Abstract
The topic of imprinting defects present in the sperm of infertile patients has been addressed by several reports in the last few years. However, whether methylation abnormalities at one or few CpGs within an imprinted locus are pathological is a matter of debate. Moreover, whether imprinting anomalies in sperm could interfere with fertility treatment outcomes is still unknown. In this report we analyze the sperm DNA methylation profile of H19-ICR, KvDMR, SNRPN-ICR, IG-DMR and MEG3-DMR by pyrosequencing in 107 infertile men series and a control population of 30 proven fertile males. DNA methylation was statistically evaluated from two points of view: first, the methylation of each CpG was analyzed in the control population and the mean, standard deviation and range were determined and compared with infertile population data; second, in order to define altered methylation patterns for each region, a hierarchical cluster analysis was performed by which individuals were grouped in different clusters according to the degree of similarity of their methylation pattern. Two pieces of data supported the results obtained in the multi-variate analysis: the classification of the vast majority of control individuals in clusters with normal methylation patterns and the significant differences in methylation levels found between individuals within the normal and abnormal clusters. Individuals included in normal and abnormal methylation clusters were compared according to seminal parameters as well as to the outcome of assisted reproduction.
Keywords: DNA methylation, assisted reproduction, imprinting, male infertility, spermatozoa
Introduction
Imprinting controls the expression of a gene on the basis of its parental origin, which results in monoallelic expression. To date, there are about 60 known imprinted genes in the human genome (http://igc.otago.ac.nz/home.html) grouped in clusters distributed in different autosomes.1 The differences in expression of these genes are controlled by epigenetic modifications. Imprinted genes play an important role in embryo, extra-embryonic tissues and neurological development. Thus, the allele-specific epigenetic marks that control the expression pattern of these genes are loci-specific and established in the male and female germline to allow proper development from early stages of embryogenesis. In particular, allele-specific DNA methylation is established at germline Differentially Methylated Regions (DMR) in the adult spermatogonia in human males2 and during oocyte maturation in human females.3,4 These germline DMRs act as Imprinting Control Regions (ICR), regulating, in cis, the monoallelic expression of the different imprinted genes within a cluster.
DNA methylation is subject to alterations that could be caused by genetic factors (variations in the DNA sequence of factors involved in the methylation establishment or maintenance),5-7 intrauterine environment,8 toxin exposure,9 hormone treatment10,11 and diet.12 Some authors have related the presence of imprinting errors in newborns to the application of assisted reproduction techniques (ART). Epidemiological data showed a moderate increase of the risk of syndromes caused by imprinting abnormalities in the assisted reproduction population.13-17 In these reports, the imprinting defect corresponds to Loss of Methylation (LOM) in the maternal allele, indicating a possible deleterious effect of ovarian stimulation or embryo in vitro culture on methylation establishment in the female germline or its maintenance at early stages of development. Even though the reported epidemiological data lead one to consider maternal methylation defects exclusively, a relationship between male infertility and imprinting abnormalities in the sperm has also been described.18-26 Since paternal imprinted genes mainly contribute to extra-embryonic tissue development, it is not discarded that loss of imprinting (LOI) at paternal methylated loci could cause phenotypes that prevent placenta development or could cause intrauterine growth retardation (IUGR). IUGR has been described as being associated with imprinting defects8 and also related to assisted reproduction.27
In this study we have analyzed the DNA methylation profile of the two ICR of 11p15.5 (H19-ICR and KvDMR), the ICR of 15q11-q13 (SNRPN-ICR), the IG-DMR and MEG3-DMR of 14q32.2 in human sperm of infertile men after establishing a reference threshold of normal methylation status in a proven fertile male population by pyrosequencing. KvDMR and SNRPN-ICR were included in our study in order to identify possible errors in imprinting erasure of maternal imprinting DMRs. MEG3-DMR corresponds to a secondary or somatic paternal imprinted DMR and it was included to confirm that paternal imprinting is also erased during imprinting reprogramming in the male germline.
The analysis of a control population is crucial to identify abnormal methylation patterns considered as being pathological. Furthermore, with the aim of achieving a detailed correlation between imprinting abnormalities and male infertility, infertile patients were further divided into several sub-groups according to sperm concentration, mobility and morphology. To identify possible interferences with the assisted reproduction treatment outcome, sperm methylation results were correlated with the rate of fertilization, embryo development, pregnancy, abortion and birth, as well as with embryo quality and birth weight.
Results
CpG to CpG analysis
Beginning with the values obtained in the control population for each CpG analyzed, a range of reference methylation was calculated, using the value of the mean ± 2 SD (Tables S1–5).
The mean and 95% confidence interval (CI 95%) methylation in the H19-ICR and IG-DMR populations was established at 88.49% (78.16–98.82) and 78.95% (61.70–95.24), respectively, while KvDMR, SNRPN-ICR and MEG3-DMR were 0.99% (0.10–1.89), 1.03% (0.26–1.86) and 1.18% (0.26–2.13), respectively (Table 1). Furthermore, within a region, only specific CpGs presented equivalent values. In this sense, the SNRPN-ICR region was showed the most uniform results, with 80% of CpG with equivalent values. In the remaining regions analyzed, the results were grouped between 40% and 53.8% (Table 1).
Table 1. Methylation means (MM) and number of equivalent CpG for each region in the control population.
| Loci | Analyzed CpGs | MM (%) | Equivalent CpG |
|---|---|---|---|
|
H19-ICR |
13 |
88.49 |
7/13 (53,8%) |
|
KVDMR |
21 |
0.99 |
11/21 (52,4%) |
|
SNRPN-ICR |
20 |
1.03 |
16/20 (80%) |
|
IG-DMR |
5 |
78.95 |
2/5 (40%) |
| MEG3-DMR | 21 | 1.18 | 11/21 (52,4%) |
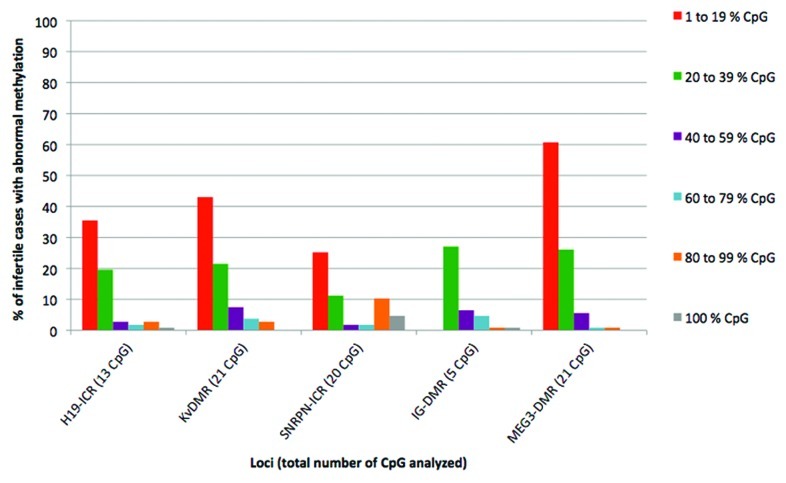
In order to analyze the results of methylation obtained in the infertile population, all CpGs that had methylation values outside of the reference range established for the control population were identified. The percentage of infertile individuals showing at least one CpG with methylation values outside of the range of normality obtained from the control series was 40.1% for IG-DMR, 55.1% for SNRPN-ICR, 63.5% for H19-ICR, 78.5% for KvDMR and 94.4% for MEG3-DMR (Fig. 1). The majority of individuals showed methylation anomalies in at least 20% of the CpGs and, as the percent of altered CpGs increased, the number of affected individuals decreased significantly (Fig. 1).
Figure 1. Percentage of infertile individuals showing abnormal methylation values at every single locus. The total percentage of infertile cases with abnormal methylation is broken down according to the percentage of altered CpG. Red, 1 to 19% of abnormal CpG; green, 20 to 39% of abnormal CpG; lilac, 40 to 59% of abnormal CpG; blue, 60 to 79% of abnormal CpG; orange, 80 to 99% of abnormal CpG; gray, 100% of abnormal CpG.
Analysis of the methylation pattern
Starting from the methylation values obtained for each CpG in the series of control and infertile individuals, a multi-variate analysis was performed. From this analysis, a dendrogram was obtained per region where the relationships of proximity between individuals were represented by means of clusters. Four individuals (95, 24, 97 and 107) were excluded from the clusters analysis due to showing a very different behavior from the rest. Individual 95 presented a mean methylation of 9.30% (CI 95% 6.20–12.00) in H19-ICR and individual 24 presented a mean methylation of 38.6% (CI 95% 16.53–60.67) in IG-DMR. Cases 97 and 107 were considered outliers due to presenting very low methylation values in CpG1 in locus IG-DMR (13% and 41%, respectively). Table 2 summarizes the results obtained in the grouping of the individuals in clusters.
Table 2. Results of the multi-variate analysis.
| Loci | Variable | Cluster 1 |
Cluster 2 |
Cluster 3 |
|||
|---|---|---|---|---|---|---|---|
| C | I | C | I | C | I | ||
|
H19-ICR |
N |
30 |
79 |
0 |
16 |
0 |
11 |
|
MM ± SD (%) |
88 ± 6,9 |
94,16 ± 5,26 |
80 ± 10,7 |
||||
|
KvDMR |
N |
29 |
79 |
1 |
16 |
0 |
12 |
|
MM ± SD (%) |
1,07 ± 1,08 |
2,58 ± 2,63 |
2,06 ± 1,82 |
||||
|
SNRPN-ICR |
N |
26 |
67 |
4 |
22 |
0 |
18 |
|
MM ± SD (%) |
0,95 ± 1,08 |
2,32 ± 1,97 |
8,12 ± 5,26 |
||||
|
IG-DMR |
N |
28 |
55 |
0 |
12 |
2 |
37 |
|
MM ± SD (%) |
78 ± 12,84 |
85,95 ± 11,34 |
74,81 ± 15,89 |
||||
| MEG3-DMR |
N |
28 |
52 |
2 |
55 |
||
| MM ± SD (%) | 1,21 ± 0,89 | 2,03 ± 2,54 | |||||
N: total number of individuals; C: control; I: infertile; MM: methylation mean; SD: standard deviation. Abnormal clusters are bolded.
In the five loci analyzed, the mean methylation in individuals from the normal and altered clusters showed significant differences (p < 0.05). This analysis permitted the establishment of a methylation mean (MM) of normal and altered clusters and a confidence interval of 95%. Beginning with the 95% CI for each region, a range was established based on which a MM could be considered within the range of normality or abnormality (Table 3).
Table 3. Variables that determine the differences between the normal and abnormal clusters.
| Loci | MMNC | MMAC | CI 95% range | Range of normal methylation levels | Range of abnormal methylation levels |
|---|---|---|---|---|---|
|
H19-ICR |
88,87 |
80,01 |
[87,3/72,6] |
[100/87,3] |
[87,3/0] |
|
KvDMR |
1,07 |
2,36 |
[1,4/3,2] |
[0/1,4] |
[1,4/100] |
|
SNRPN-ICR |
1,24 |
8,12 |
[4,7/11,5] |
[0/4,8] |
[4,8/100] |
|
IG-DMR |
79,79 |
74,85 |
[80,6/69,1] |
[100/80,6] |
[80,6/0] |
| MEG3-DMR | 1,21 | 2,03 | [1,3/2,7] | [0/1,3] | [1,3/100] |
MMNC, methylation mean of normal clusters; MMAC, methylation mean of abnormal clusters; CI 95% range, range of methylation mean that includes 95% of individuals within abnormal clusters.
The number of individuals with genetic imprinting anomalies per region was noticeably inferior to the percent observed in the CpG to CpG analysis (Table 2). Overall, a total of 91 infertile individuals (91/107; 85%) and three control individuals (3/30; 10%) presented altered methylation patterns at least in one of the regions analyzed. It is important to highlight that specific individuals presented methylation anomalies in more than one region. In this sense, one control individual (1/30; 3.3%) and 34 infertile individuals (34/107; 31.8%) presented hypermethylation anomalies in regions that normally do not show methylation in spermatozoa and hypomethylation in regions where methylation is normally established. Of these latter ones, three infertile individuals (3/34; 8.8%) showed alterations in four or more of the six analyzed regions. In one control individual (1/30; 3.3%) and 44 infertile individuals (44/107; 41.1%) alterations were identified in regions associated with anomalies of methylation erasure (KvDMR, SNRPN-ICR, MEG3-DMR). Of these, five infertile individuals (5/44; 11.4%) showed alterations in the three regions. Lastly, one control individual (1/30; 3.3%) and 13 infertile individuals (13/107; 12.1%) presented a hypomethylation pattern in regions where methylation is established. Of these 13, two infertile individuals (2/13; 15.4%) showed alterations in the two regions that were analyzed with paternal-specific methylation (H19-ICR and IG-DMR).
Relationship between methylation, seminal parameters and age of individuals
A Chi-square test (χ2) to establish the relation between methylation patterns and sperm analysis components was performed. Despite the fact that the different deviations from the reference range for each parameter were found represented in the clusters with normal and altered methylation levels, significant associations were found between the N component and normal methylation patterns in H19-ICR, KvDMR, SNRPN-ICR and MEG3-DMR. Component O showed associations with altered methylation patterns in SNRPN-ICR. Finally, altered methylation patterns were associated with component T in H19-ICR, SNRPN-ICR and MEG3-DMR.
In four of the five loci analyzed (H19-ICR, KvDMR, SNRPN-ICR and IG-DMR), the mean age of the individuals in the normal and altered clusters did not show significant differences (p > 0.05). On the contrary, age differed significantly among patients classified in the normal and abnormal clusters for the MEG3-DMR locus (p = 0.013). Moreover, we observed a positive correlation between age and presence of abnormal methylation at KvDMR (p = 0.0009) and MEG3-DMR (p = 0.0001).
Relationship between methylation patterns and assisted reproduction outcomes
No significant differences were observed for the parameters analyzed with the exception of indicators of embryo quality, which showed differences when comparing the results of individuals within the normal and altered clusters. In the case of KvDMR region, a significantly higher number of type A embryos (best quality) were observed in normal cluster compared with individuals of the abnormal cluster. On the other hand, an increase in type A embryos was observed in the cluster of individuals with anomalies at IG-DMR, compared with individuals of the normal cluster. In both KvDMR and SNRPN-ICR regions, individuals of the abnormal clusters showed significantly higher numbers of embryos (poor quality), when compared with the normal clusters (Table 4).
Table 4. Comparison of assisted reproduction treatment results between infertile patients that belong to normal and abnormal methylation pattern clusters.
| |
Loci |
||||
|---|---|---|---|---|---|
| H19-ICR | KvDMR | SNRPN-ICR | IG-DMR | MEG3-DMR | |
| |
p value |
p value |
p value |
p value |
p value |
| Fertilization rate |
0.5741 |
0.5189 |
0,1282 |
0.2312 |
0.3604 |
| A embryos/total |
0.2490 |
0.0161* |
0,1763 |
0.0252* |
0.6589 |
| B embryos/total |
0.8353 |
0.3069 |
0,0961 |
0.2557 |
0.2143 |
| C embryos/total |
0.2042 |
0.8261 |
0,2962 |
0.1394 |
0.6006 |
| D embryos/total |
0.5909 |
0.0387* |
0,034* |
0.1366 |
0.4539 |
| Development rate |
0.7103 |
0.7567 |
0,2826 |
0.2387 |
0.5430 |
| Discarded embryos/total |
0.8504 |
0.0947 |
0,078 |
0.2753 |
0.2713 |
| Pregnancy rate/Transfer |
0.3183 |
0.8179 |
0,8722 |
0.9030 |
0.6409 |
| Abortions/gestations |
- |
- |
0,2482 |
0.2482 |
0.2482 |
| Birth/informative gestation |
0.8940 |
0.6504 |
0,3388 |
0,2286 |
0.3121 |
| XX:XY proportion |
1.0000 |
0.6206 |
0,4062 |
0.2673 |
0.1707 |
| Mean of weight (mg) |
0.9939 |
0.9831 |
0,9997 |
0.9894 |
0.9888 |
| Mean of weight 1 fetus (mg) |
0.8428 |
0.2437 |
0,8023 |
0.7769 |
0.1916 |
| Mean of weight 2 fetus (mg) | 0.0528 | 0.1387 | - | 0.7479 | 0.3980 |
Statistically significance (p ≤ 0,05)
Discussion
Methylation analysis
Methylation analysis of the control population permitted the establishment of a reference range of methylation values for the CpGs in the analyzed regions, to which the values measured in the infertile population were compared. This analysis clearly showed that, within the same region, (1) CpGs differ in methylation levels, (2) regions regulated by genetic imprinting do not show individual methylation mean values equivalent to those theoretically expected and, (3) the differences vary depending on the analyzed region. Previous studies have not analyzed a control population but have instead compared the methylation results in the infertile population to the expected theoretical values of 0% or 100%.18,19,22 Our results show the necessity of the analysis of a control population in order to be able to discriminate between normal and anomalous variations.
When comparing methylation values obtained in infertile individuals to the reference methylation range established for the specific CpGs, we found a great proportion of patients showing at least one altered CpG among the analyzed loci. Nevertheless, the biological significance that the presence of specific hypo- or hypermethylated CpGs has in the overall behavior of a region can be irrelevant if we consider that, as has been indicated in the literature, imprinted regions do not show homogenous values of methylation in the totality of CpGs,28 an observation that was corroborated by the methylation values of our control population. Additionally, a higher incidence of syndromes caused by imprinting anomalies inherited from the paternal genome would be expected in offspring conceived by ART if the presence of few CpGs with abnormal methylation levels brought on alterations. On the contrary, this increase is not observed, and the identified syndromes have mainly been linked to imprinting alterations inherited from the maternal genome.29
Faced with these considerations, it was considered that not all of the infertile individuals who showed methylation errors in at least one CpG could be evaluated as individuals with anomalies. The soundness of our multi-variate analysis lies in the classification of the vast majority of control individuals in clusters with normal methylation patterns (Table 2), and the existence of significant differences in the methylation levels between individuals within the normal and abnormal clusters in the five regions analyzed.
The grouping of individuals in normal and altered clusters also permitted the establishment of a mean methylation range to which the mean methylation of each analyzed region could be compared (Table 3). This calculation was performed for each one of the regions analyzed. As an example, an individual with a mean methylation of 80% in H19-ICR could be considered as an individual with an altered methylation pattern for this region (range of normal methylation: 83.3–100%).
In terms of types of alterations, errors in the regions that do not normally present methylation in spermatozoa and errors in regions of paternal methylation have been identified. The erasure of methylation originates by an active process that is independent of DNA replication.30 The activation of the repair pathway by the excision of bases during the period in which epigenetic reprogramming in primordial germ cells (PGC) is produced suggests that demethylation is performed by this pathway.31 In this sense, the deficient or anomalous functioning of some of the factors involved in the repair pathway by the excision of bases could involve an insufficient deletion of the pre-existing imprinting marks in PGC, detectable in the form of a hyper-methylation pattern at the end of spermatogenesis. Moreover, we observe a positive correlation between age and presence of abnormal methylation at MEG3-DMR and KvDMR. Considering that imprinting is erased in PGC, i.e., during the fetal period, the correlation between the presence of maternal-specific methylation in male gametes and age could only be explained by a failure in the maintenance of those unmethylated loci. This hypothetical fact could be due to the damage or loss of histone marks, such us H3K4 methylation, or caused by anomalies in proteins that read these marks and contribute to the unmethylated state.
With reference to the establishment of methylation, the adequate functionality of the family of DNA methyltransferases (DNMT) as well as of other factors involved in the epigenetic reprogramming of the masculine germinal line (such as CTCFL) is essential for the establishment of imprinting during spermatogenesis. Kobayashi et al.7 demonstrated the existence of variants in homozygosis in the sequence of the DNMT3L co-factor, which is necessary for the activity of the de novo DNMT3, in individuals with methylation anomalies in spermatozoa; however, these abnormalities were not present in all of the regions studied. The authors suggested that each of the regions present a different vulnerability to present alterations in function of the degree of tandem repetitions that they contain. In mice, different studies have demonstrated that anomalies of the genes that codify for DNMT32-37 or variations in the functionality of these proteins due to interaction with environmental factors10,38-40 produce methylation alterations in regions regulated by genetic imprinting, causing infertile phenotypes.
On the other hand, during spermatogenesis and in stages previous to the substitution of histones by protamines, regions regulated by genetic imprinting are marked by specific combinations of histone modifications, in which cases substitution by protamines would not occur, indicating regions of imprinting establishment.41 Thus, the presence of histones in regions regulated by genetic imprinting could yield epigenetic information essential for the adequate functioning of spermatozoa,42 and alterations of this process could generate modifications in epigenetic information.
Relationship between methylation patterns and sperm analysis components
Several studies indicate that methylation anomalies in infertile individuals in regions regulated by genetic imprinting are directly related to the presence of oligozoospermia.18-20,24 Furthermore, in these studies it is emphasized that as the severity of oligozoospermia increases, the frequency of methylation alterations increases. Nevertheless, the analysis only mentioned that those individuals who present anomalies in genetic imprinting are oligozoospermic, but does not specify that they are also teratozoospermic and/or astenozoospermic in many cases. Additionally, it is not indicated that, typically, as one of the sperm analysis components deteriorates, another one also becomes worse.
In order to be exhaustive in the establishing of a relation between methylation patterns and sperm analysis components, each of the components was separately valued. The finality of this separation allowed us to determine if a specific component was preferentially linked to individuals with normal or altered methylation patterns and in which region or regions. The results indicated that the presence of altered methylation patterns is not exclusively related to oligozoospermia, but that other alterations could also be involved.
A possible nexus between methylation anomalies in regions regulated by genetic imprinting and oligozoospermia could reside in the meiotic blockages that would be produced in spermatozoa with altered methylation patterns. Recombination hot-spots have been described in chromosomal regions regulated by imprinting.43,44 The presence of methylation anomalies could cause alterations in the meiotic pairing of chromosomes, leading to the activation of meiotic checkpoints that would stop the cells with anomalies, reducing the final number of spermatozoa. In terms of the relationship between methylation defects and teratozoospermia, a possible explanation could be given by the effects of epigenetic alterations in the packing of the nucleus. In human spermatozoa, the nucleus retains approximately 15% of histones, which are not substituted by protamines during spermiogenesis. Evidence exists that the presence of histones is not the result of insufficient substitution by protamines, but rather that specific sequences are packed, such as genes regulated by genetic imprinting, HOX genes, and transcription and signaling factors that are important for development.41 Genes regulated by genetic imprinting that do not present methylation in spermatozoa are associated with the H3K4me3 mark, while methylated genes do not show this modification and contain a moderate presence of H3K9me3.41 If anomalies in the epigenetic modifications of genes regulated by genetic imprinting are produced (alterations in methylation and/or in the modifications of histones), nucleosomes could be miss localized or present alterations in the specific modifications. As a consequence, an abnormal packing of the chromatin could occur that could give rise to an anomalous form of the sperm head.
Relationship between altered methylation patterns and assisted reproduction outcomes
Despite the fact that no case report or epidemiological study has been published reporting a major incidence of syndromes produced by methylation alterations in the paternal allele of children conceived by ART, the presence of anomalies in the imprinting of H19-ICR in samples of umbilical cord blood in three newborns out of 61 conceived by ART has recently been described.45 It is noteworthy that (1) all three cases correspond to pregnancies of di-zygotic twins in which siblings did not present anomalies, (2) all three pregnancies were conceived by intracytoplasmic sperm injection (ICSI) because the fathers presented fertility problems and (3) none of the cases was identified as showing a Silver-Russell Syndrome phenotype, characteristic of this type of epigenetic anomaly.
It would have been of great interest to extrapolate the anomalies present in a single spermatozoon to possible alterations in newborns or in the placentas. Faced with the impossibility of performing these studies, this work concentrated on the study of the relationship between semen samples containing spermatozoa with significantly increased abnormal methylation and the available data from cycles of assisted reproduction. We considered the hypothesis that if an abnormal spermatozoon fertilizes the oocyte, embryo development could be abnormal or show an increase in IUGR, as a result of alterations in the transfer of nutrients between mother and fetus. This effect was not confirmed in our results. It was neither confirmed in the study performed by Kanber et al.,46 where the authors analyzed whether babies born by ART with low weight (< 3rd percentile) showed methylation alterations in regions regulated by genetic imprinting that were related to fetal growth.
As far as the remaining analyzed parameters are concerned, the results in individuals with altered methylation patterns were, in general, equivalent to those obtained in individuals with normal patterns. Even individuals who presented clearly altered methylation results (24, 95, 97 and 107) had methylation values that were comparable to those of individuals from normal clusters. We cannot discard the possibility that the lack of differences can be a consequence of a purely numerical fact. As we have indicated, none of the individuals with altered methylation patterns presented alterations in the totality of spermatozoa studied. Except in individual 95, who showed the more extreme abnormal values in H19-ICR, in the rest of the analyzed cases the percent of gametes with normal methylation patterns was higher than the percent of spermatozoa with anomalies and, thus, the probability of fertilization by a spermatozoon showing a normal methylation pattern was higher. Another possibility would be the natural selection of normally methylated spermatozoa, in the case of conventional in vitro fertilization (IVF), or the absence of methylation anomalies in spermatozoa with normal shape and motility selected for ICSI. Nevertheless, to the best of our knowledge, there is no data demonstrating that single spermatozoon with abnormal DNA methylation have also abnormal shape or motility.
In any case, according to the clinical data at our disposal, our results coincide with the four studies published to date,19,22,25,47 in which no evidence was found that these anomalies affect the results of assisted reproduction treatments.
In summary, and considering the results of this work, we can conclude that individuals with fertility problems generate a percentage of spermatozoa with DNA methylation anomalies in regions regulated by genetic imprinting that is higher than that of control individuals. Nevertheless, these anomalies would not condition the results of treatments of assisted reproduction.
Materials and Methods
Sperm samples
Thirty semen samples from sperm donors were selected as a control population. All controls met the following characteristics: (1) normal karyotype, (2) proven fertility, (3) more than 90 x 106 of total motile progressive sperm, in the raw ejaculates, (4) more than 14% of spermatozoa with normal morphology (strict criteria) and (5) More than 10x106 of spermatozoa with progressive motility per ml after post-thawing survival test. The average age of the control group was 26 ± 6.15 y of age (range: 19–45).
One hundred and seven samples from individuals who sought consultation for fertility problems have been analyzed: 15 normozoospermic (N), 1 oligozoospermic (O), 8 astenozoospermic (A), 30 teratozoospermic (T), 1 oligoastenozoospermic (OA), 5 oligoteratozoospermic (OT), 31 astenoteratozoospermic (AT) and 16 oligoastenoteratozoospermic (OAT). The average age of the infertile individuals was 36 ± 5.50 y of age (range: 26–53). Seminal classification was performed following the criteria of the World Health Organization48 taken from the sperm morphology analyzed by means of the strict criteria of Kruger.
In 75 of the 107 infertile individuals, data were obtained that referred to the cycles of assisted reproduction to which the couple was submitted (Tables S6): the ART treatment, the number of recovered oocytes, number of zygotes, embryo quality,49 number of discarded embryos, number of transferred embryos, clinical pregnancies, number of miscarriages, births, sex and weight of the descendant. Embryo quality classification was done according to the ASEBIR criteria,49 which are based on the morphological features of the embryo (mainly cell number, degree of fragmentation and presence of vacuoles). There are four categories: category A, top quality embryos with the maximum capacity of implantation; category B, embryos of high quality with high capacity for implantation; category C, regular embryos with low chance of implantation and category D, poor quality embryos with scarce possibilities of implantation.
Starting from these data, the following were calculated: the fertilization rate (Zygotes/mature Oocytes), the quality of the embryos obtained, the rate of embryo development (Embryos/Zygotes), the non-viable embryos, the pregnancy rate, the miscarriage rate and the birth rate, the proportion of girls:boys and the average weight of the offspring.
The spermatozoa were separated from the rest of the cells of the ejaculate by the direct swim-up technique.50 The extraction of genomic DNA of the spermatozoa was performed with the commercial extraction kit PUREGEN (Gentra Systems).
Bisulfite pyrosequencing
Bisulfite treatment of genomic DNA was performed with the EZ DNA Methylation-DirectTM kit (Zymo Research Corp.). The input DNA to perform the conversion differs between samples depending on sperm concentration. DNA samples were grouped into series according to DNA concentration (from 200 to 500 ng) and the DNA bisulfite conversion was performed on each sample series together with a similar input of a sperm control DNA and a somatic control DNA sample in order to confirm the conversion efficiency. Further, cases showing abnormal methylation levels were retested. Bisulfite converted DNA was amplified by nested PCR with AmpliTaq Gold DNA Polymerase and Buffer II (Applied Biosystems), using the conditions recommended by Tost and Gut.51 Biotinylated amplicons were conjugated with streptavidin and recovered using the PyroMark Vacuum Prep Workstation (QIAGEN). Pyrosequencing was performed with the PyroGold SQA reagent (QIAGEN) on a PSQ96MA Pyrosequence System (Biotage) and analyzed with Pyro Q-CpG Software (QIAGEN). Due to the limitation to analyzing amplicons longer than 80 bp by pyrosequencing, the H19-ICR, KvDMR and MEG3-DMR loci were analyzed by three pyrosequencing reactions covering a total of 13 (excluding CpG 5 that corresponds to the SNP rs10732516 at the 6th CTCF-binding site), 21 and 21 CpGs, respectively. SNRPN-ICR was analyzed by two pyrosequencing reactions covering 20 CpGs. The repetitive sequence of IG-DMR does not allow for the design of optimal sequential pyrosequencing primers, so it was analyzed by a single pyrosequencing reaction covering 5 CpGs. Primer sets excluded the presence of CpG and SNP (single nucleotide polymorphisms) in their sequence. Accession numbers, nucleotide positions, primer sequences, amplicon length, annealing temperature and number of CpG are specified in Table 5.
Table 5. Bisulfite-treated sperm DNA amplification and pyrosequencing conditions.
| Loci (Accession number) |
Primer name |
Primer sequence (5′-3′) |
Nucleotide | Amplicon Length (Annealing) |
Number of CpG |
|---|---|---|---|---|---|
|
H19-ICR (AF087017) |
H19Fa |
AGGTGTTTTAGTTTTATGGATGATGG |
6006–6032 |
322 bp (61°C) |
|
| H19Ra |
TCCTATAAATATCCTATTCCCAAATAACC |
6299–6328 |
|
||
| P-H19F |
AGGGTTTTTGGTAGGTATAGAGTT |
6064–6087 |
265 bp (61°C) |
|
|
| PBT-H19R |
TCCTATAAATATCCTATTCCCAAAT |
6304–6328 |
|
||
| PS1-H19F |
TCGGTTTTATCGTTTGGATG |
6133–6152 |
|
5 |
|
| PS2-H19F |
TAGTGTAGGTTTATATATTA |
6200–6219 |
|
4 |
|
| PS3-H19F |
AACGTTTCGGGTTATTTAAG |
6257–6276 |
|
4 |
|
|
KvDMR (AJ006345) |
KCNFb |
TATGAGGTATTGGTTGGGTGTG |
255249–255270 |
450 bp (62°C) |
|
| KCNRb |
AAATCCCAAATCCTCAAAAATAAAC |
255673–255698 |
|
||
| P-KCNF |
TATTGGTTGGGTGTGAGGT |
225256–225274 |
212 bp (63°C) |
|
|
| PBT-KCNR |
AAACCCACAACAATATCAAAATAC |
255444–255467 |
|
||
| PS1-KCNF |
GGTTGGGTGTGAGGT |
225260–225274 |
|
6 |
|
| PS2-KCNF |
TTTGTTGTTTTTGAGTAT |
255304–255321 |
|
9 |
|
| PS3-KCNF |
GTGTGATGTGTTTATTATTT |
255397–255416 |
|
6 |
|
|
SNRPN-ICR (NC 000015) |
SNRPNFb |
TTATTGTAATAGTGTTGTGGGG |
131151–131172 |
400 bp (53°C) |
|
| SNRPNRb |
CTCCAAAACAAAAAACTTTAAAA |
131524–131546 |
|
||
| P-SNF |
TGTGGGGTTTTAGGGGTTTAGT |
131166–131187 |
296 bp (63°C) |
|
|
| PBT-SNR |
TAACCACTCCCCAAACTATCTCTTA |
131441–131465 |
|
||
| PS1-SNF |
AGGGAGGGAGTTGGGATT |
131216–131233 |
|
11 |
|
| PS2-SNR |
GAGTTTGGAGTAGAGTGGA |
131332–131350 |
|
9 |
|
|
IG-DMR (AL117190) |
MEG3Fb |
TTAGGTTGGAATTGTTAAGAGTTTGT |
50952–50977 |
370 bp (59°C) |
|
| MEG3Rb |
AAAAACTACATTTAAACAAAAAAAA |
51296–51320 |
|
||
| P-MEF |
AGAGTTTGTGGATTTGTGAGAA |
50969–50990 |
316 bp (62°C) |
|
|
| PBT-MER |
AAAAAACGAATCCATTATAACC |
51262–51283 |
|
||
| PS1-MEF |
TTGTGGATTTGTGAGAAATGA |
50974–50994 |
|
5 |
|
|
MEG3-DMR (AL117190) |
MEG3.2Fb |
AATTTATGTTTTTGTGGGGTTGTAG |
64272–64296 |
297 bp (56°C) |
|
| MEG3.2Rb |
TTAACCACAATATTAATAACTAAAAAACAA |
64532–64561 |
|
||
| P-M2F |
ATTTATGTTTTTGTGGGGTTGTAG |
64285–64302 |
246 bp (62°C) |
|
|
| PBT-M2R |
TCAACAACCAAAAACCCCTATC |
64497–64518 |
|
||
| PS1-M2F |
GTGGGGTTGTAGGGTTGA |
64285–64302 |
|
8 |
|
| PS2-M2F |
GGCGGTTATTATAGTTTTTA |
64369–64388 |
|
8 |
|
| PS3-M2F | GTTTTCGAGAGGTTAGTAAT | 64439–64458 | 5 |
Loci, primers sequence and position, amplicon length, annealing temperature and number of CpG within each amplicon are specified. aPrimers described by Kerjean et al., 2000;2 bPrimers designed using Methprimer software (www.urogene.org/methprimer/); P primers were designed using the Pyrosequencing Assay Design Software (QIAGEN):P, nested primer; PBT, biotinylated nested primer; PS, sequencing primer.
Statistical analysis
The statistical analysis was performed with the specialized support of the Servei d’Estadistica Aplicada (Applied Statistics Service) of the Universitat Autònoma de Barcelona. The level of statistical significance was 0.05.
CpG to CpG Analysis
Starting from the percent of methylation of each CpG analyzed in the control population, the mean, standard deviation (SD) and range were determined. Starting from the mean ± 2SD, a range of reference methylation was obtained that included 95% of the values for each CpG. This value was compared with the methylation values obtained in the infertile individuals. The comparison permitted the classification of each CpG analyzed from each infertile individual within or outside the range of reference methylation. With the goal of knowing whether the analyzed CpGs presented equivalent methylation values inside of each region in the control population, the ANOVA test was performed. In the cases in which the SD showed significant differences, the Kruskal-Wallis test was applied.
Analysis of the methylation pattern
In order to define altered methylation patterns, a hierarchical cluster analysis was performed52 where the variables used were the percent of methylation of the CpG analyzed in the population studied (control and infertile). The programs used were JMP v7.0.2 (SAS Institute, Inc.) and SPAD v4.5 (Centre International de Statistiques et d’Informatique Appliquees). Distances were calculated by means of the Ward method and the groupings among individuals were represented by means of a dendrogram.
The clusters that showed a mean methylation value close to the expected theoretical value were classified as normal clusters (formed by individuals with normal methylation patterns). On the other hand, clusters with the most distant mean methylation values were considered as abnormal clusters (formed by individuals with altered patterns).
With the goal of determining whether individuals within the clusters with anomalies showed significant differences in the levels of methylation, compared with individuals belonging to clusters with normal patterns, a Mann-Whitney test was performed. The performance of this test permitted the determination of a mean methylation value and a confidence interval of 95% for each analyzed regions.
Relationship between methylation, seminal parameters and the age of the individuals
In order to evaluate the relationship between altered patterns of methylation and anomalies in sperm counting, motility and/or morphology, a contingency analysis was performed using the Chi-square test (χ2). This analysis was performed considering whether the individuals within the normal and abnormal clusters presented the variables of the sperm analysis/semen parameters: normozoospermia, non-normozoospermia; oligozoospermia, non-oligozoospermia, astenozoospermia, non-astenozoosermia, teratozoospermia and non-teratozoospermia. This analysis was specifically performed for each of the regions analyzed.
To determine the influence of age over the methylation results, two analyses were separately performed in each of the regions. First, comparing the mean values of methylation of the individuals classified in the clusters with anomalies and those individuals included in the clusters with normal patterns, a Mann-Whitney test was performed. In a second analysis, the age of every single individual (controls and patients) was correlated with their respective mean methylation level using a Spearman test.
Relationship between methylation patterns and results of the cycles of assisted reproduction
We evaluated whether significant differences existed in the results of the cycles of assisted reproduction of the infertile individuals who were grouped in the clusters with normal methylation patterns vs. those who were grouped in clusters with anomalies. A lineal model was used that is generalized for repeated measures considering a normal distribution for the fertilization rate, a binomial distribution for pregnancy rate and a Poisson distribution for the comparison of the different types of embryos (A, B, C or D), rate of embryo development, discarded embryos due to transfer, miscarriage, births, XX:XY proportion and weight of the descendant.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Javier Nadal from the Unidad de Reproducción Asistida of the Centro Médico Teknon (Barcelona, Spain) and the Laboratorio de Andrología y Banco de Semen of the Instituto Universitario IVI Valencia (Valencia, Spain) for providing the semen samples.
Glossary
Abbreviations:
- DMR
differential methylated region
- ICR
imprinting control region
- LOM
loss of methylation
- LOI
loss of imprinting
- IUGR
intrauterine growth retardation
- CI
confidence interval
- MM
methylation mean
- ART
assisted reproduction techniques
- PGC
primordial germ cells
- DNMT
DNA methyl transferase
- N
normozoospermic
- O
oligozoospermic
- A
astenozoospermic
- T
teratozoospermic
- OA
oligoastenozoospermic
- OT
oligoteratozoospermic
- AT
astenoteratozoospermic
- OAT
oligoastenoteratozoospermic
- ICSI
intracytoplasmic sperm injection
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
C.C. was involved in study conception and design, data analysis and interpretation, manuscript writing and final approval. M.P. was involved in experimental procedures, data collection and assembly, data analysis and interpretation, manuscript writing and final approval. M.G. contributed in patient selection and analysis, ART data collection and assembly and final approval of the manuscript. N.G. contributed in control selection and analysis and final approval of the manuscript. M.C.P. contributed in patient selection and analysis, ART data collection and assembly and final approval of the manuscript. J.B. was involved in study conception and design, data analysis and interpretation, manuscript writing and final approval.
Financial Disclosures
This work was supported by Projects PS09/00330 (Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, Spain) and SGR2009–282 (Agència de Gestió d’Ajuts Universitaris i de Recerca, Generalitat de Catalunya, Spain). Marta Pladevall is the recipient of a grant from the Universitat Autònoma de Barcelona (UAB2006–00213).
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/21743
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/21743
References
- 1.Arnaud P, Feil R. Epigenetic deregulation of genomic imprinting in human disorders and following assisted reproduction. Birth Defects Res C Embryo Today. 2005;75:81–97. doi: 10.1002/bdrc.20039. [DOI] [PubMed] [Google Scholar]
- 2.Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Pàldi A, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9:2183–7. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- 3.Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12:2873–9. doi: 10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- 4.Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22:26–35. doi: 10.1093/humrep/del316. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, Lee HC, Jeong YM, Chung TG, Kim HJ, Kim NK, et al. MTHFR C677T polymorphism associates with unexplained infertile male factors. J Assist Reprod Genet. 2005;22:361–8. doi: 10.1007/s10815-005-6795-0. [DOI] [PubMed] [Google Scholar]
- 6.Zogel C, Böhringer S, Gross S, Varon R, Buiting K, Horsthemke B. Identification of cis- and trans-acting factors possibly modifying the risk of epimutations on chromosome 15. Eur J Hum Genet. 2006;14:752–8. doi: 10.1038/sj.ejhg.5201602. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17:1582–91. doi: 10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim AL, Ferguson-Smith AC. Genomic imprinting effects in a compromised in utero environment: implications for a healthy pregnancy. Semin Cell Dev Biol. 2010;21:201–8. doi: 10.1016/j.semcdb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Anway MD, Skinner MK. Epigenetic programming of the germ line: effects of endocrine disruptors on the development of transgenerational disease. Reprod Biomed Online. 2008;16:23–5. doi: 10.1016/S1472-6483(10)60553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pathak S, Kedia-Mokashi N, Saxena M, D’Souza R, Maitra A, Parte P, et al. Effect of tamoxifen treatment on global and insulin-like growth factor 2-H19 locus-specific DNA methylation in rat spermatozoa and its association with embryo loss. Fertil Steril. 2009;91(Suppl):2253–63. doi: 10.1016/j.fertnstert.2008.07.1709. [DOI] [PubMed] [Google Scholar]
- 11.Pathak S, D’Souza R, Ankolkar M, Gaonkar R, Balasinor NH. Potential role of estrogen in regulation of the insulin-like growth factor2-H19 locus in the rat testis. Mol Cell Endocrinol. 2010;314:110–7. doi: 10.1016/j.mce.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Chmurzynska A. Fetal programming: link between early nutrition, DNA methylation, and complex diseases. Nutr Rev. 2010;68:87–98. doi: 10.1111/j.1753-4887.2009.00265.x. [DOI] [PubMed] [Google Scholar]
- 13.Cox GF, Bürger J, Lip V, Mau UA, Sperling K, Wu BL, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–4. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ørstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, et al. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72:218–9. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, et al. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003;40:62–4. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–41. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–60. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700–2. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–51. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 20.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 21.Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585–94. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 22.Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–9. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 24.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 25.El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, et al. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–9. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 26.Minor A, Chow V, Ma S. Aberrant DNA methylation at imprinted genes in testicular sperm retrieved from men with obstructive azoospermia and undergoing vasectomy reversal. Reproduction. 2011;141:749–57. doi: 10.1530/REP-11-0008. [DOI] [PubMed] [Google Scholar]
- 27.Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346:731–7. doi: 10.1056/NEJMoa010806. [DOI] [PubMed] [Google Scholar]
- 28.Reinhart B, Eljanne M, Chaillet JR. Shared role for differentially methylated domains of imprinted genes. Mol Cell Biol. 2002;22:2089–98. doi: 10.1128/MCB.22.7.2089-2098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23:2826–34. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 30.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–81. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–9. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 33.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 34.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 35.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–3. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 36.Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–80. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 37.Takashima S, Takehashi M, Lee J, Chuma S, Okano M, Hata K, et al. Abnormal DNA methyltransferase expression in mouse germline stem cells results in spermatogenic defects. Biol Reprod. 2009;81:155–64. doi: 10.1095/biolreprod.108.074708. [DOI] [PubMed] [Google Scholar]
- 38.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–9. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He F, Lidow IA, Lidow MS. Consequences of paternal cocaine exposure in mice. Neurotoxicol Teratol. 2006;28:198–209. doi: 10.1016/j.ntt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Oakes CC, Kelly TL, Robaire B, Trasler JM. Adverse effects of 5-aza-2′-deoxycytidine on spermatogenesis include reduced sperm function and selective inhibition of de novo DNA methylation. J Pharmacol Exp Ther. 2007;322:1171–80. doi: 10.1124/jpet.107.121699. [DOI] [PubMed] [Google Scholar]
- 41.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–7. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 43.Sandovici I, Kassovska-Bratinova S, Vaughan JE, Stewart R, Leppert M, Sapienza C. Human imprinted chromosomal regions are historical hot-spots of recombination. PLoS Genet. 2006;2:e101. doi: 10.1371/journal.pgen.0020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnheim N, Calabrese P, Tiemann-Boege I. Mammalian meiotic recombination hot spots. Annu Rev Genet. 2007;41:369–99. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- 45.Shi X, Ni Y, Zheng H, Chen S, Zhong M, Wu F, et al. Abnormal methylation patterns at the IGF2/H19 imprinting control region in phenotypically normal babies conceived by assisted reproductive technologies. Eur J Obstet Gynecol Reprod Biol. 2011;158:52–5. doi: 10.1016/j.ejogrb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Kanber D, Buiting K, Zeschnigk M, Ludwig M, Horsthemke B. Low frequency of imprinting defects in ICSI children born small for gestational age. Eur J Hum Genet. 2009;17:22–9. doi: 10.1038/ejhg.2008.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, et al. Influence of global sperm DNA methylation on IVF results. Hum Reprod. 2005;20:768–73. doi: 10.1093/humrep/deh684. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organization. 1999: WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. 4th Edition, Cambridge University Press; New York. [Google Scholar]
- 49.Ardoy M, Calderón G. Cuadernos de Embriología Clínica: Criterios de Valoración Morfológica de Oocitos, Embriones tempranos y Blastocistos Humanos. 2nd edn, 2008. ASEBIR, Madrid, Spain. [Google Scholar]
- 50.Solvas I, Grossmann M, Santaló J, Pons MC. Estudio comparativo entre dos métodos de swim-up. Revista ASEBIR. 2002;7:28–32. [Google Scholar]
- 51.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–75. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 52.Lebart L, Morineau A, Piron M. Statistique exploratoire multidimensionelle. 4th edn, 2006. Dunod Press, France. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.