Abstract
Despite their conserved functional role in sexually reproducing organisms, spermatozoa are a diverse and rapidly evolving cell type. This phenomenon is largely attributed to sexual selection in polygamous species where sperm from multiple males compete to fertilize a limited number of oocytes. Drosophila have proven to be a particularly informative model system for the study of spermatogenesis and in this review we discuss how the characterization of the Drosophila melanogaster sperm proteome has advanced our understanding of the evolutionary genomics of sperm form and function. We summarize the molecular evolutionary characteristics of sperm genes and highlight recent evidence demonstrating the importance of novel gene creation in the evolution of sperm function and competitive ability. Comparative proteomic evidence is also provided, supporting an overall functional conservation between the Drosophila and mouse sperm proteomes. This analysis reveals a diverse repertoire of proteins functioning in proteolytic pathways, as well as the presence of proteins of the complement and innate immunity systems. We propose that these pathways may have functional relevance to post-mating female immunological responses as well as coevolved interactions with pathways expressed in the female reproductive tract, including those involved in sperm-oocyte recognition and fertilization.
Keywords: proteomics, sexual selection, positive selection, sperm competition, retrogene, immunity, proteolysis
Introduction
Spermatozoa are among the most morphologically diverse cell types despite their conserved role in delivering the paternal genetic contribution to the egg at fertilization.1 The remarkable diversification of sperm is believed to be driven by sexual selection, where sperm are the focal point of the intense selective forces associated with sperm competition. The theory of sperm competition was first proposed by Geoff Parker2 and has subsequently become a major discipline of evolutionary research. Drosophila have proven to be an extremely amenable model system for the study of sperm competition and we now possess a remarkably advanced understanding of the selective forces associated with sperm competition3 and morphological diversification of sperm within this group.4 Drosophila also represent one of the most sophisticated model systems due to the development of advanced genetic tools, a carefully curated genome annotation and the availability of an enormous range of mutant strains. The use of these resources, in conjunction with complementary experimental studies of sperm, has resulted in an unparalleled understanding of the molecular and physiological impact of mating,5 as well as sperm behavior in the female reproductive tract.6 In Drosophila, spermatogenesis proceeds in an essentially linear manner from the apical to distal end of the testis, with distinct testis regions enriched for cells in mitotic, meiotic and post-meiotic stages of spermatogenesis. It has therefore been possible to isolate sperm from these regions by dissection to analyze gene expression at these primary timepoints during this carefully regulated differentiation process.7
Despite the utility of Drosophila as a model for the study of spermatogenesis, an incomplete understanding of the evolutionary genomics of male reproduction persisted for some time due to a paucity of information regarding the molecular composition of sperm. This lack of knowledge can be largely attributed to the progressive silencing of transcription as the genome is repackaged and compacted during spermatogenesis,8 which limits the utility of traditional methods of gene expression quantification as a means of inferring whether a resultant protein is in fact an integral component of mature sperm. Indirect inferences of sperm protein composition is further complicated by the fact that the testis transcriptome is extraordinarily diverse9 making gene expression profiles of the testis relatively uninformative. This gap in our knowledge base has largely been rectified by rapid advances in the field of mass spectrometry (MS) and the subsequent application of proteomic approaches to the study of sperm composition10 and reproduction more generally (reviewed in ref. 11). We now know that testis gene expression is a poor predictor of whether or not the protein product of a given gene is an integral sperm component.12 Prior to the initial characterization of the Drosophila melanogaster sperm proteome (DmSP),13 only 16 genes had characterized phenotypes specifically influencing sperm morphology or function, of which only 5 had been empirically demonstrated to encode protein components in mature sperm. A recent reanalysis of the D. melanogaster sperm proteome resulted in the robust identification of more than 1100 proteins which encode components of the mature spermatozoa.12
In this review we focus specifically on how the characterization of the sperm proteome has advanced our understanding of the evolutionary and functional genomics of reproduction in Drosophila. Wherever possible, we highlight the way in which a proteomics based perspective of sperm is being complemented by the diverse array of molecular and genetic approaches currently being used to understand sperm form and function. We begin by providing a summary of the evolutionary genomics of the sperm proteome in the context of the rapid evolution of many reproductive genes and the more recent conclusions concerning the heterogeneity of selective forces operating across reproductive systems. We follow with evidence supporting the prominent role of gene creation in sperm evolution, citing recent examples of novel sperm genes that influence sperm competitive ability. We then discuss the results of a comparative analysis of the D. melanogaster and Mus musculus (mouse) sperm proteomes, which illustrates the overall functional and compositional conservation of this cell type. This analysis also reveals the diversity of D. melanogaster sperm immunity and proteolytic proteins and we discuss their potential role in mediating post-mating female immunity responses and coevolutionary interactions with the abundant proteolytic proteins present in the female reproductive tract. Ultimately, the discussion of sperm proteome research highlights the multifaceted insights that can be inferred from this unique cell type and how the proteome continues to serve as a complementary resource for genetic and molecular investigations into sperm function.
Molecular Evolution of the Sperm Proteome
Despite widespread historical interest in sperm competition in insects and the rapid advancement of molecular studies of accessory gland proteins in Drosophila,14 little was known about the molecular composition of Drosophila sperm prior to the application of MS to the study of the sperm proteome. Molecular evolutionary studies of sperm proteins were restricted to a relatively small number of examples given the obstacles to empirical characterization discussed previously. The initial characterization of the Drosophila melanogaster sperm proteome13 (termed the DmSP-I) represented the first whole-cell catalog of integral sperm proteins and resulted in the first “whole-cell” evolutionary genomic analysis in eukaryotes. Reanalysis of the proteome, using improved methodologies, has resulted in substantial expansion of the proteome and a significant increase in the depth of coverage.12 The expanded sperm proteome (termed the DmSP-II) represents the most comprehensive database of insect sperm proteins for evolutionary genomic analyses and in this section we highlight the main observations stemming from its characterization.
Evolution of D. melanogaster sperm proteome (DmSP-I). The original molecular evolutionary analysis of the DmSP aimed to asses the selective forces acting upon the proteome as a whole.13 This was achieved using estimates of nonsynonymous (dN) and synonymous (dS) rates of divergence for 342 orthologous genes between D. melanogaster and its closely related sister taxa, D. simulans. Contrary to many studies demonstrating the rapid evolution of reproductive genes (reviewed in ref. 15), this analysis revealed a high level of functional constraint (as measured by dN/dS ratio) upon the proteome relative to genes expressed in the accessory gland (Fig. 1) and a complete absence of genes showing evidence of positive selection (as measured by a dN/dS ratio that significantly exceeded 1.0). Thus, it was concluded that the sperm proteome, as a whole, is evolving quite conservatively, presumably under the influence of functional and structural constraints. Consistent with this conclusion was the pronounced levels of functional constraint acting upon genes with cytoskeletal, central metabolic and energetic functions (Fig. 1). It is also noteworthy that the sperm proteome is significantly enriched for genes within these functional categories, many of which are widely expressed within a range of cell types and are thus likely to experience pleiotropic functional constraint.12 Despite the absence of any evidence of positive selection, it was revealed that DNA/RNA binding factors and genes with uncharacterized functions were evolving more rapidly, at a rate statistically indistinguishable from accessory gland genes. Consistent with this observation is the subsequent finding that positive selection is likely to have influenced the evolution of Drosophila protamines (Mst35Ba and Mst35Bb) which have undergone a D. melanogaster specific gene duplication event (see discussion below).16
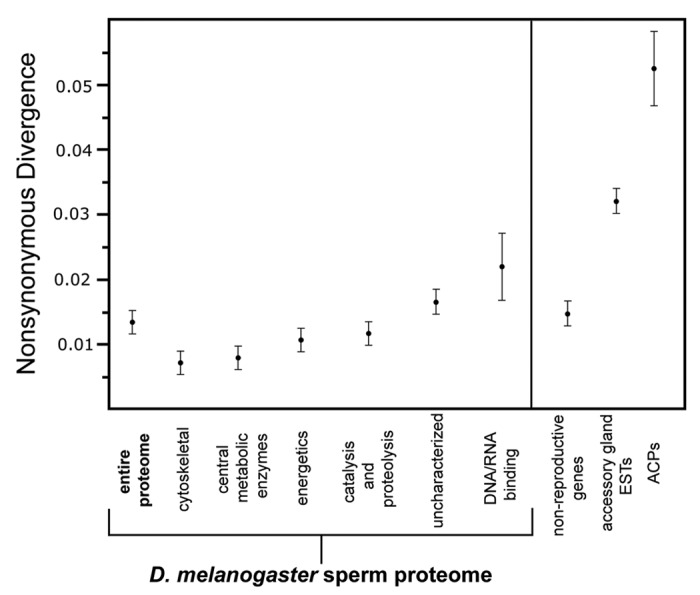
Figure 1. Molecular evolutionary analysis of the Drosophila melanogaster sperm proteome (DmSP). Average nonsynonymous substitution rates (± s.e.m.) between D. melanogaster and D. simulans are displayed for the DmSP-I (bold), individual functional categories of DmSP genes, nonreproductive genes, accessory gland transcripts and ACP genes. Substitution rates for accessory gland and ACP genes is based on Swanson et al. PNAS. The figure has been adapted from Dorus et al., 2006.
Evolution of the expanded Drosophila sperm proteome (DmSP-II). The reanalysis of the DmSP using improved prefractionation and MS techniques resulted in a 3-fold increase in the number of empirically characterized proteins (1108 in total). In addition, the genes encoding the DmSP-II have now been analyzed using maximum likelihood evolutionary models, facilitated by the recent availability of 12 genomes across the Sophophora subgenus.17 The findings of this analysis are generally consistent with the DmSP-I study and confirm that (1) purifying selection is the predominant force acting on the sperm proteome and (2) the evolution of the sperm proteome is quite distinct from the evolution of genes expressed in the accessory gland (details of this analysis are reviewed in ref. 18). One notable exception is that the use of more sensitive site-based models revealed the signature of positive selection on 8% (77 in total) of the sperm genes analyzed. A rather limited number of these genes are testis-specific in expression (24%) and few have established functions in sperm, thus restricting conclusions about the relative importance of sexual selection on these genes. One interesting example of a positively selected gene with a well-established sperm function is the sperm cation channel, Pkd2, which is implicated in sperm competition through the control of the directional motility.19 The abundance of genes with uncharacterized functions among these positively selected genes, and within the sperm proteome as a whole, highlights the potential for targeted molecular studies to establish roles in sperm that may influence competitive fertilization ability.
The expanded coverage of the DmSP-II was also extremely informative, as a greatly increased number of genes influencing sperm function and development were identified.12 In fact, a substantial proportion of these genes have sperm development phenotypes, arising either during spermatid development or sperm individualization, and are likely to have pronounced affects on sperm function. In total, 40 genes were known from either classical or molecular genetic studies to directly affect sperm development or function with the majority functionally related to sperm motility. This includes well-characterized sperm structural proteins such as βTub85D, βTub56D and αTub84B,20-23 as well as a range of proteins that bind microtubules or have functions in microtubule motor activity. Of particular interest among these are the Y-linked fertility factors, kl-3 and kl-5,24-26 which encode dynein heavy chains essential to the axoneme outer arm structure.24-26 Despite the heterochromatic nature of the Y chromosome, Y-linked genetic variation has been demonstrated to impact testis-specific gene expression27 and it is important to determine if fertility factor variation may impact sperm characteristics.
Another informative observation stemming from the analysis of the DmSP-II was the confirmation that many sperm genes are widely expressed across non-reproductive cell types, and that approximately 25% (271/1108) were expressed at least 2-fold higher in non-reproductive tissues than in the testis. An inverse correlation between expression breadth and evolutionary rates has been well-documented and is thought to be caused by increasing pleiotropic constraints.28 Given the enrichment of widely expressed structural, central metabolic and energetic proteins within the DmSP, pleiotropic constraints acting on some sperm genes appears likely to explain the evolutionary rate heterogeneity observed between Drosophila sperm genes and other male reproductive genes. It is also noteworthy that evolutionary analyses of the sperm proteome have focused on the melanogaster and obscura groups and have not yet been expanded to include more recently available genomic resources in species exhibiting sperm gigantism. Additional analyses are required to determine if accelerated evolution of the sperm proteome may be concentrated on lineages where the phenotypic evolution of sperm has been most pronounced.
Gene Creation and the Functional Evolution of Drosophila Sperm
Duplication of existing genetic material is an essential process in the evolution of genome complexity and the creation of genes with novel functions.29,30 Studies of novel genes have been conducted in a wide range of taxa revealing that many gene duplicates have acquired novel functions and that this process may occur in association with positive selection (reviewed by ref. 31). Of relevance is the observation that many novel genes acquire testis gene expression, an observation which is particularly common among new genes created via retrotransposition, a mechanism whereby a processed mRNA molecule is reverse transcribed and reinserted into the genome.32 Despite the demonstration that novel genes frequently acquire testis expression, the relevance of this process to sperm evolution and function was largely unknown prior to the characterization of the sperm proteome. Comparative genomic analyses of the DmSP-I revealed that gene creation, by both DNA- and RNA-based mechanisms, has played a prominent role in the evolution of sperm composition.16 In the following section, we review the influence of gene creation on the evolution of the sperm proteome with an emphasis on recent observations regarding the relevance of gene duplication to the process of sperm competition and possibly the evolutionary diversification of sperm morphology.
Protamine evolution in D. melanogaster. Tandem duplication of genes is a common phenomenon and there are several characterized examples of such duplicates which encode integral sperm components.16 One notable example is the lineage-specific duplication of protamines (Mst35Ba and Mst35Bb) in D. melanogaster.16 Protamines are highly basic molecules responsible for replacing histones during genome condensation and repackaging.33 Molecular evolutionary analysis in Drosophila revealed the signature of positive selection on these gene duplicates during their recent divergence with a concentration of evolutionary changes within the DNA binding high-mobility group box domain (HMG). While the functional implications of this observation remain unclear, it is interesting to speculate that this divergence may alter the temporal or spatial localization of these molecules during genome repackaging. Protamines are believed to be influenced by sexual selection in both primates34 and mice, and it has been suggested that protamine evolution in mice might influence head hydrodynamics and therefore motility.35 However, unlike mammals, Drosophila protamine mutants are not haploinsufficient and fertility is maintained in double homozygote knockout strains, although nuclear morphology is often abnormal.36 The non-essential nature of Drosophila protamines is intriguing in light of their recent duplication and targeted studies will be required to determine whether this has influenced sperm chromatin characteristics relevant to sperm competition and fertilization.
The origination of X-linked sperm gene clusters. In this section we summarize evidence supporting the evolution of novel X-linked sperm gene clusters and their role in male fertility. X-linked sperm gene clusters are somewhat unexpected in light of the extensive trafficking of male-based genes off the X chromosome, as well as a paucity of X-linked testis-expressed genes in both Drosophila32,37 and mammals.38,39 These observations have been attributed to two factors. The first is meiotic sex chromosome inactivation (MSCI) whereby the X chromosome is progressively silenced during spermatogenesis, thus restricting X-linked expression during later stages of spermatogenesis. While MSCI has been clearly documented in mammals,40 its presence in Drosophila remains controversial (see ref. 7 but also refs. 41 and 42). The second is the influence of sexually antagonistic selection upon X-linked male-biased genes, ultimately favoring the transfer of male-biased functions to the autosomes.43,44 Regardless of the ultimate cause, the presence of new and expanding X-linked sperm gene clusters is unusual and may provide insights into the role of the X chromosome in male fertility.
The most comprehensive characterization of novel sperm gene creation is the Sperm dynein intermediate chain (Sdic) gene family, which arose in the D. melanogaster lineage within the last 5.4 million years. Sdic is the product of a chimeric gene fusion of Annexin X (AnnX) and short wing (sw) that has subsequently been duplicated into a tandem array of 8–10 copies.45,46 Sdic1 is testis-specific and its protein product has been visualized in maturing spermatocytes, and the flagellum of mature sperm using a Sdic::GFP fusion protein under control of the Sdic promoter. It is noteworthy that Sdic, for reasons that are yet unclear, has not been identified in the proteomic analysis of D. melanogaster sperm12,13 despite the fact it has been visualized in mature sperm. Based on its subcellular localization, the well-characterized structural role of dyneins in the sperm axoneme and a population genetic signature of recent selective sweeps, it has been proposed that Sdic is involved in sperm motility and, therefore, may be impacted by sexual selection associated with sperm competition.47 This proposition is supported by the recent analysis of strains harboring Sdic deletions which found a significant decrease in sperm competitive ability in comparison to wild-type sperm.46 Thus, strong support exists for an evolutionarily acquired role for the X-linked Sdic gene cluster in sperm function that has direct relevance to sperm competition.
A second example, with clear similarities to the evolution of the Sdic gene cluster, is the recent evolutionary expansion of the X-linked tektin gene cluster.13 Tektins are cytoskeletal proteins that directly interact with microtubules in the sperm axoneme48 and a total of five tektins have been identified in the DmSP. Three tektins (CG17450, CG32820 and CG32819) reside in an X-linked cluster which originated very recently on the D. melanogaster lineage through sequential tandem duplication events. A recent analysis of the relationship of X-linked genetic variation with sperm competitive ability identified three distinct mutations in CG17450 which influence reproductive phenotypes in competitive mating experiments.49 This finding provides strong support for the functional importance of the tektin gene cluster in sperm function and highlights the importance of the X-chromosome variation in male fertility.
The characterization of two recently expanded structural sperm gene clusters on the X chromosome suggests that these findings may apply to other X-linked genes. For example, a recent study demonstrated that a testis-specific, tandemly duplicated pair of X-linked polyubiquitin genes also influence male fecundity.50 As stated previously, these observations do not necessarily conform to current models regarding the genomic distribution of male-biased genes. Furthermore, they may also suggest that MSCI, if it exists in Drosophila, may have regionally distinct properties across the X chromosome that favor the creation of co-localized copies of genes in certain locations whose expression is required during later stages of spermatogenesis. Lastly, this pattern of spatial clustering is reminiscent of ampliconic regions on the mouse X chromosome that harbor postmeiotically expressed testis genes.51 Understanding the manner in which the X chromosome harbors genetic variation associated with male fertility may depend on understanding the selective forces responsible for the heterogeneous physical distribution of spermatogenesis genes on the X chromosome.
Novel sperm retrogenes. Analysis of the DmSP revealed that a number of retrogenes encode integral sperm components16 and the importance of retrotransposition, as a generator of functional genetic diversity, has been confirmed in a recent survey of retrogenes in the D. melanogaster genome (Dorus and Rettie, unpublished data). This analysis revealed that 19.8% (20 out of 101) encode integral sperm components (Table 1) and that 75% of these (15 out of 20) function in specific mitochondrial metabolic pathways, including the tricarboxlic acid cycle, electron transport and oxidative phosphorylation. An abundance of mitochondrial gene duplicates encoding sperm proteins has previously been attributed to selection associated with motility based energetic requirements.44 However, several lines of evidence suggest that insect sperm motility is largely dependent on glycolysis generated ATP. The role of insect sperm mitochondrial derivatives as energy-producing organelles in mature sperm has thus been called into question (reviewed in ref. 52). Although it is possible that motility is enhanced by residual ATP generated by the nebenkern (the fused giant mitochondria that restructure in parallel with the developing axoneme), recent work demonstrates that mitochondrial processes in Drosophila sperm are closely linked with flagellum development. Importantly, the association between microtubule dynamics and nebenkern elongation is structurally essential for uniaxial spermatid tail development and the disruption of mitochondria-microtubule linkage results in developmental defects of the flagellum.53 Based on the evidence that mitochondrial derived ATP is unlikely to be the predominant energetic source for motility, we propose that the selective retention of metabolic sperm retrogenes in D. melanogaster may be associated with their role in the nebenkern as a specialized, energy producing organizing center for microtubules during sperm flagellum elongation. The enhancement of these mitochondrial pathways required for flagellum development may thus be associated with the remarkable diversification of flagellum length in these taxa.4 This hypothesis is also consistent with the observation that numerous cytoskeletal flagellum components have also originated recently during D. melanogaster sperm evolution, including (1) retrogene Cdlc2, whose parent, Ctp (also known as Ddlc1) is an intermediate dynein binding protein required for actin filament assembly in the developing flagellum54 and (2) the previously discussed X-linked Sdic (sperm dynein) and tektin gene clusters. Although further investigation will be required to establish the specific functions of these novel sperm genes, the evolutionary timing of their origination is intriguing in light of the dramatic sperm flagellum length diversification during Drosophilidae evolution.
Table 1.D. melanogaster sperm retrogenes.
| Phylogenetic distribution |
Sperm retrogenes |
X to autosome | Parental Symbol | |
|---|---|---|---|---|
| Retrogene symbol1 | Metabolic process (GO:0008152) |
|||
| Sophophora subgenus |
CG17856 |
yes |
yes |
CG3560 |
|
CG5265 |
yes |
no |
CG1041 |
|
|
CG7514 |
no |
no |
CG1907 |
|
|
Prosα6T |
yes |
no |
Pros35 |
|
| Drosophilidae |
CG4706 |
yes |
no |
Acon |
|
S-LAP7 |
yes |
no |
S-LAP3 |
|
|
CG5718 |
yes |
no |
Scs-fp |
|
|
Act87E |
no |
no |
Act57B |
|
|
CG14508 |
yes |
no |
CG4769 |
|
|
Cdlc2 |
no |
yes |
Ctp |
|
|
Ef1α48D |
yes |
no |
Ef1α100E |
|
|
Pglym87 |
yes |
no |
Pglym78 |
|
|
CG6255 |
yes |
no |
Scsα |
|
|
Vha36 |
no |
yes |
CG8310 |
|
|
Hsp60B2 |
yes |
yes |
Hsp60 |
|
|
Gskt (mojoless)
2
|
yes |
yes |
Sgg |
|
|
CG10749 |
yes |
no |
CG7998 |
|
| Diptera |
Hsc70–4 |
yes |
no |
Hsc70–1 |
|
CG11913 |
yes |
no |
CG1970 |
|
| CG6180 | no | no | CG17919 | |
Expansion of the sperm leucyl-aminopeptidase gene family (S-LAPs). Comparative genomic analysis in Drosophila has revealed that gene families with functions in reproduction evolve rapidly through the creation and attrition of family members.55 An excellent example of this is the S-LAP gene family that has experienced a dramatic expansion during Drosophila evolution. This gene family is of great functional interest because it is testis-specific and encodes the most abundant proteins, by mass, in Drosophila sperm.56 Due to the age of the S-LAPs the exact mechanisms and sequence of duplication events resulting in the family are unclear. However, the intron/exon pattern and the occurrence of genes in tandem suggest that there have been two cases of tandem duplications (S-LAP1 and S-LAP2; S-LAP3 and S-LAP4) and S-LAP7, that has been characterized as a retrogene. Perhaps the most unusual aspect of S-LAP evolution is that the most dramatic period of gene family expansion occurred after the ancestral gene underwent a series of amino acid substitutions that radically altered critical residues within the catalytic site, most likely abolishing enzymatic activity. Therefore, it appears that the S-LAP gene family has evolved a novel, but yet to be determined, sperm specific function and that this neofunctionalization may have been selectively involved in the retention of newly created S-LAP gene copies during early Drosophila evolution. The loss of catalytic activity among the S-LAP family is reminiscent of the loss of enzymatic activity and acquisition of structural roles in other ubiquitously expressed metabolic enzymes, such as the crystallin family of lens proteins. We have therefore proposed that the selective retention of enzymatically inactive S-LAPs may be related to the establishment of structural functionality, which may in turn be associated with sperm morphological diversification in these taxa.56
Therefore, novel gene creation has thus been an influential force in the evolution of the sperm proteome and recent analyses have begun to identify links between these loci and sperm function. A precise understanding of the functional ramifications of this process still requires additional, targeted molecular studies. Nonetheless, patterns of novel gene creation in Drosophila suggest that the enhancement and expansion of both cytoskeletal and mitochondrial energetic systems may be critical to the morphological diversification of Drosophila sperm, as well as sperm competitive ability. In addition, the physical distribution and localized expansion of X-linked sperm gene families may provide useful insights into how selection has shaped genetic variation on the X chromosome associated with male fertility.
Comparative Sperm Proteomics and Genomics
Whole-cell comparative sperm proteomic analyses are now possible with the application of MS based proteomics to sperm from a range of species, including Drosophila and an increasing number of mammalian taxa (reviewed by ref. 11). This approach, in conjunction with comparative genomics, has already provided novel insights into the conserved, as well as highly diverged, molecular aspects of sperm function. For example, the comparison of
D. melanogaster and the mouse sperm proteomes has revealed that greater than 40% of mouse axoneme proteins have homology to the DmSP,13 and that greater than 65% of DmSP genes possess a mouse homolog, of which 20% have also been identified within the mouse sperm proteome.12 In this section, we reanalyze conservation between the Drosophila and mouse sperm proteome and discuss functional parallels between the two, including the abundance of immunity and proteolytic proteins within the DmSP.
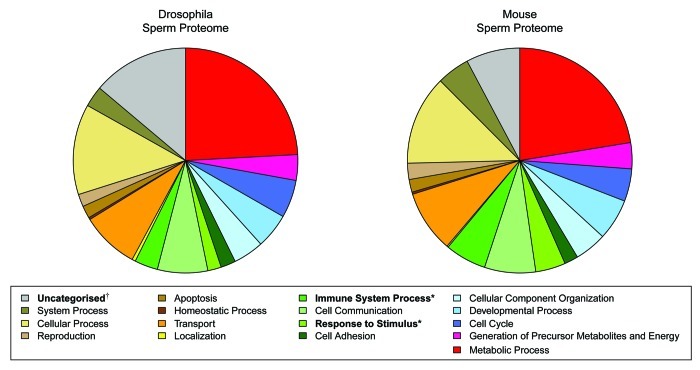
Conservation of sperm proteome functional composition. Next to D. melanogaster, the mouse has the most comprehensively characterized sperm proteome as it has been interrogated using a variety of approaches. These include (1) whole-cell shotgun proteomics,57,58 (2) proteomic analysis of membrane enriched samples and cell surface proteins59 and (3) 2D-gel analysis of biochemically purified accessory structures of the axoneme60and sperm membrane lipid rafts.61 To assess conservation of the functional composition of sperm, we have compared the combined Mus musculus sperm proteome (MmSP) from these studies with the DmSP utilizing PANTHER functional categories (Fig. 2). Statistical analysis of the functional composition of the proteomes reveals substantial similarities in the functional composition of insect and mammalian sperm proteomes with only 3 out of 18 functional categories identified as being statistically distinguishable in their abundance between the two. These include a significantly higher proportion of uncharacterized genes in the DmSP, consistent with the higher proportion of uncharacterized genes in the Drosophila genome as a whole, and a significantly higher proportion of immunity and response to stimuli annotated proteins observed within the MmSP. The relative underrepresentation of immunity proteins in the DmSP is attributable in part to the absence of adaptive immunity in insects, whereas proteins functioning in adaptive immunity contribute substantially to the total repertoire of immunity pathways present in mammalian sperm (reviewed by ref. 62). As such, with the exception of immunity and stimulus response processes, the sperm proteome of insects and mammals show a high level of evolutionary conservation at the level of their functional composition.
Figure 2. Functional comparison of the Drosophila and mouse sperm proteomes. Gene ontology information was obtained from the PANTHER database (http://www.pantherdb.org/) and genes without annotated functions are labeled as uncharacterized. Proportions are based upon the total number of annotated functions, permitting genes to be present in multiple functional categories. Significant differences in the number of genes in Drosophila melanogaster and Mus musculus sperm proteomes for each functional category was determined using a two-tailed Fisher’s Exact Test with Bonferroni correction for multiple testing (*indicates overrepresentation in the mouse sperm proteome; † indicates overrepresentation in Drosophila sperm proteome).
Immunity proteins in the DmSP. The evolutionary trade-offs between reproduction and immunity have been investigated extensively and female immunological responses to mating, based on changes in the expression of female immunity genes, have been well characterized in Drosphila.63-67 Despite variation in experimental approaches, antimicrobial genes appear to be consistently upregulated, while innate immunity genes within the Toll and IMD pathways are more variable in their response.67 Female survival against infection also decreases in the first day after mating, although it is not known how regulatory responses in immunity pathways may ultimately be associated with this phenomenon.65 Interestingly, experimental approaches designed to disentangle the relative regulatory effects of sperm and accessory gland proteins (ACPs) determined that ACPs exclusively upregulate antimicrobial peptides (AMPs) (including members of the Attacin and Cecropin gene families), while sperm impact the expression of a more widespread set of genes across a range of functional classes, including the downregulation of certain key immune response genes.64 Therefore sperm, in the absence of ACPs, have the capacity to mediate female immunity responses, although little attention has been focused specifically on how components of the Drosophila sperm proteome may function in this manner.
A great deal more is known about the function of immunity proteins in mammalian sperm and proteomic studies have been useful in revealing the full diversity of such pathways, which include a diverse repertoire of complement, adaptive immunity and antimicrobial proteins (reviewed in ref. 62). Functional characterization of immunity proteins in mammalian sperm also indicates that some have acquired sperm-specific functions during their evolution. For example, acrosome associated complement regulator CD46 influences acrosome reaction dynamics69 and its presence in sperm is believed to be related to variation in the intensity of sperm competition across species.70,71 Another notable example is DEFENSIN-β 126 (DEFB126), an AMP which modulates sperm penetration of the cervical mucus in the female reproductive tract.72 Common mutations in human DEFB126 have been shown to result in subfertility.73 Sperm are coated with DEFB126 in the epididymis, and immunological studies have revealed that DEFB126 inhibits female immune recognition and thus acts as a cloaking device while sperm traverse the female reproductive tract.74 It is noteworthy that this process is quite reminiscent of the coating of Drosophila sperm by accessory gland protein Sex-peptide (SP), a protein which also influences female immune response.75 The widespread involvement of immunity pathways in mammalian sperm function raises the question of whether parallel functional systems may be present in Drosophila.
To explore this possibility, we have surveyed the DmSP for all immunity-related proteins present in mature sperm. This analysis identified a total of 20 proteins functioning across three general categories: innate immunity response, antimicrobial function and antiviral function (Table 2). An additional 26 genes have annotated functions in phagocytosis and three are involved in hemocyte development or function. Of interest is the identification of Dscam 1, 2 and 3 in the DmSP, which are members of the Immunoglobulin-superfamily. Dscam genes have an unusual genomic structure containing thousands of exons that can be variably spliced, resulting in the potential translation of more than 18,000 protein isoforms.76 This phenomenon, although mechanistically distinct, has parallels to somatic diversification of immune receptors in the adaptive immune systems of other taxa. Dscam 1 has been studied intensively in relation to its role in immunity77 and neurogenesis,76 but a function in reproduction has yet to be characterized. Consistent with the composition of mammalian sperm, the DmSP contains TepII and TepIV, which are opsonin thioester-containing proteins homologous to mammalian complement proteins. Although neither have identified functions in reproduction, they were also recently found to be upregulated in the female following mating.67 DmSP constituents were also identified across a diverse range of antimicrobial functions, including peptidoglycan recognition and binding, regulation of AMPs, regulation of symbiont growth and antimicrobial humoral response. Notable among these are Hel89B, an ATP dependent DNA helicase, and alphaTub84B, a cytoskeletal structural protein, which are implicated in sperm function based on the identification of male sterile alleles.
Table 2. DmSP genes with functions in the immunity pathways.
| Gene Symbol |
GO Accession |
GO Term |
|---|---|---|
| Innate immune response and Toll pathway | ||
| Toll-4,1Tollo1 |
45087, 45089 |
Innate immune response |
|
Galpha49B,1norpA1 |
02385 |
Mucosal immune response |
|
psidin
2
|
02253, 06959 |
Humoral Immune response |
|
Nos
1
|
06952 |
Defense response |
|
Dscam,1Dscam2, Dscam3, Hel89B,2,3Spz3 |
08063 | Toll signaling pathway |
| Antimicrobial functions | ||
|---|---|---|
|
kis,1lola,1mask,1Hel89B,2,3Dscam,1Pvr,1par-11, TepII,1TepIV, alphaTub84B1,3 |
19730, 19731, 08348 |
Antimicrobial humoral response |
|
Dscam,1TepII1 |
42742, 50829, 50830 |
Defense response to bacterium |
|
Dscam
1
|
61057, 42834, 16019 |
Peptidoglycan recognition/binding |
|
Hel89B,2,3Dscam1 |
02805, 06963 |
Regulation of AMPs |
| Vps4 1 | 44130 | Regulation of symbiont growth |
Innate immunity is dependent on the Toll and IMD pathways and our survey reveals that proteins in the Toll pathway, but not IMD, are present in D. melanogaster sperm, including Toll-4, Tollo (Toll-8), spz3 and Hel89B. Activation of the Toll pathway occurs via the cleavage of spätzle, which binds to the Toll receptor, by a serine protease cascade resulting in the activation of the Toll receptor; this in turn leads to a cascade of protein interactions, culminating in the transcription of antimicrobial peptides. It is therefore plausible that Toll-4, Tollo and spz3 may be interacting proteins in sperm, if the Toll pathway in sperm operates in a manner consistent with its operation in immunity. In this context, it is interesting to note that both Toll-4 and spz3 are recently evolved genes and may therefore have male-specific functions which distinguish them from stereotypical members of the Toll pathway. Interestingly, the elicitation of AMP genes by the seminal peptide, SP, is also dependent on the Toll pathway.75 Although the Toll-pathway is dependent upon serine protease activity, the functional significance of these proteins (and immunity proteins more generally) in the DmSP remains unclear. Given the abundance of serine protease pathways in both mammalian and Drosophila reproductive systems it is plausible that their presence may be related to regulation of the Toll pathway.
Enrichment of protease and protease inhibitors in the sperm proteome. Regulators of proteolysis are functionally relevant to sperm biology because of their ubiquitous abundance in both the male and female reproductive tract14,78,79 and demonstrated ability to regulate the activity of other reproductive proteins, such as SP.75,80 Proteases and peptidases have also been found to be significantly enriched within the DmSP,12,13 which includes the highly abundant proteins encoded by the recently expanded S-LAP gene family.57 A survey of the DmSP identifies a total of 60 proteins with peptidase domains (primarily metallopeptidases and serine peptidases), including 8 proteins with proteinase inhibitory domains (Table 3). Despite the fact that a number of these are over- or specifically expressed in the testis, none have documented functions in sperm function or fertility. Among the metalloproteases, stl and Tace possess the Peptidase M12B and ADAM/reprolysin domains found in the mammalian A-disintegrin and metalloproteinase (Adam) gene family. This is of interest as Adam proteins are abundant in the mouse sperm acrosome (8 are present in the MmSP) and function in sperm-egg recognition and fusion (reviewed by ref. 81). These may be excellent candidates for further functional characterization as relatively little is known about the molecular basis of Drosophila sperm-egg recognition.
Table 3. DmSP genes with peptidase or proteinase inhibitor domains.
| Peptidase domain |
Gene symbols |
|---|---|
| Metallopeptidases | |
| Peptidase M1 |
CG5849, Psa,2Taf21 |
| Peptidase M12B |
Ppn, stl,1Tace2 |
| Peptidase M13 |
CG15485, CG31918, CG5527, Nep2,1Nep3, Nep52 |
| Peptidase M14 |
CG17633, NnaD,1east1 |
| Peptidase M16 |
CG10588,2CG3731,1CG4169, CG8728 |
| Peptidase M17 |
Dip-B1, S-Lap1–52, S-Lap6 (loopin-1),2S-Lap7–82 |
| Peptidase M24 or M24A |
dre4, und
2
|
| Peptidase M41 | CG2658,2CG65122 |
| Serine peptidases | |
|---|---|
| Chemotrypsin (S1A or S1/S6) |
CG32269, CG34350, CG4927, CG5390,1Corin, CG16321 |
| Non-chemotrypsin (S8, S9, S10, S14,S16, S26A, S28) | Fur1,1CG17684,2CG9059, CG4572,1CG5045,2CG8798, CG13991,2CG9240,2CG37342 |
| Other peptidases | |
|---|---|
| Peptidase A2A |
rngo
2
|
| Peptidse C19 |
CG12082,1CG30421,1CG5794,1CG88302 |
| Peptidase C48 | CG10107 2 |
| Proteinase inhibitor | |
|---|---|
| Proteinase inhibitor I2 (Kunitz) |
axo, Ppn |
| Proteinase Inhibitor I1 (Kazal) |
CG13830, CG7173, magu
1
|
| Proteinase inhibitor I7 (squash) |
dp |
| Serpin domain | Spn43Ab, Spn47C2 |
Sexually antagonistic coevolution has been invoked to explain the rapid evolution of male and female proteins associated with sperm competition, storage and utilization.79 One possible hallmark of such a coevolving system is the involvement of unique male and female genes that can respond efficiently to male- and female-specific selective forces. If this is the case, one might predict that male and female contributed proteolytic proteins might be generally distinct despite the requirement that they function in closely related pathways. Consistent with this prediction is the observation that the repertoire of serine proteases present within the DmSP (Table 3) is encoded by a completely distinct set of genes than the serine proteases highly expressed in the two primary sperm storage organs in Drosophila, the spermatheca and seminal receptacles.79,80 Furthermore, the presence of serine protease inhibitors (Serpins) within the DmSP could also be indicative of coevolved male-female interactions and it would be informative to determine if these Serpins do in fact regulate serine proteases in the female reproductive tract. To this end, it would be useful to determine if Serpins are localized to the sperm cell membrane where they would be accessible to direct molecular interactions. Although little is known about their reproductive functions, targeted proteomic analysis of the mouse sperm cell membrane and acrosome have identified a remarkable diverse family of Serpins,59 of which several show evidence of rapid evolution or positive selection.58 The shared presence of Serpins in Drosophila and mouse sperm not only supports their functional relevance to sperm function but also suggests that similar pathways (e.g., serine proteases and their inhibitors) may be generally involved in coevolved male-female molecular interactions relevant to sperm competition and fertilization.
Conclusions and Future Perspectives
Sexual selection, as a primary selective force in evolution, was first discussed by Charles Darwin82 and has been a major focus of both theoretical and empirical investigations by evolutionary biologists ever since. The amount of phenotypic data relevant to the evolution of reproductive traits is tremendous, yet despite the advancement of relevant genetic theories, a nuanced understanding of the evolutionary relationship between genotype and phenotype in reproductive systems has yet to be achieved. Molecular evolutionary analyses of reproductive genes has revealed widespread evidence of rapid evolution, although few examples have been documented where the underlying selective forces are well understood. Sperm may ultimately be an ideal system for exploring how sexual selection operates on genotype-phenotype relationships because they are amenable to experimental manipulation and we possess well-developed theoretical predictions concerning the impact and outcomes of sperm competition. In this review, we have thus set out to describe our current understanding of the D. melanogaster sperm proteome and how it has informed our understanding of sexual selection and the evolutionary genomics of sperm.
Characterization of the D. melanogaster sperm proteome has provided the necessary knowledge base for investigating the genetic basis of sperm evolution. These analyses identified an unexpected level of functional constraint upon the proteome as a whole and also revealed significant evolutionary rate heterogeneity within male reproductive systems. Ultimately, a more detailed understanding of compartmentalized adaptation in response to sexual selection has been achieved through additional studies in both mammals and insects.58,83 Analysis of the sperm proteome has also revealed that gene creation is a prominent mechanism in the evolutionary diversification of the sperm proteome. More significant perhaps is the observation that the selective retention of novel sperm genes appears to be non-random, as novel Drosophila genes are enriched within mitochondrial energetic pathways and proteins functioning in the development and structure of the sperm flagellum. Although the ultimate ramifications of this trend have yet to be determined, recent progress has been made in associating sperm competitive ability with genetic variation in some of these novel sperm components. We anticipate that the sperm proteome will be a valuable asset as more studies attempt to elucidate the association between intra- and interspecific genetic variation and a range of sperm phenotypes, including the independent origination of sperm gigantism in multiple Drosophilidae species.84
Previous molecular analyses of sexual antagonistic coevolution have naturally focused on the unique complement of secreted proteins of the male accessory gland and, more recently, genes upregulated within the female reproductive tract. Prominent among these are a diverse set of proteolytic and immunity genes, many of which have been demonstrated to evolve rapidly.14,78,79,85 Analysis of both the Drosophila and mouse sperm proteomes reveal that these functional classes are abundant in sperm and, in the case of the mouse, that many of these proteins are localized to the cell membrane or acrosome and evolve rapidly.58 Although the true abundance of immunity protein in sperm has only recently been revealed through proteomic analysis, it has long been recognized that male immunity proteins may be influential in mediating female immune responses post-mating.86 Despite the observation that sperm induce a range of gene expression changes in the female reproductive tract, including some immunity genes, almost nothing is known about the function of the innate immunity or complement system in Drosophila sperm. Given the central role of serine proteases in the Toll pathway and their presence within both male and female reproductive systems, the sperm proteome provides a foundation for exploring the functional coevolution of immunity and proteolytic pathways in reproduction, particularly those related to male-female interactions influencing sperm utilization and fertilization.
Glossary
Abbreviations:
- ACPs
accessory gland proteins
- AMPs
antimicrobial peptides
- ATP
adenosine triphosphate
- DmSP
Drosophila melanogaster sperm proteome
- GFP
green fluorescent protein
- GO
gene ontology
- HMG
high mobility box group
- MmSP
Mus musculus sperm proteome
- MS
mass spectrometry
- MSCI
meiotic sex chromosome inactivation
- Serpin
serine protease inhibitor
Footnotes
Previously published online: www.landesbioscience.com/journals/spermatogenesis/article/21748
References
- 1.Pitnick S, Hosken DJ, Birkhead TR. Sperm morphological diversity. In: Birkhead TR, Hosken DJ, Pitnick S, eds. Sperm biology: An evolutionary perspective. USA: Academic Press, 2009:69-149. [Google Scholar]
- 2.Parker GA. Sperm competition and its evolutionary consequences in insects. Biol Rev Camb Philos Soc. 1970;45:525–67. doi: 10.1111/j.1469-185X.1970.tb01176.x. [DOI] [Google Scholar]
- 3.Snook RR. Sperm in competition: not playing by the numbers. Trends Ecol Evol. 2005;20:46–53. doi: 10.1016/j.tree.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Karr TL, Pitnick S. The ins and outs of fertilization. Nature. 1996;379:405–6. doi: 10.1038/379405a0. [DOI] [PubMed] [Google Scholar]
- 5.Wolfner MF. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity (Edinb) 2002;88:85–93. doi: 10.1038/sj.hdy.6800017. [DOI] [PubMed] [Google Scholar]
- 6.Manier MK, Belote JM, Berben KS, Novikov D, Stuart WT, Pitnick S. Resolving mechanisms of competitive fertilization success in Drosophila melanogaster. Science. 2010;328:354–7. doi: 10.1126/science.1187096. [DOI] [PubMed] [Google Scholar]
- 7.Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–61. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 9.Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lü JN, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliva R, de Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–17. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- 11.Findlay GD, Swanson WJ. Proteomics enhances evolutionary and functional analysis of reproductive proteins. Bioessays. 2010;32:26–36. doi: 10.1002/bies.200900127. [DOI] [PubMed] [Google Scholar]
- 12.Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, et al. The Drosophila melanogaster sperm proteome-II (DmSP-II) J Proteomics. 2010;73:2171–85. doi: 10.1016/j.jprot.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Dorus S, Busby SA, Gerike U, Shabanowitz J, Hunt DF, Karr TL. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–5. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- 14.Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci U S A. 2001;98:7375–9. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–44. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 16.Dorus S, Freeman ZN, Parker ER, Heath BD, Karr TL. Recent origins of sperm genes in Drosophila. Mol Biol Evol. 2008;25:2157–66. doi: 10.1093/molbev/msn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–18. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 18.Karr TL, Dorus S. Evolutionary genomics of the sperm proteome. In: Singh RS, Xu J, Kulathinal RJ, eds. Rapidly evolving genes and genetic systems. Oxford: Oxford University Press, 2012:153-64. [Google Scholar]
- 19.Gao ZQ, Ruden DM, Lu XY. PKD2 cation channel is required for directional sperm movement and male fertility. Curr Biol. 2003;13:2175–8. doi: 10.1016/j.cub.2003.11.053. [DOI] [PubMed] [Google Scholar]
- 20.Fuller MT, Caulton JH, Hutchens JA, Kaufman TC, Raff EC. Mutations that encode partially functional beta 2 tubulin subunits have different effects on structurally different microtubule arrays. J Cell Biol. 1988;107:141–52. doi: 10.1083/jcb.107.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoyle HD, Raff EC. Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol. 1990;111:1009–26. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimble M, Dettman RW, Raff EC. The beta 3-tubulin gene of Drosophila melanogaster is essential for viability and fertility. Genetics. 1990;126:991–1005. doi: 10.1093/genetics/126.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaltschmidt B, Glätzer KH, Michiels F, Leiss D, Renkawitz-Pohl R. During Drosophila spermatogenesis beta 1, beta 2 and beta 3 tubulin isotypes are cell-type specifically expressed but have the potential to coassemble into the axoneme of transgenic flies. Eur J Cell Biol. 1991;54:110–20. [PubMed] [Google Scholar]
- 24.Goldstein LSB, Hardy RW, Lindsley DL. Structural genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982;79:7405–9. doi: 10.1073/pnas.79.23.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci U S A. 1993;90:11132–6. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci U S A. 2000;97:13239–44. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sackton TB, Montenegro H, Hartl DL, Lemos B. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci U S A. 2011;108:17046–51. doi: 10.1073/pnas.1114690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- 29.Ohno S. Gene duplication as an evolutionary force. Peeters, H (Edited by) Protides of the Biological Fluids, Proceedings of the 17th Colloquium at Bruges, Belgium Xv+542p Illus Pergamon Press, Inc: Elmsford, NY, USA 1970:39-42. [Google Scholar]
- 30.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–4. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 31.Kaessmann H, Vinckenbosch N, Long MY. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betrán E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–9. doi: 10.1101/gr.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120:1689–700. doi: 10.1242/jcs.004663. [DOI] [PubMed] [Google Scholar]
- 34.Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–9. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 35.Martin-Coello J, Dopazo H, Arbiza L, Ausió J, Roldan ERS, Gomendio M. Sexual selection drives weak positive selection in protamine genes and high promoter divergence, enhancing sperm competitiveness. Proc Biol Sci. 2009;276:2427–36. doi: 10.1098/rspb.2009.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathke C, Barckmann B, Burkhard S, Jayaramaiah-Raja S, Roote J, Renkawitz-Pohl R. Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur J Cell Biol. 2010;89:326–38. doi: 10.1016/j.ejcb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emerson JJ, Kaessmann H, Betrán E, Long MY. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–40. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 39.Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14:2911–8. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis (Drosophila-allocycly-chromosome evolution-male sterility-dosage compensation) Proc Natl Acad Sci U S A. 1972;69:182–6. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meiklejohn CD, Landeen EL, Cook JM, Kingan SB, Presgraves DC. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 2011;9:e1001126. doi: 10.1371/journal.pbio.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikhaylova LM, Nurminsky DI. Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome. BMC Biol. 2011;9:29. doi: 10.1186/1741-7007-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu CI, Xu EY. Sexual antagonism and X inactivation--the SAXI hypothesis. Trends Genet. 2003;19:243–7. doi: 10.1016/S0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 44.Gallach M, Chandrasekaran C, Betrán E. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol. 2010;2:835–50. doi: 10.1093/gbe/evq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–5. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 46.Yeh S-D, Do T, Chan C, Cordova A, Carranza F, Yamamoto EA, et al. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc Natl Acad Sci U S A. 2012;109:2043–8. doi: 10.1073/pnas.1121327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nurminsky D, Aguiar DD, Bustamante CD, Hartl DL. Chromosomal effects of rapid gene evolution in Drosophila melanogaster. Science. 2001;291:128–30. doi: 10.1126/science.291.5501.128. [DOI] [PubMed] [Google Scholar]
- 48.Steffen W, Linck RW. Evidence for tektins in centrioles and axonemal microtubules. Proc Natl Acad Sci U S A. 1988;85:2643–7. doi: 10.1073/pnas.85.8.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenspan L, Clark AG. Associations between variation in X chromosome male reproductive genes and sperm competitive ability in Drosophila melanogaster. Int J Evol Biol. 2011;2011:214280. doi: 10.4061/2011/214280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhan Z, Ding Y, Zhao R, Zhang Y, Yu H, Zhou Q, et al. Rapid functional divergence of a newly evolved polyubiquitin gene in Drosophila and its role in the trade-off between male fecundity and lifespan. Mol Biol Evol. 2012;29:1407–16. doi: 10.1093/molbev/msr299. [DOI] [PubMed] [Google Scholar]
- 51.Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JMA. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40:794–9. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werner M, Simmons LW. Insect sperm motility. Biol Rev Camb Philos Soc. 2008;83:191–208. doi: 10.1111/j.1469-185X.2008.00039.x. [DOI] [PubMed] [Google Scholar]
- 53.Noguchi T, Koizumi M, Hayashi S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr Biol. 2011;21:805–14. doi: 10.1016/j.cub.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh-Roy A, Desai BS, Ray K. Dynein light chain 1 regulates dynamin-mediated F-actin assembly during sperm individualization in Drosophila. Mol Biol Cell. 2005;16:3107–16. doi: 10.1091/mbc.E05-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hahn MW, Han MV, Han S-G. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorus S, Wilkin EC, Karr TL. Expansion and functional diversification of a leucyl aminopeptidase family that encodes the major protein constituents of Drosophila sperm. BMC Genomics. 2011;12:177–90. doi: 10.1186/1471-2164-12-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker MA, Hetherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics. 2008;8:1720–30. doi: 10.1002/pmic.200701020. [DOI] [PubMed] [Google Scholar]
- 58.Dorus S, Wasbrough ER, Busby J, Wilkin EC, Karr TL. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Mol Biol Evol. 2010;27:1235–46. doi: 10.1093/molbev/msq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stein KK, Go JC, Lane WS, Primakoff P, Myles DG. Proteomic analysis of sperm regions that mediate sperm-egg interactions. Proteomics. 2006;6:3533–43. doi: 10.1002/pmic.200500845. [DOI] [PubMed] [Google Scholar]
- 60.Cao WL, Gerton GL, Moss SB. Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol Cell Proteomics. 2006;5:801–10. doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Asano A, Nelson JL, Zhang S, Travis AJ. Characterization of the proteomes associating with three distinct membrane raft sub-types in murine sperm. Proteomics. 2010;10:3494–505. doi: 10.1002/pmic.201000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorus S, Skerget S, Karr TL. Proteomic discovery of diverse immunity molecules in mammalian spermatozoa. Syst Biol Reprod Med. 2012;58:218–28. doi: 10.3109/19396368.2012.700442. [DOI] [PubMed] [Google Scholar]
- 63.Lawniczak MKN, Begun DJ. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–10. doi: 10.1139/g04-050. [DOI] [PubMed] [Google Scholar]
- 64.McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 2004;14:1509–14. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 65.Fedorka KM, Linder JE, Winterhalter W, Promislow D. Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proc Biol Sci. 2007;274:1211–7. doi: 10.1098/rspb.2006.0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kapelnikov A, Zelinger E, Gottlieb Y, Rhrissorrakrai K, Gunsalus KC, Heifetz Y. Mating induces an immune response and developmental switch in the Drosophila oviduct. Proc Natl Acad Sci U S A. 2008;105:13912–7. doi: 10.1073/pnas.0710997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Innocenti P, Morrow EH. Immunogenic males: a genome-wide analysis of reproduction and the cost of mating in Drosophila melanogaster females. J Evol Biol. 2009;22:964–73. doi: 10.1111/j.1420-9101.2009.01708.x. [DOI] [PubMed] [Google Scholar]
- 68.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 69.Inoue N, Ikawa M, Nakanishi T, Matsumoto M, Nomura M, Seya T, et al. Disruption of mouse CD46 causes an accelerated spontaneous acrosome reaction in sperm. Mol Cell Biol. 2003;23:2614–22. doi: 10.1128/MCB.23.7.2614-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson PM, Clift LE, Andrlikova P, Jursova M, Flanagan BF, Cummerson JA, et al. Rapid sperm acrosome reaction in the absence of acrosomal CD46 expression in promiscuous field mice (Apodemus) Reproduction. 2007;134:739–47. doi: 10.1530/REP-07-0363. [DOI] [PubMed] [Google Scholar]
- 71.Clift LE, Andrlikova P, Frolikova M, Stopka P, Bryja J, Flanagan BF, et al. Absence of spermatozoal CD46 protein expression and associated rapid acrosome reaction rate in striped field mice (Apodemus agrarius) Reprod Biol Endocrinol. 2009;7:29–38. doi: 10.1186/1477-7827-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tollner TL, Yudin AI, Treece CA, Overstreet JW, Cherr GN. Macaque sperm coating protein DEFB126 facilitates sperm penetration of cervical mucus. Hum Reprod. 2008;23:2523–34. doi: 10.1093/humrep/den276. [DOI] [PubMed] [Google Scholar]
- 73.Tollner TL, Venners SA, Hollox EJ, Yudin AI, Liu X, Tang G, et al. A common mutation in the defensin DEFB126 causes impaired sperm function and subfertility. Sci Transl Med. 2011;3:92ra65. doi: 10.1126/scitranslmed.3002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yudin AI, Generao SE, Tollner TL, Treece CA, Overstreet JW, Cherr GN. Beta-defensin 126 on the cell surface protects sperm from immunorecognition and binding of anti-sperm antibodies. Biol Reprod. 2005;73:1243–52. doi: 10.1095/biolreprod.105.042432. [DOI] [PubMed] [Google Scholar]
- 75.Peng J, Zipperlen P, Kubli E. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr Biol. 2005;15:1690–4. doi: 10.1016/j.cub.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 76.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–84. doi: 10.1016/S0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 77.Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–8. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 78.Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–65. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prokupek A, Hoffmann F, Eyun SI, Moriyama E, Zhou M, Harshman L. An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution. 2008;62:2936–47. doi: 10.1111/j.1558-5646.2008.00493.x. [DOI] [PubMed] [Google Scholar]
- 80.Ravi Ram K, Sirot LK, Wolfner MF. Predicted seminal astacin-like protease is required for processing of reproductive proteins in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:18674–9. doi: 10.1073/pnas.0606228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J Proteome Res. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- 82.Darwin C. The descent of man, and selection in relation to sex. London: John Murray, 1871. [Google Scholar]
- 83.Dean MD, Clark NL, Findlay GD, Karn RC, Yi X, Swanson WJ, et al. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus) Mol Biol Evol. 2009;26:1733–43. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pitnick SS, Spicer GS, Markow TA. How long is a giant sperm? Nature. 1995;375:109. doi: 10.1038/375109a0. [DOI] [PubMed] [Google Scholar]
- 85.Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–80. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harris CL, Mizuno M, Morgan BP. Complement and complement regulators in the male reproductive system. Mol Immunol. 2006;43:57–67. doi: 10.1016/j.molimm.2005.06.026. [DOI] [PubMed] [Google Scholar]