Abstract
Human brain function is mediated by biochemical processes, many of which can be visualized and quantified by positron emission tomography (PET). PET brain imaging of monoamine oxidase A (MAOA)—an enzyme metabolizing neurotransmitters—revealed that MAOA levels vary widely between healthy men and this variability was not explained by the common MAOA genotype (VNTR genotype), suggesting that environmental factors, through epigenetic modifications, may mediate it. Here, we analyzed MAOA methylation in white blood cells (by bisulphite conversion of genomic DNA and subsequent sequencing of cloned DNA products) and measured brain MAOA levels (using PET and [11C]clorgyline, a radiotracer with specificity for MAOA) in 34 healthy non-smoking male volunteers. We found significant interindividual differences in methylation status and methylation patterns of the core MAOA promoter. The VNTR genotype did not influence the methylation status of the gene or brain MAOA activity. In contrast, we found a robust association of the regional and CpG site-specific methylation of the core MAOA promoter with brain MAOA levels. These results suggest that the methylation status of the MAOA promoter (detected in white blood cells) can reliably predict the brain endophenotype. Therefore, the status of MAOA methylation observed in healthy males merits consideration as a variable contributing to interindividual differences in behavior.
Keywords: MAOA genotype, DNA methylation, monoamine oxidase A, positron emission tomography
Introduction
The monoamine oxidase A (MAOA) gene (hg18, chrX: 43,400,353–43,491,012) is one of the most investigated brain candidate genes.1 The gene product, MAOA, oxidizes monoamines, such as serotonin, norepinephrine and dopamine, thereby exerting pleiotropic effects on mood and arousal (reviewed in ref. 2). A growing number of studies have reported that a common tandem repeat polymorphism (uVNTR) in the MAOA promoter3 that produces “high” and “low” alleles is associated with antisocial behavior,4 impulsivity,5 major depressive disorder6 and other neuropsychiatric disorders (reviewed in ref. 1). Since the functional effect of the uVNTR was originally demonstrated in cell culture and biochemical experiments,7 we questioned the extent to which the high and low MAOA alleles modulate the levels of MAOA in the brain. In an earlier study, we measured MAOA enzyme directly with positron emission tomography (PET) and [11C]clorgyline in a group of 28 normal healthy men who were also genotyped for the “high” and “low” alleles. Although we did not find a relationship between genotype and brain MAOA activity,8 we observed a high interindividual variability in brain MAOA levels that merited closer investigation.
Prior studies of the MAOA genotype have underscored the critical role of environment for the manifestation of specific phenotypes; for example, an association of “low” allele with impulsive behaviors was contingent upon exposure to developmental stressors,4,9 suggesting the involvement of factors other than sequence variation in translating the MAOA genotype to different behavioral phenotypes.
Epigenetic modifications of the genome are pivotal for proper genome functioning enabling precise control of gene expression and thereby the availability of the final protein products. Within mammalian DNA, epigenetic information is predominantly encoded by the enzymatic conversion of cytosine to 5-methyl-cytosine.10 Methylated DNA is mitotically stable and therefore this epigenetic modification is commonly considered as epigenetic marker.11 The functional consequences of methylation depend on the location of the methylated CpGs relative to the transcription start site (TSS) of the cognate gene. That is, methylation occurring at the site of transcriptional initiation often suppresses gene expression,12,13 while intragenic methylation is important for regulation of gene expression in tissue- and cell-specific manner.14 Although aberrant methylation has long been associated with cancer15 and more recently with common diseases,16 our understanding of the physiological norms of methylation variability remains limited. Indeed, only a few studies have reported non-disease methylation heterogeneity related to phenotypic discordance of twins,17,18 normal aging19 and ethnic diversity.20
In the context of brain/behavioral phenotypes, epigenetic mechanisms enable adaptations and contribute to heterogeneity and plasticity by mediating environmental and stochastic effects. For example, evidence from animal models has linked changes in locus-specific methylation to social cognition,21 learning and memory22 and stress-related behaviors.23 However, generalization from preclinical findings is challenging due to the uniqueness of the human brain. Moreover, the biological impact of DNA methylation is species-specific, so that at some loci methylation can have differential effects on gene expression even in such closely related species as humans and chimpanzees.24 At the same time, the lack of available technology hinders the comprehensive exploration of the human brain epigenome, while studies of the human brain are limited to analysis of post mortem samples with associated bias (see ref. 25 for example).
An alternative strategy to analyze the relationships between DNA methylation and brain phenotype is to use an easily accessible proxy tissue, e.g., white blood cells (WBC). The reasoning for this is that while cell and tissue specific epigenetic patterns are established during development, subsequent environmental exposure would have a global impact on epigenome. Indeed, empirical evidence indicates that specific epigenetic modifications can be shared across the genome, and thus may serve as epimarkers.14 The utility and validity of this approach is supported by reports of associations between DNA methylation patterns on WBC and various common diseases, including psychiatric disorders.26,27 For example, a recent fMRI study demonstrated that DNA methylation in the COMT gene measured in WBC predicted changes in brain activation patterns after stress exposure.28
Here we hypothesized that that epigenetic mechanisms and individual variations in the sequence of the core promoter would modulate the epigenetic landscape of the MAOA locus, thereby influencing gene expression and individual brain MAOA activity level. We tested this hypothesis by measuring brain MAOA activity using PET with [11C]clorgyline, a radiotracer with specificity for MAOA,29,30 in 34 healthy non-smoking males (tobacco smoke inhibits MAOA31) and analyzing the MAOA methylation in WBC by bisulphite conversion of genomic DNA and subsequent sequencing of cloned DNA products.
Results
MAOA genotype
In our sample, most individuals (18 out of 34) carried “high” MAOA allele. Allele distribution varied across the ethnic group; the population-specific allele frequencies were closed to previously reported32. The differences in the MAOA allele frequencies between the ethnic groups were not significant: χ2 = 3.85, df = 4 and p = 0.43. Demographic characteristics of the study sample are shown in Table 1.
Table 1. Demographic characteristics of the study sample.
| Genotype | “Low” MAOA | “High” MAOA | |
|---|---|---|---|
| |
Count |
n = 16 |
n = 18 |
| |
Age |
29 ± 7.5 |
30.9 ± 7.5 |
| Ethnicity | Caucasians |
3 |
8 |
| African-Americans |
9 |
7 |
|
| Hispanics |
2 |
2 |
|
| Asian |
1 |
0 |
|
| more-than-one race | 1 | 1 |
The differences in the MAOA allele frequencies between the ethnic groups are not significant: χ2 = 3.85, df = 4, p = 0.43. Population-specific genotype frequencies are close to previously reported.
Individual differences in the sequence of the MAOA promoter
To account for the potential impact of individual variation in DNA sequence on epigenetic differences, we obtained the complete sequence of the extended promoter region that encompasses two tandem repeat polymorphisms.33 Two VNTRs alleles were strongly intercorrelated, so that the carriers of the “low” allele (uVNTR) ultimately had a 10R-allele of the second, 10 bp VNTR, while the “high” allele uVNTR carriers had 9R-allele of 10 bp VNTR. Apart from these variances, we did not detect other sequence variations in our sample.
DNA methylation of the MAOA core promoter
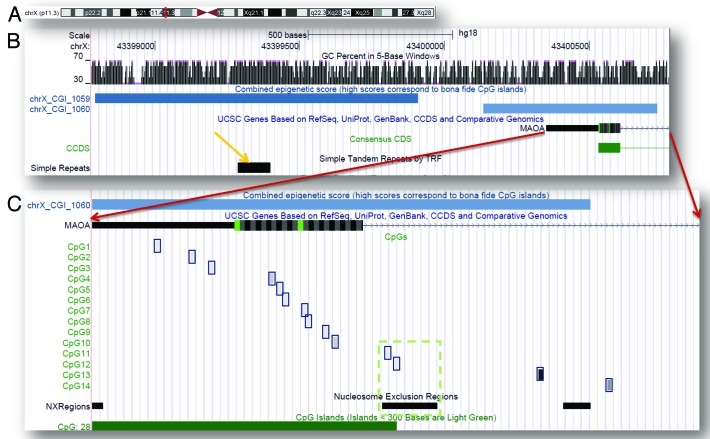
Based on our recent characterization of the epigenetic features of the MAOA gene33 and on our analysis of the methylation potential of the MAOA locus, here we focused on a subregion of the CGI (CpG island)_1060 (chrX: 43,400,353–43,400,797) encompassing 14 CpG sites (Fig. 1). We chose this region because the intermediate epigenetic score (447) of CGI_1060 indicates its propensity to differential methylation34 and considering that the 500 bp flanks of TSSs exhibit the most methylation variability associated with the gene expression.35 Furthermore, a recent study36 provided new evidence on the role short sequence modules within the proximal promoters have in defining their own methylation states. The potential functional significance of the methylation marks occurring in this region is also indicated by the spike in conservation score and co-occurrence of multiple regulatory elements (pleiadesgenes_R).33
Figure 1. MAOA genomic locus and region analyzed in the study. (A) X-chromosome ideogram with the MAOA position indicated in red; (B) overview of the MAOA promoter and its epigenetic features; (C) Amplicon overview (red arrows indicate its position relatively to the MAOA gene) represents the methylation status of individual CpG sites, averaged across the samples (shades of gray correspond to the methylation percentage). Green dashed box shows colocalization of the CpG11 and CpG12 with nucleosome exclusion region.
Aiming to obtain detailed quantitative and qualitative information on the methylation status of individual DNA molecules with a single CpG resolution, we used bisulphite conversion of genomic DNA followed by cloning and sequencing of the cloned products.37 Currently, this method is considered the most reliable in enabling single nucleotide resolution for locus-specific methylation across the DNA molecule38.
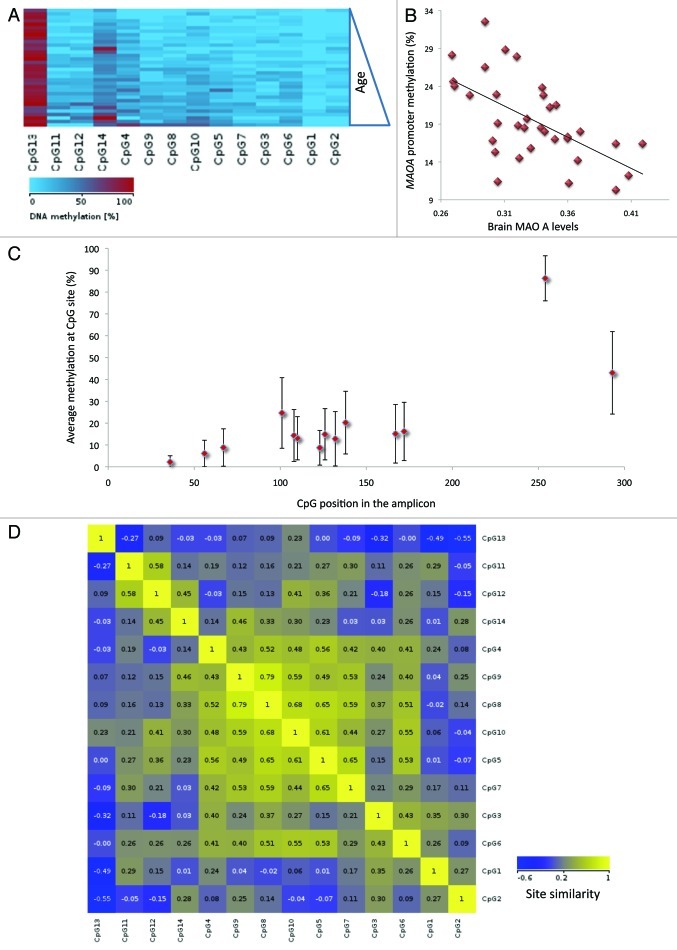
Typical CGIs overlapping with TSSs of transcriptionally active genes are methylation-free; therefore, little or no methylation was expected within the CGI_1060. In our sample, between 10.3% and 35.5% of analyzed CpGs were methylated (median 18.8%), with approximately normal distribution of the regional methylation means (Fig. S1A). The differences in DNA methylation across individuals (Fig. S2) were not related to ethnicity (F(2,29) = 0.2, p = 0.81). Age did not have an apparent effect on methylation either (Fig. 3A), (F(19,14) = 0.92; p = 0.58). We did not detect influence of the genotype on methylation; in our sample, genotype groups had similar methylation means (low: 20%, high: 19%, F(1,32) = 0.26, p = 0.6).
Figure 3. Interindividual variability of the MAOA methylation states. (A) Methylation statuses of CpG sites in individual samples (sorted by age) are compiled as methylation map. (B) Average methylation for the amplicon calculated for individual genomic samples is plotted against the respective brain MAOA levels (λk3 values). (C) Experiment-wide average (percentile) and range (± s.d.) of site-specific methylation. (D) Graphical representation of Pearson’s similarity between the methylation values of individual CpG sites.
Interindividual variation in brain levels of MAOA activity
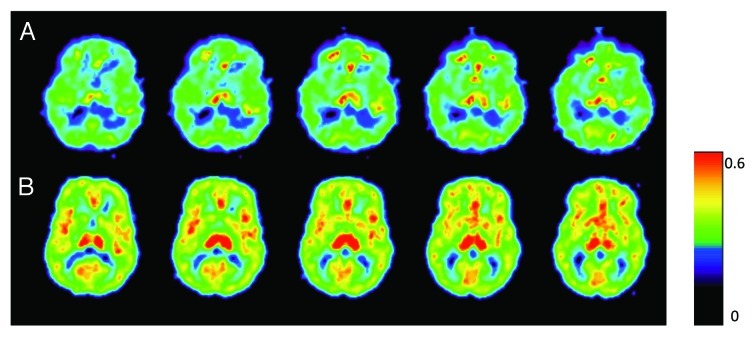
An average brain measure of enzyme levels (brain λk3) was used as an outcome measure because MAOA activities in the different brain regions are highly intercorrelated.31 On a sample level, global measures of brain MAOA activity exhibited approximately normal distribution (Fig. S1B shows distribution pattern and Table S1 present values of individual λk3 measures for the whole brain), with markedly different levels of enzyme activity among individuals visualized with PET and [11C]clorgyline (Fig. 2). Brain MAOA levels were independent of age [Pearson’s r(34) = 0.001, p = 0.9] and did not differ between genotype-based and ethnicity-based subgroups (ANOVA: F(1,32) = 1.74, p = 0.2 and F(2,30) = 0.51, p = 0.6, respectively).
Figure 2. Variability in brain MAOA levels observed in healthy males. Parametric images of the model term which is a function of MAOA activity for two healthy individuals show the same planes of the brain. Brain λk3 for those individuals are 0.27 mLplasma(mLbrain)−1min−1 and 0.4 mLplasma(mLbrain)−1min−1 (top and bottom panels, respectively). A rainbow color scale is used where red represents the highest λk3 values.
Relationship between MAOA methylation and brain MAOA activity
The average methylation levels measured across the analyzed region and brain MAOA activity were negatively correlated [Pearson’s r(34) = 0.61, p < 0.001] (Fig. 3B). This relationship was independent of genotype, so that the average methylation predicted brain λk3 in both genotype groups (low MAOA: b = 0.57, t(16) = 2.67, p = 0.018; high MAOA: b = 0.65, t(16) = 3.29, p = 0.005), explaining a significant proportion of the variance in brain λk3 (low MAOA: R2 = 0.322, F(1,15) = 7.11, p = 0.018; high MAOA: R2 = 0.42, F(1,15) = 10.82, p < 0.01).
To assess the unique contribution of age, ethnicity and genotype along with methylation and explain variance in the brain measures, we conducted multiple regression analysis. Regression model was significant (F4,29 = 11.81, p = 0.002), with methylation explaining 25.5% of the outcome measure. However, the addition of the other variables did not improve our accuracy in predicting values for the MAOA brain activity.
Analysis of the methylation patterns
Providing a good epigenetic marker, the average value of subclonal DNA methylation variances can also mask differences in position-specific methylation. Therefore, we next explored individual methylation patterns, i.e., the combination of methylation statuses of contiguous CpG nucleotides in a DNA strand39. Overall, there was a slight gradient in site-specific methylation (Fig. 3C), with the lowest level detected in TSS-proximal CpG1 (in 24 out of 34 samples it was methylation free). CpGs2–7 exhibited low (median 11.4%) and CpG8–14 exhibited medium (median 27.9%) methylation levels. Most methylation was detected at CpG13 (averaging 83.5%; it was fully methylated in 6 samples). The distribution of the methylation values at individual CpG units across samples was not uniform: for some sites (e.g., CpG4, CpG14), it was close to normal (Fig. S3A), for others (e.g., GpG1, CpG2), it was skewed toward zero (Fig. S3B). For CpG13 it was skewed toward 1 (Fig. S3C).
Based on the combined data set, we next computed the correlations of DNA methylation at individual CpG units between individual samples and between amplicon averages (graphed as similarity plot in Fig. 3D). Pearson's correlation coefficients for the pairwise combinations of amplicons ranged from −0.03 to 1 (median 0.57). Spearman's correlation coefficients for the pairwise combinations of CpG units ranged from −0.55 to 0.79 (median 0.26).
There were differences in variability of the DNA methylation values measured at individual CpG sites wherein the variance of methylation values for a given CpGs across the individuals was less pronounced than the differences in methylation values across the analyzed CpG units. We also detected some constraints to the variability of the methylation pattern, for example, methylation states of the CpG8 and CpG9 remain significantly different from CpG1 and CpG2 states across all DNA samples (Kolmogorov-Smirnoff test: p < 0.02).
A closer look at between-pair methylation differences for CpG pairs revealed a complex pattern of the CpG clustering and marked discordance between the methylation states of proximally positioned CpG sites. A high degree of methylation discordance within a relatively short region is unexpected; notably, even tandem CpG5 and CpG6 (5′-TATCGCGGG-3′) displayed only limited similarity (Pearson’s correlation coefficient 0.53). The negative values of the correlation coefficient point out to dissimilarity between neighboring CpG sites; for example, CpG11 vs. CpG13: Pearson’s coefficient = -0.27; and CpG2 vs. CpG5: Pearson’s coefficient = -0.07. The degree of within-pair resemblance of methylation at individual CpG sites is displayed as a dendrogram in Figure S4.
Next, we compared the methylation patterns of the subclones derived from unrelated input DNA samples. Along with marked variation in average DNA methylation among individuals, there were notable differences in the methylation patterns of the subclones derived from the same sample of genomic DNA. Figure S5 displays clonal methylation patterns of 3 individual genomic DNA samples. Most individual genomic DNA samples produced methylation-free clones (samples 1 and 2 but not sample 3); however, having analyzed about 1,200 clonal sequences, we did not detect a single fully methylated DNA molecule. Several clones with concordant methylation pattern were observed in the same sample (see sample 3 where patterns of the clones 1 and 2, 5 and 6, 13, 14 and 15 are similar—indicated with green brackets). The patterns shared across the samples (purple brackets) were very simple (i.e., only CpG13 is methylated).
Relationships between the site-specific methylation and brain MAOA activity
We speculated that site-specific methylation levels might be better predictors of the brain MAOA measures because the observed difference in methylation state and in extent of methylation variability of individual CpGs most likely reflects their specific localization relative to the functional elements of the MAOA gene promoter. A correlation analysis revealed a significant (negative) relationship between the enzyme level and methylation state of several CpG sites: CpG 5 [Spearman’s r(32) = -0.52, p = 0.002], CpG6 [r(32) = -0.42, p = 0.017], CpG8 [r(34) = -0.44, p = 0.01], CpG10 [r(34) = -0.5, p = 0.003], CpG11 [r(34) = -0.62, p < 0.001], and CpG12 [r(34) = -0.62, p < 0.001]. The significance of the model with the methylation value of CpG12 as predictor variable (adjusted Rsq. = 0.398; F1,32 = 17.3; p < 0.0001. β = -0.593; p = < 0.0001) highlights its potential value as a single epigenetic marker.
Principal component analysis
In order to reduce the dimension space of the epigenetic variability, we performed a principal component analysis. The combination of four principal components (PCs) accounted for 70% of the variance: PC1 captured most (23.1%) variation, PC2, PC3 and PC4 accounted for 19.1%, 13.9% and 13.8% of the variance, respectively. PC-based analysis underscored position-dependency of methylation variability: PC1–3 largely represented methylation states of the CpG clusters in the beginning, middle and end of the analyzed region, respectively, while PC4 represented CpG11 and CpG12. The correlation between PC1 and brain MAOA level (λk3) was significant [Spearman’s r(33) = -0.364, p = 0.037], though PC2 and PC3 were not informative with respect to the brain endophenotype. A strong negative correlation between PC4 and λk3 [Spearman’s r(33) = -0.507, p = 0.003] seems rather intriguing because the CpG11 and CpG12 sites, represented by PC4, reside within the nucleosome exclusion region (Fig. 1).
Discussion
The MAOA gene and its protein product, MAOA enzyme, are among the most studied in psychiatry.1,9,40 The ability to image and quantify human brain MAOA with PET has enabled the investigation of relationships between MAOA levels and behavioral and disease phenotypes in healthy volunteers and in patients.41-43 For example, PET studies have revealed elevations in brain MAOA activity in major depressive disorder (MDD)44 and mood disorders,43 whereas low MAOA activity has been related to maladaptive behavioral traits, such as aggression.45 Interestingly, PET imaging has also demonstrated a modulating effect of the environment on brain enzyme levels by showing that current cigarette smokers have reduced brain MAOA.31
The present study was stimulated by a prior study in our laboratory that revealed that MAOA levels vary widely between healthy non-smoking men and that this variability was not explained by the MAOA polymorphism (uVNTR genotype).8 We speculated that epigenetic modifications may mediate the normal variability in brain MAOA levels and set out to investigate the relationship between the methylation status of the MAOA gene and brain MAOA levels using a combination of imaging and epigenetic analysis.
The major findings of the present study are the following: First, with this PET imaging study with [11C]clorgyline, we were able to replicate, in an independent sample, our prior finding of a insignificant effect of the MAOA genotype on brain MAOA activity.8 Second, our results suggest that in normal populations, the MAOA promoter exhibits variable DNA methylation pattern wherein methylation variability can be largely attributed to non-genetic factors. Third, the observed non-randomness of methylation levels and methylation variability, where methylation discordance is constrained to few CpG sites (e.g., CpG5 and CpG11), points out to its potential physiological relevance. Fourth, a significant association between MAOA methylation and brain MAOA enzymatic activity supports the idea that regional DNA methylation measured on WBC can be a useful epigenetic biomarker for brain endophenotype.
Replication of prior study
In a prior study8 we reported large inter-subject variability in brain MAOA in a relatively homogenous group of healthy male subjects, irrespective of MAOA genotype. This finding was in need for replication because of study limitations arising from the small sample size and ethnic diversity of study population. Here, we replicated the prior finding in a new, larger population sample, thereby corroborating the notion that heterogeneity of the MAOA brain endophenotype is likely to be driven by non-genetic factors (aka, environmental exposure).
Variable methylation
Our analysis revealed extensive variation in DNA methylation of the MAOA core promoter, with discordance of the methylation patterns both within the region and between the individuals. The overall distribution of DNA methylation exhibited some directionality, being the lowest in the vicinity of TSS and gradually increasing toward the 3′ end of the region, with a variable degree of methylation heterogeneity at individual CpG sites therein. Our analysis revealed only small differences between ethnic groups and we did not detect differences between the MAOA genotype groups. Considering relative sequence uniformity of the analyzed locus, it appears that the contribution of genetic background to DNA methylation is rather small. Thus, the majority of methylation variance can be attributed to non-genetic factors, including stochastic variations during developmental epigenetic programming.46
Methylation discordance restricted to a few sites
Differences in methylation patterns of the subclones derived from individual input DNA samples suggest variable methylation of the core MAOA promoter.16 A difference in the range of methylation levels detected at individual CpG sites (Fig. 1C) implies that there are certain constraints facilitating “physiologically permitted” variability in methylation levels which are likely to be physiologically relevant. Because the observed difference in methylation state and in extent of methylation variability of individual CpGs most likely reflects their specific localization relative to the functional elements of the MAOA gene promoter, it is likely that site-specific methylation levels detected in individual DNA can be related to the brain MAOA measures.
Association between the MAOA methylation and MAOA
Deciphering the role of DNA methylation and human behavior is difficult, largely due to inaccessibility of the human brain tissue.25 Here, we tested a correlation between methylation state of the MAOA locus and respective levels of brain MAOA activity in a living brain using WBC as a proxy tissue for methylation analysis. The design of this imaging genetic study does not allow making an inference on causality; nonetheless, the significance of the relationships points to a negative impact of MAOA promoter methylation on MAOA expression (in a proxy tissue), which is measured at the brain level. If confirmed by further studies, this finding may be used for the development of new diagnostics.
Study limitations
This is the first report on the predictive value of peripheral MAOA methylation for the MAOA brain endophenotype and, as such, its findings are in need for replication. We also want to note several limitations of our study: (1) In order to minimize the biological complexity of the variables analyzed in this study, we did not include females in our analysis, which involved only healthy males. This reduced possible bias but limited the generalizability of the findings; (2) The assay employed in our study to analyze DNA methylation, i.e., cloning and sequencing of BS-converted DNA, has its technical limitations and, therefore, it is important to validate our findings using alternative analytical technologies; (3) Similar to most PET imaging studies, our sample size is relatively small; (4) In this study, we analyzed only part of the large methylation potential of the MAOA gene, which comprises two CGIs and encompasses 161 CpGs. It is possible that future methylation analyses covering the entire promoter would be able to detect more robust and reliable epimarkers related to MAOA regulation.
Conclusion
We found significant interindividual differences in core MAOA promoter methylation status and pattern, which were not influenced by the VNTR genotype. However, we found a strong association between regional and CpG site-specific MAOA promoter methylation and brain MAOA levels. These results suggest that the methylation status of the MAOA promoter detected in white blood cells can reliably predict the brain endophenotype and that the status of MAOA methylation observed in healthy males merits consideration as a variable contributing to interindividual differences in behavioral and disease phenotypes. Since our knowledge on the extent of normal epigenetic variability and the functional relevance of epigenetic polymorphisms remains very limited, our study contributes to the understanding of interindividual variations in DNA methylation and its functional implications at the level of the gene product in the living human brain.
Methods
Subjects
Research was approved by the local institutional review board [Committee on Research in Human Subjects (CORIHS), Stony Brook University]. Healthy male subjects (n = 34) were enrolled after informed consent was obtained and confidentiality of information was assured. Subjects were comprised of two different study populations for studies of MAO inhibitor drugs30 in which each volunteer had a baseline PET scan and provided a blood sample for DNA analysis. The baseline PET scan and DNA analysis for each subject was used for the present study. Inclusion criteria allowed for healthy males able to understand and give informed consent, age 18–64 y, with Body Mass Index (BMI) ≥ 21 and ≤ 30. Health status was confirmed by on site physician on the day of PET scan. We excluded smokers, since cigarette smoke inhibits MAOA31 as well as subjects with a prior history of substance abuse or with positive urine drug screens, prior or current treatment with psychotropic medications, psychiatric co-morbidities, neurological disease, medical conditions that may alter cerebral function (e.g., cardiovascular, endocrinological, oncological or autoimmune diseases) or head trauma with loss of consciousness (greater than 30 min). All subjects had a clinical blood test and a urine drug screen (Biosite) on the day of PET scan. Age of participants ranged from 20 to 49 y (average: 30 ± 7.3 y). Ethnicity was self-identified. The demographic characteristics of the study sample are shown in Table 1.
Genomic DNA extraction and MAOA genotyping
Genomic DNA was isolated from the venous blood using PAXgene kit (Qiagen) according to the manufacturer’s instructions. The uVNTR MAOA genotyping was performed by PCR analysis as described elsewhere;3 amplified PCR products were resolved using QIAxel capillary electrophoresis and individual genotypes were assigned based on the amplicons sizes. All Individuals (n = 34) were partitioned onto the “high”(n = 18) and the “low”(n = 16) groups, as previously suggested.7
Sequencing of the MAOA extended promoter in genomic samples
The region encompassing extended MAOA promoter (chrX: 43,398,598–43,400,807) was PCR amplified using the following primers: forward, 5′-AATTGCCTGGTCTCCCCCAA-3′ and reverse, 5′-ATTGTGTCTGCCTCCCCCAG-3′. PCR reaction was set up in 25 μl volume with final concentrations of 1× Platinum Taq PCR Buffer (Invitrogen), 1.5 mM MgSO4, 0.2 mM each of dATP, dTTP, dCTP and 7-deaza dGTP, 0.64 μM of each primer, 1 unit of Platinum Taq polymerase (Invitrogen) and 1.6 μl (~100−200 ng) of DNA template. Amplification was performed with Touchdown PCR protocol with the following conditions: initial denaturation at 95°C for 2 min, 15 cycles of Touchdown at 95°C for 30 sec, 63°C (decreasing 1°C each cycle) for 45 sec, 72°C for 2:30, followed by 20 cycles at 95°C for 30 sec, 48°C for 45 sec, 72°C for 2:30 and a final extension at 72°C for 7 min. PCR products were gel purified using a column purification kit (Zymo Reaseach) and eluted in 20 μl TE buffer. Sequencing of the amplicons was performed using an ABI 3130xl (Applied Biosystems). After the initial editing with the Sequencher program (Gene Codes), individual sequences were aligned against the reference genomic sequence (hg18) using CLUSTALW program (www.ebi.ac.uk/Tools/msa/clustalw2).
Bisulfite cloning and sequencing
Aliquots of 500 ng of genomic DNA were treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen). Bisulfite-treated DNA was PCR amplified using the following primers: forward, 5′-GGGGAGTTGATAGAAGGGTTTTTTTTAT-3′ and reverse, 5′-TATATCTACCTCCCCCAATCACACC-3′. In a set of optimizing experiments, we established a PCR protocol with enhanced amplification efficiency for methylated DNA by increasing the annealing temperature for PCR to overcome potential PCR bias in quantitative methylation analysis47. Amplification was performed using touchdown protocol: initial denaturation at 95°C for 2 min, 15 cycles of Touchdown at 95°C for 30 sec, 63°C (decreasing 1°C each cycle) for 45 sec, 72°C for 2 min 30 sec, followed by 20 cycles at 95°C for 30 sec, 48°C for 45 sec, 72°C for 2 min 30 sec, with a final extension at 72°C for 30 sec. PCR products were gel purified using a column purification kit (ZymoResearch) and ligated into the pCR2.1-TOPO vector (Invitrogen, Life technologies) as suggested by the manufacturer’s protocol.
Reaction products were electroporated into Top 10 cells using BioRad Gene Pulser. After addition of SOC Media, each sample was transferred to a microtube and allowed to express for 1 h and 37°C. A 50 μl aliquot of transformance mix was plated on 2 × YT agar supplemented with 50 μg kanamycin and agar plates were grown overnight at 37°C. Individual colonies were isolated from the growing plates and transferred into the Deep Well 96 well plate to grow as 0.75 ml cultures in 2 × YT media supplemented with 50 μg of kanamycin.
All cultures were screened using a Boil PCR method (see above). From the samples containing an insert plasmids were isolated using alkaline lysis followed by column purification (ZymoResearch). Fifteen to 40 clones (average: 26.5 ± 4.1) per individual sample were prepared for the sequencing to ensure a representative view on the methylation status of the MAOA promoter. Clones were sequenced using sequencing primers flanking the cloning site: M13 forward, (-20) 5′-GTAAAACGACGGCCAG-3′ and M13 reverse, 5′-CAGGAAACAGCTATGAC-3′.
Sequencing was performed using an ABI 3130xl (Applied Biosystems). Initial sequence editing was done with the Sequencher software program (Gene Codes), followed by analysis with BISMA. On average, 371.1 CpGs were analyzed over all clonal sequences obtained from each input DNA.
Quality control
To ensure the accuracy and reliability of our evaluation of the MAOA methylation levels, we used stringent quality control to select the sub-clonal sequences for the analysis. The quality control tool is integrated into the BISMA sequence processing; it was designed to resolve cases of deletion/insertions of thymine by PCR artifacts. The quality control assessment includes the following parameters: The degree of identity to the reference sequence, the conversion rate, the appearance of insertions and deletions and the number of unresolved nucleotides. Only sequences which pass the default parameters of the quality control were considered in downstream statistical tests.
Methylation data analysis and visualization
BISMA analyzer software (biochem.jacobs-university.de/BDPC/BISMA) was used to calculate average methylation values and to derive methylation patterns from the sequencing results. The BDPC (Bisulfite sequencing Data Presentation and Compilation) program was used to present the methylation pattern of the individual amplicons, and to prepare the methylation maps and hit plots.
Visualization of the genomic and epigenetic data.
USCS Genome Browser (http://genome.ucsc.edu/index.html?org=Human&db=hg18&hgsid=246324863) was used to visualize the genomic region under analysis. Methylation states of the single CpG sites of the amplicon are shown as a Custom Tack in the Genome Browser display.
[11C]Clorgyline PET scans
All subjects had PET scans with [11C]clorgyline using the radiopharmaceutical synthesis procedure and PET scanning and arterial sampling described previously45,48 . Briefly, PET scans were obtained on a whole body, high resolution positron emission tomograph (Siemen’s HR+ 4.5 × 4.5 × 4.8 mm at center of field of view) in 3D dynamic acquisition mode for 60 min. Injected radioactivity averaged 7.1 ± 0.6 mCi/scan; specific activity ~250 mCi/µmol at time of injection). An arterial plasma input function for [11C]clorgyline was measured using the same solid phase extraction procedure reported previously49.
PET image analysis
After emission data were attenuation corrected and reconstructed using filtered back projection, time frames from dynamic images taken from 0 to 60 min were summed and then manually re-sliced along the AC-PC line. Planes were added in groups of two, placing the thalamus at plane 12, to obtain 23 planes for region of interest (ROI) placement using an atlas for reference. The regions were then projected to the dynamic scans to obtain time-activity data. ROIs for the following brain areas were obtained: frontal cortex, parietal cortex, occipital cortex, visual cortex, temporal cortex, insula, orbitofrontal cortex, anterior cingulate gyrus, rectal gyrus, thalamus, caudate nucleus and putamen. ROIs were identified in at least two contiguous slices and the weighted average was obtained. For regions occurring bilaterally, the right and left regions were averaged.
Data analysis and estimation of brain MAOA activity
For quantification of MAO-A binding, PET time-activity data for [11C]clorgyline from different brain regions and time-activity data in arterial plasma were used to calculate the model term K1, the plasma to brain transfer constant, which is related to blood flow and λk3 (a function of the concentration of catalytically active MAO-A molecules).31 The model term k3 is related to the binding or trapping of [11C]clorgyline by MAO-A; is defined as K1/k2 and is independent of blood flow50 and k2 is related to the efflux of tracer from brain to blood. For each subject, a λk3 value for each of the 21 ROIs was determined and a composite global λk3 value was determined from an average of the individual λk3 values. Individual λk3 values along with average methylation values sample-wide are shown in Table S1.
PET data processing for parametric images
The parametric images in Figure 1 were generated by applying an irreversible 2-tissue compartment model with model parameters K1 (blood to tissue), k2 (tissue to blood) and k3 (contains the free enzyme concentration) to each voxel. To reduce the number of parameters to be estimated, a clustering algorithm which groups voxels with similar kinetics was applied prior to the voxel analysis. In estimating the model parameters for each voxel, the ratio of K1/k2 (λ) was fixed at the cluster value so that only two parameters were estimated. The results are reported as λk3. This combination of parameters has been found to be a more stable estimate of the MAOA level than the single parameter k3 similar to the use of the distribution volume VT in reversibly binding tracers rather than k3. Individual λk3 values along with average methylation values sample-wide are shown in Table S1.
Statistical analysis
Demographic and imaging data were analyzed using the Statistical Package for Social Sciences (SPSS) version 15 (www.spss.com). Specific analytical techniques were chosen based on the outcome measurement (binary/categorical vs. continuous) and normality of the data (skewness and kurtosis). The Kolmogorov-Smirnov test was used to test whether all continuous variables followed normal distribution. The Student’s t-test for independent samples was used to compare the continuous variables. ANOVA was used to explore group differences (sample partitioning onto genotype- and ethnic subgroups) in demographics and brain measures and to make a statistical inference on the difference of the imaging- and methylation measures data sets between different factors.
The relationship between the DNA methylation levels in and respective brain MAOA activity was visualized with scatter plots. A linear regression was calculated to infer the dependence of λk3 value on average methylation in the amplicon and percentage of methylation at a single CpG site. The goodness of fit measure (R2) was used to quantify this relation. The p value was calculated by ANOVA to determine the significance of R2. For the normally distributed data we used Pearson’s correlation coefficient and for the data which distribution deviates from normality we used Spearman’s correlation coefficient to assess validity of the test statistics.
Multiple regressions (hierarchical and step-wise) were used to test whether the inclusion of other independent variables (age, ethnicity and MAOA genotype) along with MAOA methylation values will improve the accuracy in predicting values for the independent variable (brain MAOA activity).
A principal component analysis (PCA) on the 14 items (single CpG sites methylation values) was conducted with orthogonal rotation (Varimax). The Kaiser-Mayer-Olkin measure verifying the sampling adequacy for the analysis, KMO = 0.58 (acceptance level is 0.5). Barlett’s test for sphericity χ2(91) = 209.8, p < 0.001, indicated that correlation between the items were sufficiently large for PCA. An initial analysis was run to obtain eigenvalues for each component of the data. Four components had eigenvalues over Kaiser’s criterion of 1 and, in combination, explained 70% of the variance. The screen plot was slightly ambiguous and showed inflexions that would justify four components. Four components were retained in the final analysis. Table S2 shows the factor loading after rotation.
Supplementary Material
Acknowledgments
This study was performed at Brookhaven National Laboratory under contract DE-AC02–98CH10886 with infrastructure support from its Office of Biological and Environmental Research. Support for the MAOA studies came in part from CeNeRx BioPharma and Valeant Pharmaceuticals International and in part by the National Institutes of Health: NIDA awards KO1 DA02580 for ES and K05DA020001 for JSF; the NIAAA intramural program at Brookhaven National Laboratory (NDV) and by NIH grant MO1RR10710 from the General Research Clinical Centers (Stony Brook University). We are grateful to Dr. Beth Milligan, Alice Shanklin and Alexander Scalia for technical assistance in conducting experimental tests, Joan Terry and Hai-Dee Lee for CRC operations, to Donald Warner, Michael Schueller and David Schlyer for PET and cyclotron operations, Payton King and David Alexoff for plasma analysis and to Colleen Shea, Lisa Muench and Youwen Xu for radiotracer synthesis. We are also grateful to the individuals who volunteered for these studies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/21976
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/21976
References
- 1.Craig IW. The importance of stress and genetic variation in human aggression. Bioessays. 2007;29:227–36. doi: 10.1002/bies.20538. [DOI] [PubMed] [Google Scholar]
- 2.Bortolato M, Chen K, Shih JC. Monoamine oxidase inactivation: from pathophysiology to therapeutics. Adv Drug Deliv Rev. 2008;60:1527–33. doi: 10.1016/j.addr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabol SZ, Hu S, Hamer D. A functional polymorphism in the monoamine oxidase A gene promoter. Hum Genet. 1998;103:273–9. doi: 10.1007/s004390050816. [DOI] [PubMed] [Google Scholar]
- 4.Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–4. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 5.Meyer-Lindenberg A, Buckholtz JW, Kolachana B, R Hariri A, Pezawas L, Blasi G, et al. Neural mechanisms of genetic risk for impulsivity and violence in humans. Proc Natl Acad Sci U S A. 2006;103:6269–74. doi: 10.1073/pnas.0511311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutiérrez B, Arias B, Gastó C, Catalán R, Papiol S, Pintor L, et al. Association analysis between a functional polymorphism in the monoamine oxidase A gene promoter and severe mood disorders. Psychiatr Genet. 2004;14:203–8. doi: 10.1097/00041444-200412000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Denney RM, Koch H, Craig IW. Association between monoamine oxidase A activity in human male skin fibroblasts and genotype of the MAOA promoter-associated variable number tandem repeat. Hum Genet. 1999;105:542–51. doi: 10.1007/s004390051143. [DOI] [PubMed] [Google Scholar]
- 8.Fowler JS, Alia-Klein N, Kriplani A, Logan J, Williams B, Zhu W, et al. Evidence that brain MAOA activity does not correspond to MAOA genotype in healthy male subjects. Biol Psychiatry. 2007;62:355–8. doi: 10.1016/j.biopsych.2006.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, Craig IW, et al. MAOA, maltreatment, and gene-environment interaction predicting children’s mental health: new evidence and a meta-analysis. Mol Psychiatry. 2006;11:903–13. doi: 10.1038/sj.mp.4001851. [DOI] [PubMed] [Google Scholar]
- 10.Bird A, Macleod D. Reading the DNA methylation signal. Cold Spring Harb Symp Quant Biol. 2004;69:113–8. doi: 10.1101/sqb.2004.69.113. [DOI] [PubMed] [Google Scholar]
- 11.Feng YQ, Desprat R, Fu H, Olivier E, Lin CM, Lobell A, et al. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2006;2:e65. doi: 10.1371/journal.pgen.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–7. doi: 10.1016/S0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 13.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–6. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 14.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versteeg R. Aberrant methylation in cancer. Am J Hum Genet. 1997;60:751–4. [PMC free article] [PubMed] [Google Scholar]
- 16.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–5. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 18.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, et al. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5:516–26. doi: 10.4161/epi.5.6.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–9. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth TL, Sweatt JD. Annual Research Review: Epigenetic mechanisms and environmental shaping of the brain during sensitive periods of development. J Child Psychol Psychiatry. 2011;52:398–408. doi: 10.1111/j.1469-7610.2010.02282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–69. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 23.McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farcas R, Schneider E, Frauenknecht K, Kondova I, Bontrop R, Bohl J, et al. Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol Biol Evol. 2009;26:1379–89. doi: 10.1093/molbev/msp046. [DOI] [PubMed] [Google Scholar]
- 25.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:60–6. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 26.Kuratomi G, Iwamoto K, Bundo M, Kusumi I, Kato N, Iwata N, et al. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry. 2008;13:429–41. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- 27.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, et al. Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J Neurosci. 2011;31:6692–8. doi: 10.1523/JNEUROSCI.6631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fowler JS, Volkow ND, Logan J, Franceschi D, Wang GJ, MacGregor R, et al. Evidence that L-deprenyl treatment for one week does not inhibit MAOA or the dopamine transporter in the human brain. Life Sci. 2001;68:2759–68. doi: 10.1016/S0024-3205(01)01079-7. [DOI] [PubMed] [Google Scholar]
- 30.Fowler JS, Logan J, Azzaro AJ, Fielding RM, Zhu W, Poshusta AK, et al. Reversible inhibitors of monoamine oxidase-A (RIMAs): robust, reversible inhibition of human brain MAO-A by CX157. Neuropsychopharmacology. 2010;35:623–31. doi: 10.1038/npp.2009.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fowler JS, Volkow ND, Wang GJ, Pappas N, Logan J, Shea C, et al. Brain monoamine oxidase A inhibition in cigarette smokers. Proc Natl Acad Sci U S A. 1996;93:14065–9. doi: 10.1073/pnas.93.24.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg S, Templeton AR, Feigin PD, Lancet D, Beckmann JS, Selig S, et al. The association of DNA sequence variation at the MAOA genetic locus with quantitative behavioural traits in normal males. Hum Genet. 2006;120:447–59. doi: 10.1007/s00439-006-0198-x. [DOI] [PubMed] [Google Scholar]
- 33.Shumay E, Fowler JS. Identification and characterization of putative methylation targets in the MAOA locus using bioinformatic approaches. Epigenetics. 2010;5:325–42. doi: 10.4161/epi.5.4.11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock C, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet. 2006;2:e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–7. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 37.Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–64. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- 38.Patterson K, Molloy L, Qu W, Clark S. DNA methylation: bisulphite modification and analysis. J Vis Exp. 2011;56 doi: 10.3791/3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh CL. Dynamics of DNA methylation pattern. Curr Opin Genet Dev. 2000;10:224–8. doi: 10.1016/S0959-437X(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 40.Moffitt TE, Harrington H, Caspi A, Kim-Cohen J, Goldberg D, Gregory AM, et al. Depression and generalized anxiety disorder: cumulative and sequential comorbidity in a birth cohort followed prospectively to age 32 years. Arch Gen Psychiatry. 2007;64:651–60. doi: 10.1001/archpsyc.64.6.651. [DOI] [PubMed] [Google Scholar]
- 41.Fowler JS, Logan J, Volkow ND, Wang GJ. Translational neuroimaging: positron emission tomography studies of monoamine oxidase. Mol Imaging Biol. 2005;7:377–87. doi: 10.1007/s11307-005-0016-1. [DOI] [PubMed] [Google Scholar]
- 42.Meyer JH, Wilson AA, Sagrati S, Miler L, Rusjan P, Bloomfield PM, et al. Brain monoamine oxidase A binding in major depressive disorder: relationship to selective serotonin reuptake inhibitor treatment, recovery, and recurrence. Arch Gen Psychiatry. 2009;66:1304–12. doi: 10.1001/archgenpsychiatry.2009.156. [DOI] [PubMed] [Google Scholar]
- 43.Sacher J, Wilson AA, Houle S, Rusjan P, Hassan S, Bloomfield PM, et al. Elevated brain monoamine oxidase A binding in the early postpartum period. Arch Gen Psychiatry. 2010;67:468–74. doi: 10.1001/archgenpsychiatry.2010.32. [DOI] [PubMed] [Google Scholar]
- 44.Meyer JH, Ginovart N, Boovariwala A, Sagrati S, Hussey D, Garcia A, et al. Elevated monoamine oxidase a levels in the brain: an explanation for the monoamine imbalance of major depression. Arch Gen Psychiatry. 2006;63:1209–16. doi: 10.1001/archpsyc.63.11.1209. [DOI] [PubMed] [Google Scholar]
- 45.Alia-Klein N, Goldstein RZ, Kriplani A, Logan J, Tomasi D, Williams B, et al. Brain monoamine oxidase A activity predicts trait aggression. J Neurosci. 2008;28:5099–104. doi: 10.1523/JNEUROSCI.0925-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–64. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen L, Guo Y, Chen X, Ahmed S, Issa JP. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques. 2007;42:48–50, 50, 52 passim. doi: 10.2144/000112312. [DOI] [PubMed] [Google Scholar]
- 48.MacGregor RR, Halldin C, Fowler JS, Wolf AP, Arnett CD, Langström B, et al. Selective, irreversible in vivo binding of [11C]clorgyline and [11C]-L-deprenyl in mice: potential for measurement of functional monoamine oxidase activity in brain using positron emission tomography. Biochem Pharmacol. 1985;34:3207–10. doi: 10.1016/0006-2952(85)90173-X. [DOI] [PubMed] [Google Scholar]
- 49.Alexoff DL, Shea C, Fowler JS, King P, Gatley SJ, Schlyer DJ, et al. Plasma input function determination for PET using a commercial laboratory robot. Nucl Med Biol. 1995;22:893–904. doi: 10.1016/0969-8051(95)00042-V. [DOI] [PubMed] [Google Scholar]
- 50.Logan J, Dewey SL, Wolf AP, Fowler JS, Brodie JD, Angrist B, et al. Effects of endogenous dopamine on measures of [18F]N-methylspiroperidol binding in the basal ganglia: comparison of simulations and experimental results from PET studies in baboons. Synapse. 1991;9:195–207. doi: 10.1002/syn.890090306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.