswered. (1) Are there other proteins besides SNM and MNM in the conjunction complex? TEF is an obvious candidate but there could be others, including proteins that could not be identified in classical genetic screens because of essential roles in other processes. (2) Where do SNM and MNM localize on autosomes? Identifying sites of enrichment of conjunction proteins could be a major breakthrough in understanding the conjunction mechanism. (3) How are homologs segregated? An important unanswered question in this area is what role, if any, Separase has in releasing homolog connections. If it proves to be required for homolog segregation, identification of its targets could help to elucidate the conjunction mechanism. In addition, both condensin II and dTopors may provide fruitful avenues.86,90 A critical issue for both sets of mutants is to determine whether the unresolved connections between homologs are just random tangles or represent failure to release conjunction. (4) What are the molecular functions of SOLO and ORD? Both solo and ord mutations are phenotypically similar to rec8 mutations, and the possibility of ORD or SOLO being functional Rec8 homologs has been raised.34,103 Plausible alternatives are roles in regulating cohesin and/or in mediating cohesion independently of cohesin. Determining how SOLO and ORD interact with cohesin is a high priority. (5) Are there other core meiotic cohesion proteins besides ORD, SOLO and the canonical cohesin components? Remarkably, the answer appears to be yes. A mutation, mei(3)M20, recovered from a natural population in Japan in a screen for meiotic mutations was reported to cause a NDJ spectrum much like that of ord and solo, but from its chromosome location, it cannot be allelic to ord, solo or any cohesin gene.132 Identification of mei(3)M20 is clearly of considerable importance. (6) Do any of the known cohesin regulatory proteins play roles in meiosis? The yeast and Sordaria Pds5/Spo76 homologs have roles in multiple aspects of meiosis and the Drosophila homolog is probably required at least for recombination in female meiosis.108,109,133,134 The Eco1 acetyl transferase is required for sister centromere mono-orientation during meiosis I in fission yeast.135 It would thus not be surprising to find significant roles of cohesin cofactors in Drosophila male meiosis. (7) What role does the Passenger Complex play in regulating cohesion? Is it required for stable localization of cohesin, ORD or SOLO? (8) Does Drosophila have a “Monopolin”? We know that SOLO/ORD-dependent cohesin is required for sister centromere mono-orientation but other proteins are surely involved since SOLO/ORD-dependent cohesin is present on centromeres in both divisions but mono-orientation is limited to meiosis I. The mode of action of such a protein could be direct, binding to and clamping sister centromeres together, like Monopolin, or indirect, e.g., by modifying a cohesion protein or complex to enable it to accumulate efficiently at the kinetochore-forming region, like Moa1.17,21 Identifying such a protein would be important as next to nothing is known about the mechanism of mono-orientation in higher eukaryotes. (9) What is the molecular basis for cohesion and pairing in early prophase I? A consistent and surprising theme of the homolog pairing and cohesion stories outlined above is that the proteins critical for controlling segregation of both sister chromatids and homologs appear to be completely dispensable for pairing and cohesion prior to S3 (for arm cohesion and pairing) or S5 (for centromere cohesion). A key part of the story is clearly missing so far. (10) How are chromosome territories established and maintained? Territory integrity and resolution have been shown to depend on three key chromosomal protein complexes—cohesin, condensin II, and the conjunction complex—but this does not explain why they form in the first place.
Conclusions
Chromosome segregation in both mitosis and meiosis requires creation of stable connections that prevent the chromosomes from separating prematurely and provide the resistance that enables them to achieve bipolar orientation on the spindle. Drosophila has evolved an unusual meiotic system, used in spermatogenesis only, in which recombination and synapsis are absent and there are no chiasmata to hold homologs together. Nevertheless homologs pair intimately throughout the genome in early prophase just as they do in the standard type of meiosis. This genome-wide pairing serves as the major determinant of chromosome segregation patterns. The sex chromosomes are unusual in that homology is restricted to a single locus, the repeated rRNA genes, and pairing is confined to a defined sequence element repeated multiple times within the rRNA genes of the X and Y. Stable interhomolog connections are provided by three novel proteins, at least two of which, SNM and MNM, form a “conjunction complex” that is expressed only in male meiosis I and localizes to all four chromosome pairs until disappearing at anaphase I. Absence of any of the three conjunction proteins leads to premature homolog separation and random segregation at meiosis I.
Sister centromere cohesion in Drosophila male meiosis involves the core cohesin protein SMC1, but meiotic cohesin otherwise remains undefined. Rec8, a meiosis-specific cohesin that replaces the mitotic subunit Rad21 in other eukaryotes is absent from the fly genome. Instead, two novel meiosis-specific proteins, SOLO and ORD, with no cohesin homology are required for sister centromere cohesion and mono-orientation and for localization of SMC1 to meiotic centromeres. The precise molecular functions of these proteins remain to be determined.
Acknowledgments
The authors wish to thank Hiro Yamada for excellent technical assistance. The authors were supported by research grant # R01 GM40489 from the National Institutes of Health.
Glossary
Abbreviations:
- SC
synaptonemal complex
- PSCS
premature sister chromatid separation
- NOR
nucleolar organizing region
- rRNA
ribosomal RNA
- rDNA
ribosomal DNA
- FISH
fluorescence in situ hybridization
- RNAi
RNA interference
- GFP
green fluorescent protein
- NDJ
nondisjunction
Footnotes
Previously published online: www.landesbioscience.com/journals/spermatogenesis/article/21800
References
- 1.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–9. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 2.Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–40. doi: 10.1016/S0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 3.Hassold TJ, Hunt PA. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 4.Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–7. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 5.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 6.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 7.Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–98. doi: 10.1016/S0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 8.Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell. 2003;11:571–5. doi: 10.1016/S1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter AT. Chiasma function. Cell. 1994;77:957–62. doi: 10.1016/0092-8674(94)90434-0. [DOI] [PubMed] [Google Scholar]
- 10.McKee BD. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta. 2004;1677:165–80. doi: 10.1016/j.bbaexp.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 11.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–58. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 12.Székvölgyi L, Nicolas A. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 2010;277:571–89. doi: 10.1111/j.1742-4658.2009.07502.x. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe Y. A one-sided view of kinetochore attachment in meiosis. Cell. 2006;126:1030–2. doi: 10.1016/j.cell.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein LS. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- 15.Parra MT, Viera A, Gómez R, Page J, Benavente R, Santos JL, et al. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J Cell Sci. 2004;117:1221–34. doi: 10.1242/jcs.00947. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Dawe RK. Fused sister kinetochores initiate the reductional division in meiosis I. Nat Cell Biol. 2009;11:1103–8. doi: 10.1038/ncb1923. [DOI] [PubMed] [Google Scholar]
- 17.Tóth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–68. doi: 10.1016/S0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 18.Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, et al. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–48. doi: 10.1016/S1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 19.Corbett KD, Yip CK, Ee L-S, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–67. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–73. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–17. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–8. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- 23.Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–56. doi: 10.1016/0092-8674(95)90166-3. [Kitajima TS] [DOI] [PubMed] [Google Scholar]
- 24.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–7. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 25.Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 26.Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–72. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Clift D, Marston AL. The role of shugoshin in meiotic chromosome segregation. Cytogenet Genome Res. 2011;133:234–42. doi: 10.1159/000323793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper KW. Normal spermatogenesis in Drosophila. In: Demerec M, ed. Biology of Drosophila. New York NY: Wiley 1950:1–61. [Google Scholar]
- 29.Lindsley D, Tokuyasu TL. Spermatogenesis. In: Ashburner M, Wright T, ed. New York, NY: Academic Press, 1980:225-294. [Google Scholar]
- 30.Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, ed. The Development of Drosophila, Cold Spring Harbor, NY: Cold Spring Harbor Press 1993:71–147. [Google Scholar]
- 31.Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J Cell Sci. 1994;107:3521–34. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- 32.Tsai JH, Yan R, McKee BD. Homolog pairing and sister chromatid cohesion in heterochromatin in Drosophila male meiosis I. Chromosoma. 2011;120:335–51. doi: 10.1007/s00412-011-0314-0. [DOI] [PubMed] [Google Scholar]
- 33.Bonaccorsi S, Pisano C, Puoti F, Gatti M. Y chromosome loops in Drosophila melanogaster. Genetics. 1988;120:1015–34. doi: 10.1093/genetics/120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan R, Thomas SE, Tsai JH, Yamada Y, McKee BD. SOLO: a meiotic protein required for centromere cohesion, coorientation, and SMC1 localization in Drosophila melanogaster. J Cell Biol. 2010;188:335–49. doi: 10.1083/jcb.200904040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper KW. Meiotic conjunctive elements not involving chiasmata. Proc Natl Acad Sci U S A. 1964;52:1248–55. doi: 10.1073/pnas.52.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein LSB. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. Chromosoma. 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- 37.Meyer GF. The fine structure of spermatocyte nuclei of Drosophila melanogaster. In: Houwinck AL, Spit BJ, ed. Proceedings of the European Regional Conference on Electron Microscopy. Delft: Die Nederlandse Verening voor Electronmicroscopic Delft 1960:951–954. [Google Scholar]
- 38.Rasmussen SW. Ultrastructural studies of spermatogenesis in Drosophila melanogaster Meigen. Z Zellforsch Mikrosk Anat. 1973;140:125–44. doi: 10.1007/BF00307062. [DOI] [PubMed] [Google Scholar]
- 39.Morgan TH. No crossing over in the male of Drosophila of genes in the second and third pairs of chromosomes. Biol Bull. 1914;26:195–204. doi: 10.2307/1536193. [DOI] [Google Scholar]
- 40.Stevens N. A study of the germ cells of certain Diptera, with reference to the heterochromosomes and phenomena of synapsis. J Exp Zool. 1908;5:359–74. doi: 10.1002/jez.1400050304. [DOI] [Google Scholar]
- 41.Metz CW. Observations on spermatogenesis in Drosophila. Z Zellforsch und mikroskop Anat. 1926;4:1–28. doi: 10.1007/BF02628169. [DOI] [Google Scholar]
- 42.Vazquez J, Belmont AS, Sedat JW. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol. 2002;12:1473–83. doi: 10.1016/S0960-9822(02)01090-4. [DOI] [PubMed] [Google Scholar]
- 43.Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong WJ, McKim KS, Hawley RS. All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 2005;1:e67. doi: 10.1371/journal.pgen.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherizen D, Jang JK, Bhagat R, Kato N, McKim KS. Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics. 2005;169:767–81. doi: 10.1534/genetics.104.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fritsch C, Ploeger G, Arndt-Jovin DJ. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 2006;14:451–64. doi: 10.1007/s10577-006-1068-z. [DOI] [PubMed] [Google Scholar]
- 48.Williams BR, Bateman JR, Novikov ND, Wu CT. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics. 2007;177:31–46. doi: 10.1534/genetics.107.076356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322:1384–7. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]
- 50.Joyce EF, Williams BR, Xie T, Wu CT. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 2012;8:e1002667. doi: 10.1371/journal.pgen.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A. 2000;97:716–21. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–9. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wellauer PK, Dawid IB. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977;10:193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- 54.Gershenson S. Studies on the genetically inert region of the X chromosome of Drosophila. I. Behavior of an X chromosome deficient for a part of the inert region. J Genet. 1933;28:297–312. doi: 10.1007/BF02981776. [DOI] [Google Scholar]
- 55.Sandler L, Braver G. The meiotic loss of unpaired chromosomes in Drosophila melanogaster. Genetics. 1954;39:365–77. doi: 10.1093/genetics/39.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peacock WJ. Nonrandom segregation of chromosomes in Drosophila males. Genetics. 1965;51:573–83. doi: 10.1093/genetics/51.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKee B, Lindsley DL. Inseparability of X-heterochromatic functions responsible for X:Y pairing, meiotic drive, and male fertility in Drosophila melanogaster males. Genetics. 1987;116:399–407. doi: 10.1093/genetics/116.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKee BD, Karpen GH. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell. 1990;61:61–72. doi: 10.1016/0092-8674(90)90215-Z. [DOI] [PubMed] [Google Scholar]
- 59.McKee BD, Habera L, Vrana JA. Evidence that intergenic spacer repeats of Drosophila melanogaster rRNA genes function as X-Y pairing sites in male meiosis, and a general model for achiasmatic pairing. Genetics. 1992;132:529–44. doi: 10.1093/genetics/132.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merrill CJ, Chakravarti D, Habera L, Das S, Eisenhour L, McKee BD. Promoter-containing ribosomal DNA fragments function as X-Y meiotic pairing sites in D. melanogaster males. Dev Genet. 1992;13:468–84. doi: 10.1002/dvg.1020130609. [DOI] [PubMed] [Google Scholar]
- 61.McKee BD. The license to pair: identification of meiotic pairing sites in Drosophila. Chromosoma. 1996;105:135–41. doi: 10.1007/BF02509494. [DOI] [PubMed] [Google Scholar]
- 62.Ren X-J, Eisenhour L, Hong C-S, Lee Y, McKee BD. Roles of rDNA spacer and transcription unit-sequences in X-Y meiotic chromosome pairing in Drosophila melanogaster males. Chromosoma. 1997;106:29–36. doi: 10.1007/s004120050221. [DOI] [PubMed] [Google Scholar]
- 63.Ault JG, Lin HP, Church K. Meiosis in Drosophila melanogaster. IV. The conjunctive mechanism of the XY bivalent. Chromosoma. 1982;86:309–17. doi: 10.1007/BF00292259. [DOI] [PubMed] [Google Scholar]
- 64.Thomas SE, Soltani-Bejnood M, Roth P, Dorn R, Logsdon JM, Jr., McKee BD. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell. 2005;123:555–68. doi: 10.1016/j.cell.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 65.McKee BD. Homolog pairing and segregation in Drosophila meiosis. Genome Dyn. 2009;5:56–68. doi: 10.1159/000166619. [DOI] [PubMed] [Google Scholar]
- 66.Tsai JH, McKee BD. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci. 2011;124:1955–63. doi: 10.1242/jcs.006387. [DOI] [PubMed] [Google Scholar]
- 67.Yamamoto M. Cytological studies of heterochromatin function in the Drosophila melanogaster male: autosomal meiotic paring. Chromosoma. 1979;72:293–328. doi: 10.1007/BF00331091. [DOI] [PubMed] [Google Scholar]
- 68.Hilliker AJ, Holm DG, Appels R. The relationship between heterochromatic homology and meiotic segregation of compound second autosomes during spermatogenesis in Drosophila melanogaster. Genet Res. 1982;39:157–68. doi: 10.1017/S0016672300020851. [DOI] [PubMed] [Google Scholar]
- 69.McKee BD, Lumsden SE, Das S. The distribution of male meiotic pairing sites on chromosome 2 of Drosophila melanogaster: meiotic pairing and segregation of 2-Y transpositions. Chromosoma. 1993;102:180–94. doi: 10.1007/BF00387733. [DOI] [PubMed] [Google Scholar]
- 70.Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167:203–6. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wakimoto BT, Lindsley DL, Herrera C. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics. 2004;167:207–16. doi: 10.1534/genetics.167.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomkiel JE, Wakimoto BT, Briscoe A., Jr. The teflon gene is required for maintenance of autosomal homolog pairing at meiosis I in male Drosophila melanogaster. Genetics. 2001;157:273–81. doi: 10.1093/genetics/157.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arya GH, Lodico MJ, Ahmad OI, Amin R, Tomkiel JE. Molecular characterization of teflon, a gene required for meiotic autosome segregation in male Drosophila melanogaster. Genetics. 2006;174:125–34. doi: 10.1534/genetics.106.061556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas SE, McKee BD. Meiotic pairing and disjunction of mini-X chromosomes in drosophila is mediated by 240-bp rDNA repeats and the homolog conjunction proteins SNM and MNM. Genetics. 2007;177:785–99. doi: 10.1534/genetics.107.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Büchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, et al. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–57. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dorn R, Krauss V. The modifier of mdg4 locus in Drosophila: functional complexity is resolved by trans splicing. Genetica. 2003;117:165–77. doi: 10.1023/A:1022983810016. [DOI] [PubMed] [Google Scholar]
- 77.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–97. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 78.Zollman S, Godt D, Privé GG, Couderc J-L, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci U S A. 1994;91:10717–21. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beaster-Jones L, Okkema PG. DNA binding and in vivo function of C.elegans PEB-1 require a conserved FLYWCH motif. J Mol Biol. 2004;339:695–706. doi: 10.1016/j.jmb.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 80.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soltani-Bejnood M, Thomas SE, Villeneuve L, Schwartz K, Hong CS, McKee BD. Role of the mod(mdg4) common region in homolog segregation in Drosophila male meiosis. Genetics. 2007;176:161–80. doi: 10.1534/genetics.106.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–42. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coelho PA, Queiroz-Machado J, Sunkel CE. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J Cell Sci. 2003;116:4763–76. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- 84.Chan RC, Severson AF, Meyer BJ. Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol. 2004;167:613–25. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hirano T. Condensins: organizing and segregating the genome. Curr Biol. 2005;15:R265–75. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 86.Hartl TA, Sweeney SJ, Knepler PJ, Bosco G. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 2008;4:e1000228. doi: 10.1371/journal.pgen.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brito IL, Yu HG, Amon A. Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics. 2010;185:55–64. doi: 10.1534/genetics.110.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pungaliya P, Kulkarni D, Park HJ, Marshall H, Zheng H, Lackland H, et al. TOPORS functions as a SUMO-1 E3 ligase for chromatin-modifying proteins. J Proteome Res. 2007;6:3918–23. doi: 10.1021/pr0703674. [DOI] [PubMed] [Google Scholar]
- 89.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, et al. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 2004;279:36440–4. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 90.Matsui M, Sharma KC, Cooke C, Wakimoto BT, Rasool M, Hayworth M, et al. Nuclear structure and chromosome segregation in Drosophila male meiosis depend on the ubiquitin ligase dTopors. Genetics. 2011;189:779–93. doi: 10.1534/genetics.111.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valdeolmillos A, Villares R, Buesa JM, González-Crespo S, Martínez C, Barbero JL. Molecular cloning and expression of stromalin protein from Drosophila melanogaster: homologous to mammalian stromalin family of nuclear proteins. DNA Cell Biol. 1998;17:699–706. doi: 10.1089/dna.1998.17.699. [DOI] [PubMed] [Google Scholar]
- 92.Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, et al. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–18. doi: 10.1016/S0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 93.Valdeolmillos A, Rufas JS, Suja JA, Vass S, Heck MM, Martínez-A C, et al. Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol Cell. 2004;96:457–62. doi: 10.1016/j.biolcel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 94.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–11. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–53. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–36. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 97.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–64. doi: 10.1016/S0960-9822(00)00854-X. [DOI] [PubMed] [Google Scholar]
- 98.Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–49. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 99.Vernì F, Gandhi R, Goldberg ML, Gatti M. Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics. 2000;154:1693–710. doi: 10.1093/genetics/154.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, et al. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci U S A. 2008;105:12405–10. doi: 10.1073/pnas.0801698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, et al. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–51. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jäger H, Herzig A, Lehner CF, Heidmann S. Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev. 2001;15:2572–84. doi: 10.1101/gad.207301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khetani RS, Bickel SE. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J Cell Sci. 2007;120:3123–37. doi: 10.1242/jcs.009977. [DOI] [PubMed] [Google Scholar]
- 104.Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, et al. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117:51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanneti NS, Landy K, Joyce EF, McKim KS. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol. 2011;21:1852–7. doi: 10.1016/j.cub.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 106.Manheim EA, McKim KS. The Synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr Biol. 2003;13:276–85. doi: 10.1016/S0960-9822(03)00050-2. [DOI] [PubMed] [Google Scholar]
- 107.Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–87. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- 108.Mehrotra S, McKim KS. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2006;2:e200. doi: 10.1371/journal.pgen.0020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barbosa V, Kimm N, Lehmann R. A maternal screen for genes regulating Drosophila oocyte polarity uncovers new steps in meiotic progression. Genetics. 2007;176:1967–77. doi: 10.1534/genetics.106.069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pimenta-Marques A, Tostões R, Marty T, Barbosa V, Lehmann R, Martinho RG. Differential requirements of a mitotic acetyltransferase in somatic and germ line cells. Dev Biol. 2008;323:197–206. doi: 10.1016/j.ydbio.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–41. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moore DP, Page AW, Tang TT-L, Kerrebrock AW, Orr-Weaver TL. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J Cell Biol. 1998;140:1003–12. doi: 10.1083/jcb.140.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee JY, Hayashi-Hagihara A, Orr-Weaver TL. Roles and regulation of the Drosophila centromere cohesion protein MEI-S332 family. Philos Trans R Soc Lond B Biol Sci. 2005;360:543–52. doi: 10.1098/rstb.2005.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen F, Archambault V, Kar A, Lio’ P, D’Avino PP, Sinka R, et al. Multiple protein phosphatases are required for mitosis in Drosophila. Curr Biol. 2007;17:293–303. doi: 10.1016/j.cub.2007.01.068. [DOI] [PubMed] [Google Scholar]
- 115.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 116.Resnick TD, Satinover DL, MacIsaac F, Stukenberg PT, Earnshaw WC, Orr-Weaver TL, et al. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Clarke AS, Tang TT, Ooi DL, Orr-Weaver TL. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 118.Lee JY, Dej KJ, Lopez JM, Orr-Weaver TL. Control of centromere localization of the MEI-S332 cohesion protection protein. Curr Biol. 2004;14:1277–83. doi: 10.1016/j.cub.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 119.Bickel SE, Moore DP, Lai C, Orr-Weaver TL. Genetic interactions between mei-S332 and ord in the control of sister-chromatid cohesion. Genetics. 1998;150:1467–76. doi: 10.1093/genetics/150.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mason JM. Orientation disruptor (ord): a recombination-defective and disjunction-defective meiotic mutant in Drosophila melanogaster. Genetics. 1976;84:545–72. doi: 10.1093/genetics/84.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miyazaki WY, Orr-Weaver TL. Sister-chromatid misbehavior in Drosophila ord mutants. Genetics. 1992;132:1047–61. doi: 10.1093/genetics/132.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bickel SE, Wyman DW, Orr-Weaver TL. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 1997;146:1319–31. doi: 10.1093/genetics/146.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Balicky EM, Endres MW, Lai C, Bickel SE. Meiotic cohesion requires accumulation of ORD on chromosomes prior to condensation. Mol Biol Cell. 2002;21:3890–900. doi: 10.1091/mbc.E02-06-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bickel SE, Wyman DW, Miyazaki WY, Moore DP, Orr-Weaver TL. Identification of ORD, a Drosophila protein essential for sister chromatid cohesion. EMBO J. 1996;15:1451–9. [PMC free article] [PubMed] [Google Scholar]
- 125.Balicky EM. Regulation of chromosome segregation by ORD and dRING during Drosophila meiosis. Ph.D. dissertation. Dartmouth College 2005. [Google Scholar]
- 126.Wasser M, Chia W. The EAST protein of drosophila controls an expandable nuclear endoskeleton. Nat Cell Biol. 2000;2:268–75. doi: 10.1038/35010535. [DOI] [PubMed] [Google Scholar]
- 127.Gutiérrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, Müller J. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development. 2012;139:117–27. doi: 10.1242/dev.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Balicky EM, Young L, Orr-Weaver TL, Bickel SE. A proposed role for the Polycomb group protein dRING in meiotic sister-chromatid cohesion. Chromosoma. 2004;112:231–9. doi: 10.1007/s00412-003-0266-0. [DOI] [PubMed] [Google Scholar]
- 129.Gao S, Giansanti MG, Buttrick GJ, Ramasubramanyan S, Auton A, Gatti M, et al. Australin: a chromosomal passenger protein required specifically for Drosophila melanogaster male meiosis. J Cell Biol. 2008;180:521–35. doi: 10.1083/jcb.200708072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–90. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hauf S, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 2007;26:4475–86. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hirai K, Toyohira S, Ohsako T, Yamamoto MT. Isolation and cytogenetic characterization of male meiotic mutants of Drosophila melanogaster. Genetics. 2004;166:1795–806. doi: 10.1534/genetics.166.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–71. doi: 10.1016/S0092-8674(00)81020-X. [DOI] [PubMed] [Google Scholar]
- 134.Jin H, Guacci V, Yu H-G. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–25. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, et al. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep. 2011;12:1189–95. doi: 10.1038/embor.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]