Anillin-like protein Mid1p coordinates contractile ring assembly in fission yeast by restricting precursors in nodes around the equator. Without Mid1p, contractile ring assembly is slow and unreliable because ring precursors are separated in nodes (Blt1p, Cdc12p) or strands (myosin-II, Rng2p, Cdc15p, actin filaments) scattered widely in the cortex.
Abstract
In fission yeast cells cortical nodes containing the protein Blt1p and several kinases appear early in G2, mature into cytokinetic nodes by adding anillin Mid1p, myosin-II, formin Cdc12p, and other proteins, and condense into a contractile ring by movements that depend on actin and myosin-II. Previous studies concluded that cells without Mid1p lack cytokinetic nodes and assemble rings unreliably from myosin-II strands but left open questions. Why do strands form outside the equatorial region? Why is ring assembly unreliable without Mid1p? We found in Δmid1 cells that Cdc12p accumulates in cytokinetic nodes scattered in the cortex and produces actin filaments that associate with myosin-II, Rng2p, and Cdc15p to form strands located between the nodes. Strands incorporate nodes, and in ∼67% of cells, strands slowly close into rings that constrict without the normal ∼25-min maturation period. Ring assembly is unreliable and slow without Mid1p because the scattered Cdc12p nodes generate strands spread widely beyond the equator, and growing strands depend on random encounters to merge with other strands into a ring. We conclude that orderly assembly of the contractile ring in wild-type cells depends on Mid1p to recruit myosin-II, Rng2p, and Cdc15p to nodes and to place cytokinetic nodes around the cell equator.
INTRODUCTION
The fission yeast, Schizosaccharomyces pombe, like many other fungi and animals, undergoes cytokinesis by assembling and constricting a contractile ring of actin filaments and myosin-II (Pollard and Wu, 2010). The product of the fission yeast mid1+ gene (Chang et al., 1996; Paoletti and Chang, 2000; Sohrmann et al., 1996; Celton-Morizur et al., 2004) is similar to animal anillins (Field and Alberts, 1995) and helps to position the contractile ring. During interphase, most Mid1p associates with the nucleus, but some Mid1p localizes in punctate cortical structures called interphase nodes containing protein kinases Cdr1p, Cdr2p, and Wee1p, the putative RhoGEF Gef2p, and the uncharacterized protein Blt1p (Morrell et al., 2004; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). As cells enter mitosis, the interphase nodes mature into cytokinetic nodes by accumulating cytokinesis proteins at specific times, starting with Mid1p, IQGAP protein Rng2p, conventional myosin-II (hereafter called Myo2, consisting of heavy chain Myo2p and light chains Cdc4p and Rlc1p), F-BAR protein Cdc15p and finally formin Cdc12p (Wu et al., 2003, 2006). Mid1p helps to recruit these proteins to cytokinetic nodes through two pathways linked to Rng2p and Myo2, and to Cdc15p. Both pathways help recruit Cdc12p (Almonacid et al., 2011; Laporte et al., 2011; Padmanabhan et al., 2011). Actin filaments generated by formin Cdc12p (Kovar et al., 2003) interact with Myo2, as cytokinetic nodes condense into a continuous ring around the equator (Vavylonis et al., 2008).
Fission yeast cells lacking Mid1p are viable, but cytokinesis is unreliable, with most contractile rings off-center and/or oriented obliquely (Sohrmann et al., 1996; Paoletti and Chang, 2000). Contractile ring assembly in cells lacking Mid1p depends on a signaling pathway called the septation initiation network (SIN), which normally contributes to compaction of the contractile ring and later to constriction of the ring (Hachet and Simanis, 2008). Researchers generally view this SIN-dependent pathway in cells lacking Mid1p as mechanistically distinct from the normal pathway and often state that Mid1p is required to assemble both nodes and contractile rings (Chang et al., 1996; Paoletti and Chang, 2000; Sohrmann et al., 1996; Wu et al., 2003, 2006; Celton-Morizur et al., 2004; Motegi et al., 2004; Clifford et al., 2008; Hachet and Simanis, 2008; Huang et al., 2008; Almonacid et al., 2011), but remarkably little is known about how Mid1p contributes to the high fidelity of contractile ring assembly and cytokinesis. Previous studies (Chang et al., 1996; Sohrmann et al., 1996; Motegi et al., 2004; Hachet and Simanis, 2008; Huang et al., 2008) on cells lacking Mid1p or expressing Mid1p mutants reported that contractile rings assemble from strands of myosin-II and actin filaments but left open questions about the contribution of nodes to ring assembly, why strands often form outside the equatorial region in these cells, how strands accumulate other precursors of the contractile ring, and why the process of ring assembly is unreliable in the absence of Mid1p.
To address these important questions, we tracked in live Δmid1 cells the contractile ring proteins Blt1p, Myo2, Rng2p, Cdc15p, and Cdc12p as they appeared in the cortex and cells attempted to assemble a contractile ring. Previous studies on cells lacking Mid1p did not explain how Blt1p in interphase nodes is incorporated into contractile rings (Moseley et al., 2009) in the absence of cytokinetic nodes (Huang et al., 2008), and the localization of Rng2p, Cdc15p, and Cdc12p in mitosis was unclear. Our detailed analysis focusing on the critical steps of structural assembly during cytokinesis revealed that two defects suffice to explain the slow and unreliable assembly of the contractile ring in the absence of Mid1p: ring precursors are separated in either nodes or strands, and these precursors spread out more broadly from the equator than in wild-type cells. Our results establish that Mid1p is not required for accumulation of formin Cdc12p in cytokinetic nodes, but Mid1p is essential for restriction of cytokinetic nodes to a narrow zone around the equator in wild-type cells, and that this role of Mid1p is essential for the high fidelity of cytokinesis.
RESULTS
Normal pathway of contractile ring assembly
We reevaluated cytokinesis in wild-type fission yeast cells to better understand how cells use precursors formed during interphase for cytokinesis and to appreciate the phenotypes of cells lacking anillin Mid1p (this article) or depending on deletion constructs of Mid1p (see companion article, Saha and Pollard, 2012). Cortical nodes containing Cdr2p, Cdr1p, Wee1p, and Blt1p regulate entry into mitosis and accumulate Mid1p, the earliest marker of cytokinetic nodes (Wu et al., 2003; Morrell et al., 2004; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). We examined the composition of these nodes as cells entered mitosis and studied the behavior of these nodes during contractile ring assembly.
Localization of interphase nodes.
We tracked cortical nodes during interphase with fluorescent tags on Cdr2p and Blt1p using cell length to approximate time (Figures 1, A and B, and 2A). Wild-type cells grew in length from 7 to 9 μm at birth to 13 to 16 μm before cytokinesis. Nodes containing Cdr2p concentrated in a broad band around the equator as cells grew from 7 to 13 μm long (Figure 1A, star), whereas nodes containing Blt1p initially clustered close to the new pole and later dispersed in the cortex (Figure 1B, arrowhead) before clustering in a broad equatorial band well after Cdr2p nodes (Figures 1B, star, and 2A). Cdr2p organizes other node proteins during interphase (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009), but this kinase disappeared from the equator before cytokinesis in nonseptate cells >13 μm long (Figure 2A).
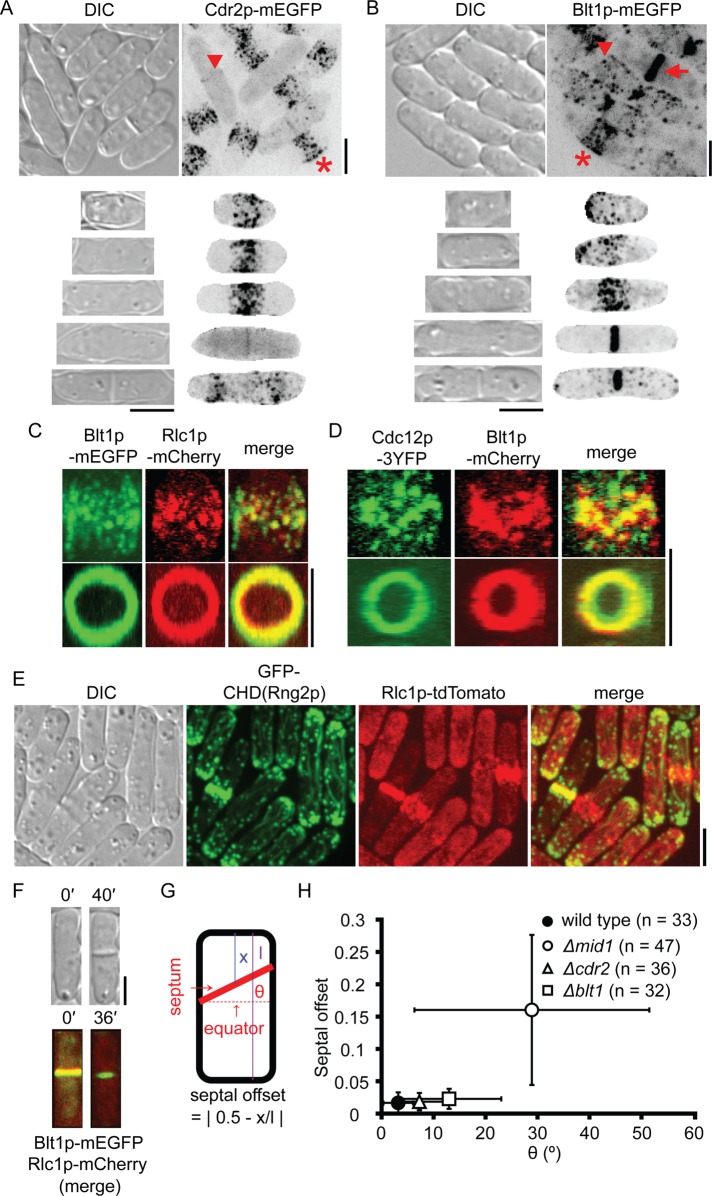
FIGURE 1:
Contractile ring assembly in wild-type cells. (A, B) Differential interference contrast (DIC) and maximum-intensity projections of negative-contrast fluorescence micrographs. Top, fields of unsynchronized cells. Rows 2–6 show individual cells of different lengths representing successive stages of the cell cycle. Regions outside the cells are masked in the fluorescence micrographs in rows 2–6. (A) Cdr2p-mEGFP concentrates in nodes that form a broad band in short cells (star) and a faint band in the middle of some long cells (arrowhead). (B) Nodes marked with Blt1p-mEGFP cluster close to the new pole just after division and then disperse in the cortex (arrowhead) and form a broad equatorial band in mitosis (star) that subsequently coalesces into contractile ring (arrow). (C, D) Two-dimensional projections of fluorescence micrographs of parts of different wild-type cells showing (C) Blt1p-mEGFP and Rlc1p-mCherry and (D) Cdc12p-3YFP and Blt1p-mCherry colocalizing in nodes (top row) and contractile ring (bottom row). (E) DIC and two-dimensional projections of fluorescence micrographs showing cells with Rlc1p-tdTomato expressed from the native locus and GFP–calponin-homology domain (CHD) of Rng2p expressed from a 41xnmt1 promoter for 24 h. (F) DIC and maximum-intensity projections of fluorescence micrographs of a cell expressing Blt1p-mEGFP and Rlc1p-mCherry. Images taken 40 min apart show constriction of the contractile ring and formation of a septum. (G) Cartoon of S. pombe defining septal offset and septum tilt angle θ. (H) Plot of septal offset vs. θ for cylindrical or near-cylindrical cells containing single septum. The larger value of θ was scored for curved septa intersecting two sides of a cell at different angles: (●, n = 33) wild-type cells, (◯, n = 47) Δmid1 cells, (∆, n = 36) Δcdr2 cells, and (□, n = 32) Δblt1 cells. Error bars, 1 SD. Scale bars, 5 μm. See also Supplemental Figure S1 and Supplemental Videos S1–S4.
FIGURE 2:
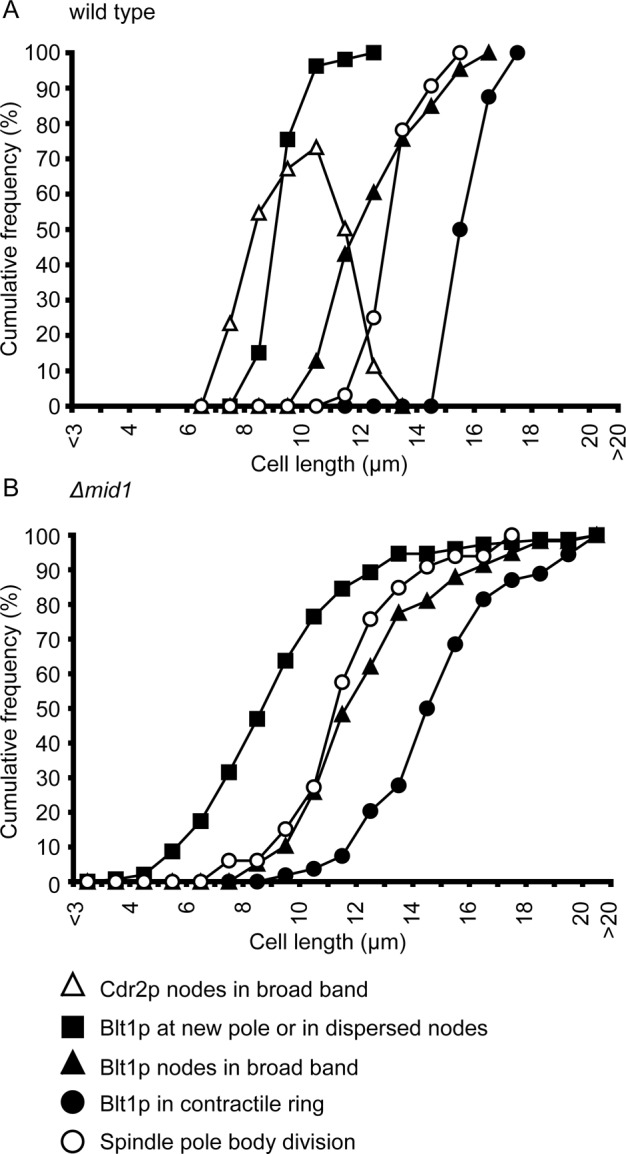
Progression of interphase node events in wild-type and Δmid1 cells. Events were measured as cumulative frequencies in cell-length bins of 1 μm. (A) Wild-type cells: (∆, n = 64) Cdr2p-mEGFP in a broad band of nodes; (■, n = 53) Blt1p-mEGFP near the new pole or in dispersed nodes; (▲, n = 86) Blt1p-mEGFP in a broad band of equatorial nodes; (●, n = 8) Blt1p-mEGFP in contractile rings; and (◯, n = 32) divided spindle pole bodies. The Cdr2p distribution was calculated by subtracting the percentage cumulative frequency of nonseptate cells lacking Cdr2p in nodes from the percentage cumulative frequency of nonseptate cells containing Cdr2p in nodes. (B) Δmid1 cells: (■, n = 149) Blt1p-mEGFP near the new pole or in dispersed nodes; (▲, n = 58) Blt1p-mEGFP in a broad band of nodes; (●, n = 54) Blt1p-mEGFP in strands or contractile rings; and (◯, n = 33) divided spindle pole bodies.
Maturation of interphase nodes into cytokinetic nodes.
Interphase nodes containing Blt1p matured by accumulating Mid1p, followed by other precursors of the contractile ring. During ∼10–20 min before spindle pole body (SPB) division at length 13.6 ± 0.9 μm (Figure 2A), Myo2 marked with regulatory light chain Rlc1p moved from a diffuse cytoplasmic pool to concentrate in equatorial nodes (Wu et al., 2003; Laporte et al., 2011) that also contained Blt1p (Figures 1C and 3A). During the 10 min after SPB division, formin Cdc12p relocated from a diffuse cytoplasmic pool and tiny cytoplasmic speckles (Coffman et al., 2009) to nodes containing Myo2 (Supplemental Video S1; Wu et al., 2003; Laporte et al., 2011) and Blt1p (Figures 1D and 3A).
FIGURE 3:
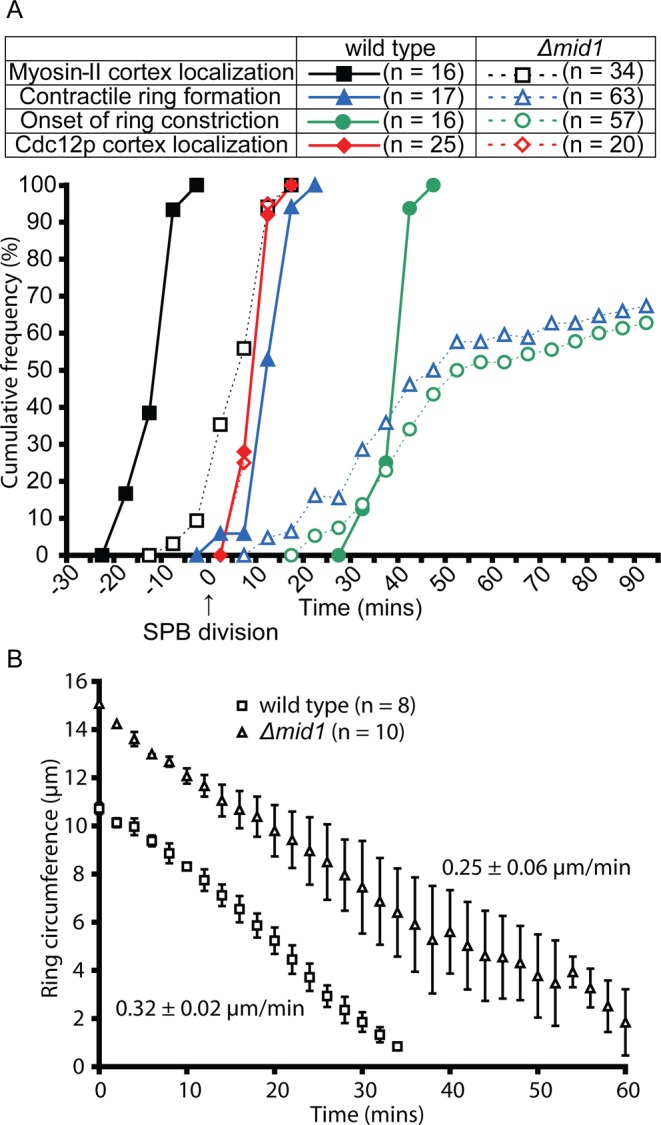
Comparison of cytokinetic parameters in the presence and absence of Mid1p. (A) Time courses of recruitment of myosin-II and formin Cdc12p to the cortex and formation and constriction of the contractile ring relative to the time of spindle pole body division tracked by Sad1p-mRFP1 or Sad1p-mEGFP defined as time zero. Appearance of Rlc1p-3GFP in the cortex in (■, n = 16) wild-type and Rlc1p-mEGFP in the cortex in (□, n = 34) Δmid1 cells, appearance of Cdc12p-3GFP in the cortex in ( , n = 25) wild-type and (
, n = 25) wild-type and ( , n = 20) Δmid1 cells, formation of contractile ring (tracked by Rlc1p-3GFP or Rlc1p-mEGFP) in (
, n = 20) Δmid1 cells, formation of contractile ring (tracked by Rlc1p-3GFP or Rlc1p-mEGFP) in ( , n = 17) wild-type and (
, n = 17) wild-type and ( , n = 63) Δmid1 cells, and onset of contractile ring constriction (tracked by Rlc1p-3GFP or Rlc1p-mEGFP) in (
, n = 63) Δmid1 cells, and onset of contractile ring constriction (tracked by Rlc1p-3GFP or Rlc1p-mEGFP) in ( , n = 16) wild-type and (
, n = 16) wild-type and ( , n = 57) Δmid1 cells. Bin width, 5 min. The sample included only cells in which Rlc1p and Cdc12p persisted in the cortex long enough to document formation of strands or rings. (B) Time course of ring constriction. Plots of contractile ring circumferences estimated in maximum-intensity projections vs. time for (□, n = 8) wild-type and (∆, n = 10) Δmid1 cells. Rates of circumferential constriction were estimated using linear regression separately in all cells and averaged. Error bars, 1 SD.
, n = 57) Δmid1 cells. Bin width, 5 min. The sample included only cells in which Rlc1p and Cdc12p persisted in the cortex long enough to document formation of strands or rings. (B) Time course of ring constriction. Plots of contractile ring circumferences estimated in maximum-intensity projections vs. time for (□, n = 8) wild-type and (∆, n = 10) Δmid1 cells. Rates of circumferential constriction were estimated using linear regression separately in all cells and averaged. Error bars, 1 SD.
During mitosis most cytokinetic nodes with Blt1p were located close to the equator, with only a small fraction persisting close to the poles. Defining the middle of the cell as position 0 and the ends as ±0.5, we found that observations with three markers all gave the mean positions ± SD of cytokinetic nodes close to 0: 0.05 ± 0.15 for nodes marked with Blt1p; 0.02 ± 0.07 for Rlc1p; and 0.01 ± 0.03 for Cdc12p (Figure 4).
FIGURE 4:
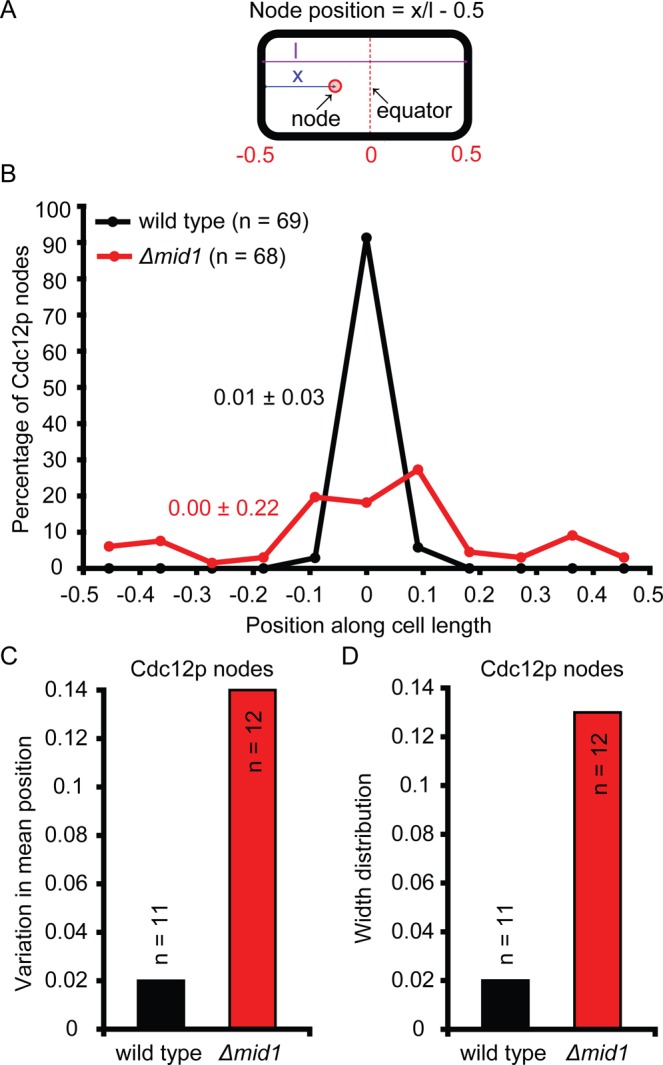
Positions of cytokinetic nodes in wild-type and Δmid1 cells. (A) Cartoon of S. pombe defining node position. (B) Histogram showing distributions of nodes marked with Cdc12p-3GFP in mitosis, scored in maximum-intensity projection fluorescence micrographs. Data from multiple cells were pooled together and distributed into bins of width 1/11 of the cell length. The frequency was normalized to the total number of nodes and plotted at the midpoint of each bin. Mean ± SD node positions are color coded to match that of the distribution of different types of nodes. •, wild-type cells (n = 69 from 11 cells);  , Δmid1 cells (n = 68 from 12 cells). (C, D) Comparison of the distribution of Cdc12p node positions among (■) 11 wild-type and (
, Δmid1 cells (n = 68 from 12 cells). (C, D) Comparison of the distribution of Cdc12p node positions among (■) 11 wild-type and ( ) 12 Δmid1 cells from data shown in B. (C) Histogram showing the variation in center of the distribution. In each cell, the mean position of nodes containing Cdc12p was calculated. The SD among cells in these mean positions is plotted. (D) Histogram showing the width of the distribution expressed as the average among cells in the SD of position of nodes containing Cdc12p. In each cell, the SD among positions of nodes containing Cdc12p was calculated. The mean of these SD values across cells is plotted.
) 12 Δmid1 cells from data shown in B. (C) Histogram showing the variation in center of the distribution. In each cell, the mean position of nodes containing Cdc12p was calculated. The SD among cells in these mean positions is plotted. (D) Histogram showing the width of the distribution expressed as the average among cells in the SD of position of nodes containing Cdc12p. In each cell, the SD among positions of nodes containing Cdc12p was calculated. The mean of these SD values across cells is plotted.
Colocalization of pairs of fluorescent fusion proteins showed that cytokinetic nodes were heterogeneous. During contractile ring assembly, movements dependent on actin filaments normally bring nodes close together and make it difficult to resolve signals from adjacent nodes. Therefore we treated cells with latrunculin A for 30 min to stop the movements and accumulate well-separated cytokinetic nodes around the equator of mitotic cells (Supplemental Figure S1, A and B). Two-thirds of these cytokinetic nodes contained both Blt1p and Rlc1p, but we detected no Rlc1p in 19% of Blt1p nodes and no Blt1p in 14% of Rlc1p nodes (Supplemental Figure S1C). Two-thirds of the nodes in these mitotic cells contained both Blt1p and formin Cdc12p, but 16% of Cdc12p-positive nodes lacked a Blt1p signal and 19% of Blt1p-positive nodes lacked a Cdc12p signal (Supplemental Figure S1D). These observations agree with the ∼70% extent of colocalization of other pairs of ring proteins in cytokinetic nodes (Wu et al., 2006; Coffman et al., 2009).
Condensation of nodes into rings.
Like nodes marked with myosin light chain Rlc1p (Vavylonis et al., 2008), nodes marked with Cdc12p or Blt1p moved intermittently as they condensed over ∼15 min into contractile rings (Supplemental Videos S1–S3). Movies of cells expressing pairs of fluorescently tagged proteins (Supplemental Videos S1–S3) confirmed that these condensing nodes were heterogeneous, including nodes with signals from only Blt1p, only Myo2, only Cdc12p, both Blt1p and Myo2, both Blt1p and Cdc12p, or both Myo2 and Cdc12p. Additional speckles of Cdc12p joined the cell equator during ring assembly from nodes (Supplemental Video S1). Latrunculin A stopped contractile ring formation but not the random, rapid movements of cytoplasmic speckles (Supplemental Video S4A), and so movements of Cdc12p nodes but not speckles depend on actin filaments. Completed rings contained Blt1p, Myo2, Cdc12p, and actin filaments (tracked with the calponin-homology domain [CHD] of Rng2p) (Figure 1, C–E) but not Cdr1p-3GFP. Some cells had a trace of Cdr2p-monomeric enhanced green fluorescent protein (mEGFP) around the equator but not in contractile rings (Figure 1A, arrowhead).
Contractile ring constriction and septation.
During contractile ring constriction, cells laid down septa in cleavage furrows (Figure 1F). Septa were oriented with high fidelity perpendicular to the long axis of the cell (mean tilt angle, 3.2°; SD = 2.9°) precisely in the middle of the cell (mean fractional offset, 0.02; SD = 0.02; Figure 1, G and H).
Cytokinesis defects in cells lacking anillin Mid1p
Strains with a deletion mutation of mid1+ are viable but suffer from cytokinesis defects (Sohrmann et al., 1996; Paoletti and Chang, 2000; Motegi et al., 2004; Hachet and Simanis, 2008; Huang et al., 2008). Many cells without Mid1p formed contractile rings containing Blt1p, IQGAP Rng2p, Myo2, F-BAR Cdc15p, formin Cdc12p, and actin filaments, tracked with the CHD of Rng2p (Figures 5, A(e), C, F, and H, and 6A), but, as observed previously (Sohrmann et al., 1996; Paoletti and Chang, 2000), most septa were misplaced from the cell center (mean fractional offset, 0.16; SD = 0.12) and oriented obliquely (mean tilt angle, 28.9º; SD = 22.6º; Figure 1H and Supplemental Video S5). It is striking that the standard deviations of both parameters were at least sixfold larger in Δmid1 cells than in wild-type cells. Due to asymmetric cytokinesis, the lengths of Δmid1 daughter cells (3–15 μm; n = 25) varied much more than did those of wild-type cells (7–9 μm; n = 20), and this heterogeneity persisted, so at the time of SPB division the lengths of Δmid1 cells varied from 8 to >16 μm, whereas wild-type cells were 12–16 μm long (Figure 2, A and B).
FIGURE 5:
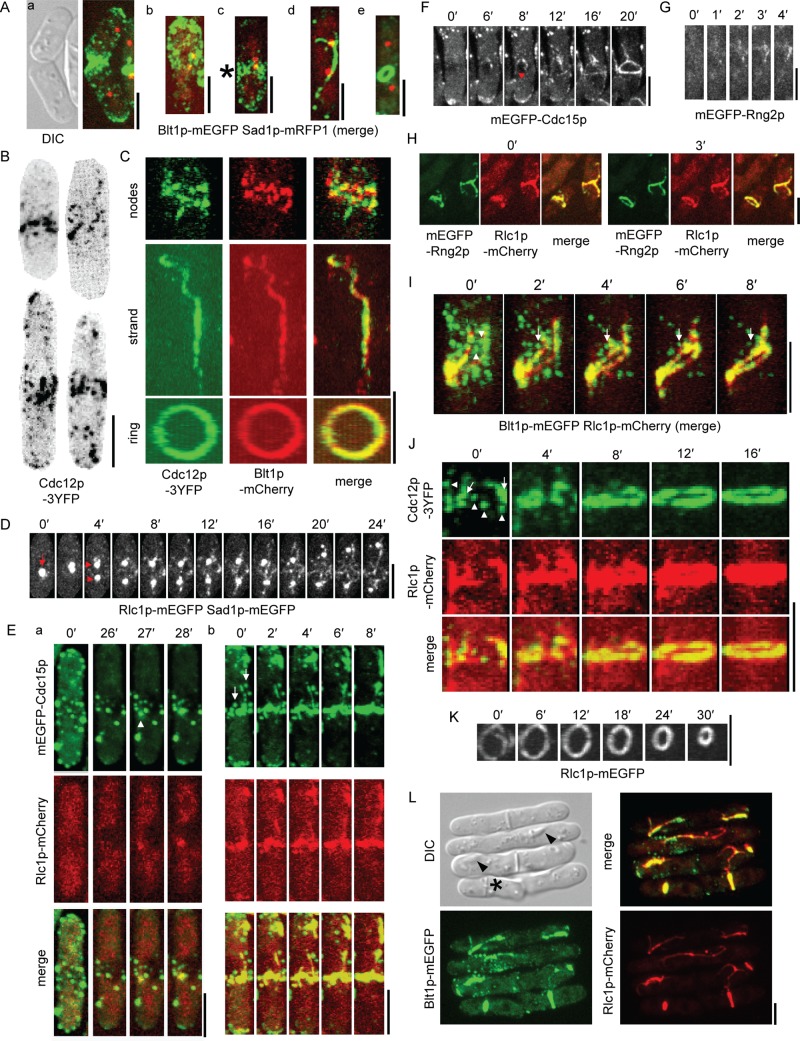
Contractile ring assembly in Δmid1 cells. (A) Localization of Blt1p-mEGFP at different stages of the cell cycle in Δmid1 cells tracked by division of Sad1p-mRFP1–marked spindle pole body. (a) DIC and maximum-intensity projection fluorescence micrographs of two cells in G2. (b–e) Maximum-intensity projection fluorescence micrographs of single cells with their long axes vertical in (b, c) mitosis immediately after spindle pole body division and (d, e) late mitosis. Blt1p-mEGFP localizes to (c, star) nodes in a broad band, (d) strand, and (e) ring. (B) Negative-contrast, maximum-intensity projection fluorescence micrographs of four different Δmid1 cells showing localization of Cdc12p-3YFP in mitosis. Regions outside the cells are masked. (C) Two-dimensional projections of fluorescence micrographs of parts of three different Δmid1 cells showing localization of Cdc12p-3YFP and Blt1p-mCherry in nodes (top), a strand (middle), and contractile ring (bottom). (D–H) Time-lapse maximum-intensity projections of fluorescence micrographs. The long axes of cells are vertical in D–G. (D) Rlc1p-mEGFP appears in strands in the cortex of a Δmid1 cell after one Sad1p-mEGFP–marked spindle pole body (arrow) divides into two (arrowheads). Regions outside the cell are masked. (E) Two different Δmid1 cells showing localization of mEGFP-Cdc15p and Rlc1p-mCherry. mEGFP-Cdc15p colocalizes with aggregates of Rlc1p-mCherry when they appear in the cortex in mitosis (a, arrowhead). During ring assembly, strands incorporate some punctate cortical structures containing mEGFP-Cdc15p (b, arrows). (F) mEGFP-Cdc15p localizes in cortical puncta and mEGFP-Cdc15p appears in strands (arrowhead) in the cortex of a Δmid1 cell. (G) mEGFP-Rng2p appears in the cortex of a Δmid1 cell. (H) mEGFP-Rng2p and Rlc1p-mCherry colocalize in strands and contractile rings in Δmid1 cells. (I–K) Two-dimensional projections of fluorescence micrographs of parts of individual Δmid1 cells. (I, J) Time course of contractile ring assembly from strands of Rlc1p-mCherry and nodes containing (I) Blt1p-mEGFP or (J, arrowheads) Cdc12p-3YFP. The long axes of cells are vertical. Some (I, arrowheads) but not all (I, arrow) nodes containing Blt1p-mEGFP were incorporated into Rlc1p-mCherry strands. Strands of Rlc1p-mCherry have already incorporated some Cdc12p-3YFP (arrows) at 0 min. (K) Time course of constriction of contractile ring marked by Rlc1p-mEGFP. (L) DIC and maximum-intensity projection fluorescence micrographs showing localization of Blt1p-mEGFP and Rlc1p-mCherry in Δmid1 cells and deposition of septal material after ring constriction (star) or along myosin-II strands, resulting in incomplete septum formation (arrowheads). Scale bars, 5 μm. See also Supplemental Figures S1 and S2 and Supplemental Videos S4–S11.
FIGURE 6:
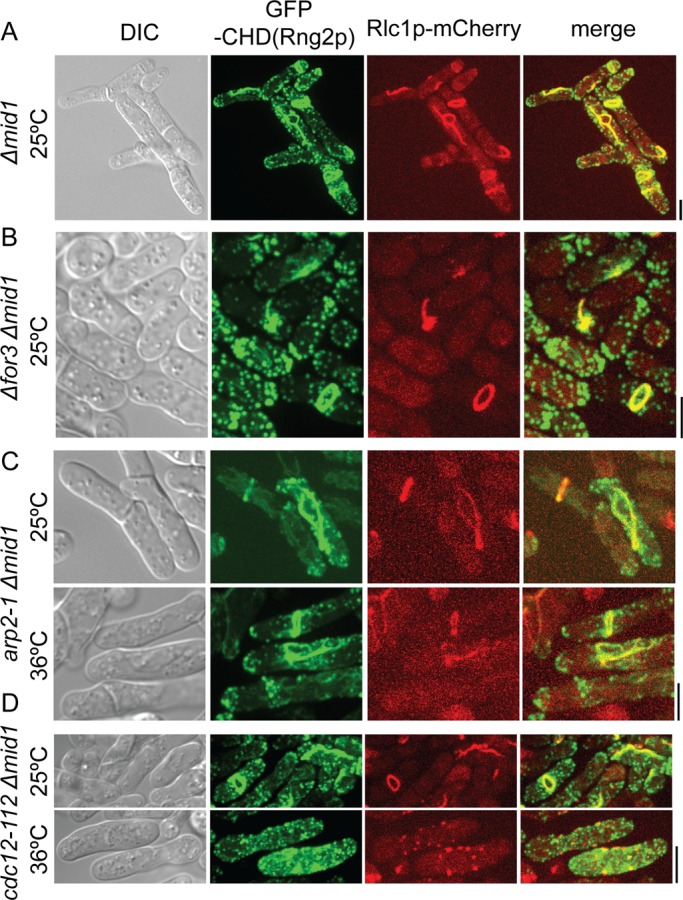
Contractile ring assembly in Δmid1 cells depends on formin Cdc12p. Cells were grown and imaged at 25ºC or grown at 36ºC for 6 h (C) and 4 h (D) before imaging at 36ºC. (A–D) DIC and maximum-intensity projections of fluorescence micrographs. Localization of GFP-calponin-homology domain (CHD) of Rng2p and Rlc1p-mCherry in (A) Δmid1 cells, (B) Δfor3 Δmid1 cells, (C) arp2-1 Δmid1 cells at 25 and 36°C, and (D) cdc12-112 Δmid1 cells at 25 and 36°C. Rlc1p-mCherry was expressed from the native locus, and GFP-CHD was expressed from a 41xnmt1 promoter for 24 h. Scale bars, 5 μm.
Cytokinetic nodes in cells lacking anillin Mid1p
Previous work concluded that Δmid1 cells lack nodes altogether (Huang et al., 2008), but close examination of contractile ring precursors in Δmid1 cells revealed the presence of cytokinetic nodes with Blt1p and formin Cdc12p. Dispersed cortical nodes containing Blt1p appeared normally during interphase in Δmid1 cells (Figure 5A(a)) and, similar to wild-type cells, accumulated Cdc12p within the 10 min after SPB division during mitosis (Figures 3A and 5C). The content of Blt1p and Cdc12p in these cytokinetic nodes was similar to that in wild-type cells, with 59% of nodes positive for both Blt1p and Cdc12p, 15% of Cdc12p-positive nodes lacking Blt1p, and 26% of Blt1p-positive nodes lacking Cdc12p (Supplemental Figure S1, B and D).
These nodes containing Blt1p and Cdc12p in Δmid1 cells had two functionally important defects: 1) they failed to accumulate Myo2, Rng2p, and Cdc15p (Figure 5, I, J, and L; note the absence of nodes in Figure 5, D–G, and that puncta in Figure 5, E and F, are not nodes); and 2) they were generally distributed more widely around the equator than in wild-type cells (Figure 4B; Figure 5, A(b) and B, and Supplemental Video S6). About fourfold more nodes (tracked by Blt1p) persisted close to the poles of mitotic Δmid1 cells (Figures 1B [compare cell next to the star] and 5A(c)), so their mean positions were 0.13 ± 0.20 in cells of normal length (12–16 μm) and 0.11 ± 0.21 in cells of abnormal lengths (<12 or >16 μm). The mean positions and widths of the distributions of cytokinetic nodes (tracked by Cdc12p) also varied much more in Δmid1 cells than in wild-type cells (Figure 4).
Formation of strands in cells lacking anillin Mid1p
Cytokinetic nodes in cells without Mid1p lacked contractile ring proteins Myo2, IQGAP Rng2p, and F-BAR Cdc15p, but all three proteins accumulated from the cytoplasm into cortical strands during mitosis (Figure 5, D, F [arrowhead], and G). Cdc15p also joined strands from some punctate structures dispersed in the cortex (Figure 5E(b), arrows). These cortical puncta differed from endocytic actin patches. Cdc15p in actin patches turned over in <1 min, whereas the puncta with Cdc15p were more stable and often persisted for >30 min as strands appeared in the cortex (Figure 5E(a)).
We used Myo2 to track strands, which also contained Rng2p and Cdc15p (Figure 5, E and H). Strands with Myo2 appeared in the cortex after SPB division (Figures 3A and 5D) at random locations throughout the cortex from pole to pole (Figure 5, D and L, Supplemental Figure S2A, and Supplemental Video S7), generally between nodes marked by Blt1p (Figure 5I) and Cdc12p (Figure 5J). In many but not all cells strands slowly merged into contractile rings.
Three lines of evidence showed that formation of strands and contractile rings in Δmid1 cells depended on actin filaments generated by formin Cdc12p, the formin that normally generates actin filaments for contractile rings (Chang et al., 1997). First, strands of Myo2 appeared in the cortex around the same time that Cdc12p appeared in nodes, ∼10–20 min later than the appearance of Myo2 in nodes in wild-type cells (Figure 3A). Second, strands of Myo2 contained actin filaments (tracked by the CHD of Rng2p; Figure 6A). Treatment of Δmid1 cells with latrunculin A prevented formation of strands and contractile rings and resulted in Myo2 forming spots rather than strands in the cortex (Supplemental Video S8). Third, Δmid1 cells depending on the temperature-sensitive allele cdc12-112 (Chang et al., 1997) failed to assemble strands or contractile rings at the restrictive temperature (36ºC), and Myo2 aggregated in punctate structures rather than strands in the cortex (Figure 6D). On the other hand, actomyosin strands and contractile rings formed in Δmid1 cells lacking the formin For3p (Figure 6B) or expressing the temperature-sensitive allele arp2-1 (Morrell et al., 1999), which encodes a subunit of the Arp2/3 complex, at both permissive (25ºC) and restrictive (36ºC) temperatures (Figure 6C).
Contractile ring proteins Blt1p and Cdc12p were initially absent from strands in Δmid1 cells, but over time Cdc12p from nodes and speckles and Blt1p from most, but not all, nodes were incorporated into strands (Figure 5, I and J). Nodes containing Cdc12p moved toward and bidirectionally along strands containing Myo2 and actin filaments, eventually distributing along the strands (Supplemental Figure S2, B–E, and Supplemental Videos S9–S11). Bidirectional movements of Cdc12p along strands showed that actin filaments are antiparallel in these strands. Speckles of Cdc12p were not as bright as nodes, and in contrast to nodes, moved randomly independent of actin filaments (Supplemental Video S4B; Coffman et al., 2009).
Contractile ring assembly in cells lacking anillin Mid1p
Faced with defects in both the composition and positions of nodes, Δmid1 cells used a variation of the normal pathway to assemble contractile rings from Blt1p, Rng2p, Myo2, Cdc15p, and actin filaments produced by Cdc12p. Wild-type cells assemble a compact cytokinetic ring from a broad band of nodes within 10–15 min after SPB division (Figure 3A; Wu et al., 2003), whereas Δmid1 cells required from 6 to >100 min after SPB division to make a contractile ring (Figure 3A). After originating at random locations, cortical actomyosin strands grew longer, moved randomly, and fused laterally and end to end, eventually forming a closed, continuous ring in two-thirds of Δmid1 cells (Supplemental Video S7). Owing to the abnormal initial conditions and assembly from strands rather than nodes, contractile rings in Δmid1 cells were often oriented obliquely, with circumferences larger than the circular rings of wild-type cells.
Contractile ring constriction in cells lacking anillin Mid1p
Wild-type cells wait ∼25 min after the completion of ring assembly before beginning constriction (Figure 3A; Wu et al., 2003), but rings began constricting in Δmid1 cells soon after strand(s) of Myo2 closed into a complete ring (Figures 3A and 5K). Rings in Δmid1 cells constricted circumferentially at 0.25 ± 0.06 μm/min, close to the rate of 0.32 ± 0.02 μm/min in wild-type cells (Figure 3B), but the constriction rate was three times more variable in Δmid1 cells than in wild-type cells. Constriction corrected imperfections in some but not all oblique rings (Supplemental Video S7). Septum formation followed ring constriction (Figure 5L, star).
One-third of Δmid1 cells failed to assemble a closed ring from Myo2 strands within 1.5 h (Figure 3A). Some of these strands persisted for >3 h. Cells deposited septal material along these oblique strands, resulting in incomplete septa and failure of cytokinesis (Figure 5L, arrowheads). Some Δmid1 cells assembled additional septa before completing a previous attempt at cytokinesis (Supplemental Video S5).
Contributions of anillin Mid1p to cellular morphology and polarity
As illustrated but not discussed previously (Paoletti and Chang, 2000), cells lacking Mid1p had serious defects in morphology in addition to the problems with cytokinesis. Wild-type cells are cylindrical, whereas >70% of Δmid1 cells had gross morphological defects, such as curves, branches, or bulges of the surface (Figure 7). Some cells formed bulges as they grew parallel to curved or disoriented septa (Supplemental Video S5), and these abnormal septa seemed to prolong the time required for the daughter cells to separate.
FIGURE 7:
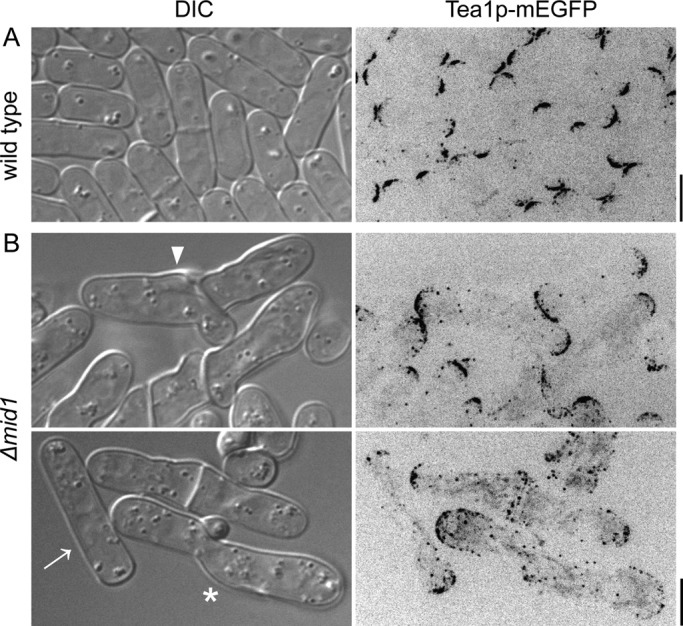
Defects in cell morphology and polarity in Δmid1 cells. DIC and negative-contrast, maximum-intensity projection fluorescence micrographs of cells expressing Tea1p-mEGFP. (A) Wild-type cells, (B) Δmid1 cells with curves, branches (arrowhead), ectopic bulges (star), and more disperse localization of Tea1p in cylindrical Δmid1 cell (arrow). Scale bars, 5 μm.
We also found polarity defects in cells lacking Mid1p. The polarity marker Tea1p–mEGFP concentrated in spots at the poles of wild-type cells (Mata and Nurse, 1997) but was divided between the poles and discrete dots scattered along the sides of Δmid1 cells (Figure 7), even in those with normal morphology (Figure 7B, arrow).
DISCUSSION
We investigated the inefficiencies in contractile ring assembly in cells lacking anillin Mid1p, to understand how Mid1p contributes to the reliability of cytokinesis in wild-type cells. Without Mid1p, cells attempt to assemble contractile rings from the same proteins as wild-type cells, but contractile ring precursors are separated in two phases—nodes and strands—scattered widely in the cortex. Ring assembly is slow without Mid1p, because strands depend on random encounters with other strands to merge into a ring. Ring assembly is unreliable in the absence of Mid1p, because strands are dispersed widely in the cortex. Previous studies on Δmid1 cells overlooked the contribution of cytokinetic nodes to contractile ring assembly (Huang et al., 2008) and the importance of placing ring precursors near the equator on the reliability of cytokinesis. Problems in cells lacking Mid1p suggest that the interphase node protein Cdr2p, although helpful in determining the plane of cell division (Almonacid et al., 2009), cannot substitute for Mid1p in coordinating reliable assembly of the contractile ring at the cell equator.
Contractile ring assembly is reliable in wild-type cells because Mid1p concentrates contractile ring precursors together in nodes in a narrow zone around the equator, which favors the transient connections between Myo2 and actin filaments that allow a search, capture, pull, and release mechanism to condense reliably nodes into a ring (Vavylonis et al., 2008). The pathway of ring assembly from strands in Δmid1 cells is almost as reliable as ring formation via condensation of nodes in wild-type cells as long as strands are restricted to a narrow zone around the equator (see accompanying paper, Saha and Pollard, 2012).
Defects in preparation for contractile ring assembly in cells lacking anillin Mid1p
We used Blt1p to follow how interphase nodes mature into cytokinetic nodes and participate in ring assembly. Blt1p is a reliable marker of cytokinetic nodes, because in wild-type cells interphase nodes containing Blt1p mature directly into cytokinetic nodes by accumulating Myo2 (Moseley et al., 2009) and Cdc12p (this study) and undergo the same intermittent movements during ring assembly (this study) as nodes marked with Myo2 (Vavylonis et al., 2008).
Three lines of evidence show that the punctate assemblies containing Cdc12p in the cortex of mitotic Δmid1 cells are bona fide cytokinetic nodes. First, Cdc12p concentrates in these puncta with Blt1p at the same time in the presence or absence of Mid1p. This colocalization confirms that at least some of the punctate “places” where Cdc12p accumulates in mitotic Δmid1 cells (Coffman et al., 2009) are nodes. Second, Cdc12p in nodes generates actin filaments that support formation of strands of Myo2 and subsequently the contractile ring. Third, nodes containing Cdc12p are incorporated into strands containing actin filaments and Myo2 by undergoing directed movements toward and along the strands.
The absence of Myo2, IQGAP Rng2p, and F-BAR Cdc15p from nodes in Δmid1 cells can be explained by the established roles of Mid1p in recruiting these proteins to nodes through two pathways: Mid1p → (Cdc4p, Rng2p) → (Myo2p, Rlc1p) → Cdc12p and Mid1p → Cdc15p → Cdc12p (Almonacid et al., 2011; Laporte et al., 2011; Padmanabhan et al., 2011; Figure 8). Given that interaction with Cdc15p (Carnahan and Gould, 2003) normally helps to recruit Cdc12p to cytokinetic nodes (Laporte et al., 2011), it is surprising that Cdc15p is absent from nodes with Cdc12p in Δmid1 cells. Therefore Cdc12p must interact with node molecules in addition to Cdc15p (Figure 8, question mark), consistent with the lack of Cdc15p in 63% of nodes with Cdc12p in wild-type cells (Laporte et al., 2011).
FIGURE 8:
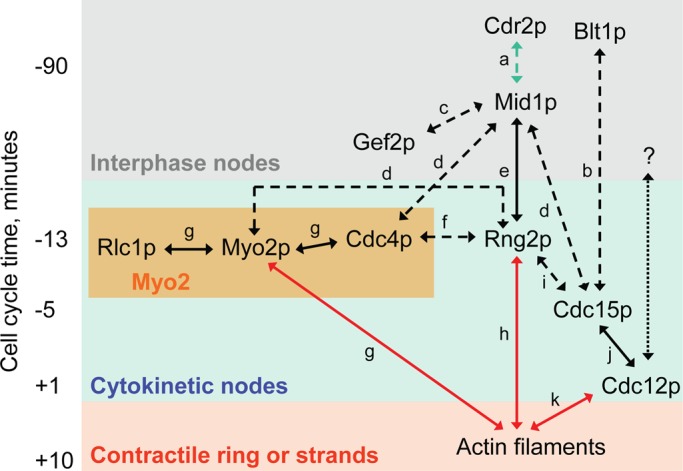
Map of interactions among proteins involved in cytokinesis expanded from a figure in Laporte et al. (2011). The vertical axis approximates time in wild-type cells relative to spindle pole body separation at time zero. Protein names are placed at the site and time of their initial appearance: gray box, interphase nodes; blue box, cytokinetic nodes; or red box, contractile rings or strands (bundles of actin filaments and Myo2 that merge to form contractile rings in cells lacking Mid1p). All proteins in interphase nodes are retained in cytokinetic nodes, and all proteins in cytokinetic nodes except Cdr2p are retained in contractile ring or strands. The yellow box contains the three subunits of myosin-II (Myo2). Arrows denote interactions among proteins: blue arrows, interactions only in nodes; black arrows, nodes, strands, and contractile ring; and red arrows, only strands and contractile ring. Broken lines represent evidence from coimmunoprecipitation experiments, and solid lines represent evidence with purified protein constructs. The black question mark and dotted line denote one or more unknown binding partners that allow Cdc12p to concentrate in cytokinetic nodes in the absence of Mid1p. References to work that established each interaction: (a) Almonacid et al. (2009), (b) Moseley et al. (2009), (c) Ye et al. (2012), (d) Laporte et al. (2011), (e) Almonacid et al. (2011), (f) D'Souza et al. (2001), (g) Lord and Pollard (2004), (h) Takaine et al. (2009), (i) Roberts-Galbraith et al. (2010), (j) Carnahan and Gould (2003), and (k) Kovar et al. (2003). Times of localization of proteins to nodes and the contractile ring were obtained from Wu et al. (2003) and Laporte et al. (2011).
Contractile ring assembly in cells lacking anillin Mid1p
We found that cells lacking Mid1p assemble contractile rings from strands of actomyosin, as observed previously (Chang et al., 1996; Sohrmann et al., 1996; Motegi et al., 2004; Hachet and Simanis, 2008; Huang et al., 2008), and offer a simple mechanism for their assembly. Our movies show that in Δmid1 cells strands containing Myo2 accumulate nodes marked with Blt1p and Cdc12p through movements similar to those of nodes in wild-type cells (Vavylonis et al., 2008). Given that latrunculin inhibits this process, a simple explanation is that Myo2 captures actin filaments emanating from nodes containing Cdc12p and pulls together the elements of the ring. This process may be similar to the ability of adjacent contractile rings to merge together, as observed experimentally (Daga and Chang, 2005) and recapitulated by computer simulations (Vavylonis et al., 2008). Actomyosin strands in Δmid1 cells eventually accumulate most contractile ring proteins except for Mid1p, so we find no need to postulate biochemical reactions other than the normal network of extensive pairwise interactions among cytokinesis proteins (Figure 8).
The impressive redundancy in this network complicates the interpretation of experiments to establish dependences. For example, in the absence of its natural partner, Mid1p, Rng2p associates with actomyosin strands, likely through interactions with actin filaments (Takaine et al., 2009), Cdc4p, or Myo2p (Laporte et al., 2011). Similarly, without its normal receptor, Mid1p, Cdc15p accumulates in strands where it may interact with Rng2p (Roberts-Galbraith et al., 2010) and Cdc12p (Carnahan and Gould, 2003). Thus only a fraction of the extensive network of pairwise interactions among node proteins appears to suffice for function. Much more work is required to sort out the hierarchy of these interactions.
Assembly of rings from strands in Δmid1 cells differs from the “leading cable” hypothesis, in which under some circumstances cables of actin filaments and Myo2 grow from a singular spot containing Cdc12p and form a contractile ring (Chang et al., 1997; Chang, 1999; Arai and Mabuchi, 2002). Without Mid1p many nodes containing Cdc12p are scattered beyond the equator, and so strands of actin and Myo2 form between nodes over a larger area of cortex than normal.
During the unreliable, prolonged course of ring formation from strands in Δmid1 cells, septal material may deposit along strands, arresting them in abnormal configurations that complicate formation of a complete ring. Huang et al. (2008) found that inactivation of Cps1p, a 1,3-β-glucan synthase involved in septum assembly, increased the formation of complete contractile rings in Δmid1 cells. We suggest that this is due to fewer actomyosin strands being arrested by attachments to the septum. We do not yet understand why contractile ring assembly is more dependent on the SIN pathway in Δmid1 cells than in wild-type cells (Hachet and Simanis, 2008).
Contractile ring constriction in cells lacking anillin Mid1p
In wild-type cells contractile rings form by ∼10–15 min after SPB separation and then mature for ∼25 min before beginning to constrict, whereas the timing of contractile ring constriction in Δmid1 cells is remarkable in two ways. On one hand, Δmid1 cells with slowly forming contractile rings can delay the constriction of strands of actomyosin well beyond the time when contractile rings constrict in wild-type cells. On the other hand, rings begin to constrict soon after strands close into rings rather than maturing normally for 25 min. The mechanisms that delay contraction of incomplete rings past 40 min after SPB separation and sense the formation of a complete contractile ring are mysteries worthy of further investigation. The near-normal rate of constriction of contractile rings in Δmid1 cells argues that the architecture of these rings is similar to that of wild-type contractile rings, in spite of differences in size, shape, orientation, and assembly pathway. Additional defects in the contractile rings of Δmid1 cells give rise to curved septa, which can support separation of the daughter cells (Supplemental Video S5).
Influence of anillin Mid1p on cell shape and polarity
Mid1p influences cell shape directly through its role in cell polarity and indirectly through its role in cytokinesis. Cells lacking Mid1p have defects in cell polarity that produce abnormal growth, whereas defects in cytokinesis similar to those in cells with mutations in sep genes (Sipiczki et al., 1993; Grallert et al., 1999) result in bulges next to curved or abnormal septa. Some Δmid1 cells look normal because they assemble a properly oriented septum, giving rise to one or two cylindrical daughter cells, or adjust their growth to correct minor bends and bulges. The abnormal localization of the polarity marker Tea1p in many morphologically normal Δmid1 cells indicates a direct role of Mid1p in cell polarity.
We suggest that the irregular shapes of Δmid1 cells contribute to defects in contractile ring assembly on top of the problems caused by the absence of Myo2 in nodes and the lack of focus of nodes around the equator. Cells without Mid1p grow abnormally during interphase due to defects in polarity, resulting in a population of cells that vary in size and shape. This sets up a vicious cycle because cells enter mitosis with a wide range of lengths and shapes, which compromises the mechanics of contractile ring assembly and the ability of the Pom1p gradient to define a narrow zone in the middle of the cell for cytokinesis (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). Mid1p may have yet-to-be-discovered influences on the Pom1p gradient or parallel pathways that specify the cleavage site.
MATERIALS AND METHODS
Generation of yeast strains
Standard molecular biology techniques in S. pombe were used to delete a gene or to tag a gene at the C-terminus using pFA6a-based constructs (Bahler et al., 1998). Genetic crosses between S. pombe cells of opposite mating type were used to generate other strains (Table 1).
TABLE 1:
S. pombe strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| SSP2-3 | h+ Δmid1::kanMX6 leu1-32 ura4-D18 his3-D1 ade6-M210 | This study |
| SSP40-12 | h+ Δmid1::natMX6 leu1-32 ura4-D18 his3-D1 ade6-M210 | This study |
| SSP48-1 | h+ tea1-mEGFP-kanMX6 leu1-32 ura4-D18 his3-D1 ade6-M210 | This study |
| SSP49-1 | h+ tea1-mEGFP-kanMX6 Δmid1::natMX6 leu1-32 ura4-D18 his3-D1 ade6-M210 | This study |
| SSP68-2 | blt1-mEGFP-kanMX6 rlc1-mCherry-natMX6 leu1-32 ura4-D18 | This study |
| SSP70-3 | Δmid1::natMX6 blt1-mEGFP-kanMX6 rlc1-mCherry-natMX6 leu1-32 ura4-D18 | This study |
| SSP78-1 | h+ rlc1-mEGFP-kanMX6 Δmid1::natMX6 leu1-32 ura4-D18 ade6-M210 | This study |
| SSP86-11 | h- cdc12-3YFP-kanMX6 blt1-mCherry-natMX6 leu1-32 ura4-D18 | This study |
| SSP87-1 | cdc12-3YFP-kanMX6 blt1-mCherry-natMX6 Δmid1::natMX6 leu1-32 ura4-D18 | This study |
| SSP88-12 | cdc12-3YFP-kanMX6 rlc1-mCherry-natMX6 leu1-32 ura4-D18 | This study |
| SSP89-3 | cdc12-3YFP-kanMX6 rlc1-mCherry-natMX6 Δmid1::natMX6 leu1-32 ura4-D18 | This study |
| SSP90-4 | rlc1-mEGFP-kanMX6 Δmid1::natMX6 sad1-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| SSP91-4 | h- Δmid1::natMX6 sad1-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| SSP92-4 | h- rlc1-mCherry-natMX6 Δmid1::natMX6 P41xnmt1-GFP-CHD(rng2)-leu1+ ura4-D18 | This study |
| SSP101-9 | cdc12-112 Δmid1::kanMX6 rlc1-mCherry-natMX6 P41xnmt1-GFP-CHD(rng2)-leu1+ ura4-D18 | This study |
| SSP102-4 | h- Δfor3::kanMX6 Δmid1::natMX6 rlc1-mCherry-natMX6 P41xnmt1-GFP-CHD(rng2)-leu1+ ura4-D18 | This study |
| SSP104-3 | h- arp2-1 rlc1-mCherry-natMX6 Δmid1::natMX6 P41xnmt1-GFP-CHD(rng2)-leu1+ ura4-D18 | This study |
| SSP106-3 | cdc12-3GFP-kanMX6 sad1-mRFP1-kanMX6 | This study |
| SSP107-5 | Δmid1::natMX6 cdc12-3GFP-kanMX6 sad1-mRFP1-kanMX6 | This study |
| SSP109-8 | blt1-mEGFP-kanMX6 sad1-mRFP1-kanMX6 leu1-32 ura4-D18 | This study |
| SSP110-6 | Δmid1::natMX6 blt1-mEGFP-kanMX6 sad1-mRFP1-kanMX6 leu1-32 ura4-D18 | This study |
| SSP115-1 | Δmid1::natMX6 kanMX6-Pcdc15-mEGFP-cdc15 rlc1-mCherry-natMX6 leu1-32 ura4-D18 | This study |
| QC139 | h+ rlc1-3GFP-kanMX6 sad1-mEGFP-kanMX6 | Chen and Pollard (2011) |
| AR140 | h+ leu1-32 ura4-D18 ade6-M210 kanMX6-Pcdc15-mEGFP-cdc15 | Arasada and Pollard (2011) |
| IRT89 | Δmid1::natMX6 kanMX6-Prng2-mEGFP-rng2 rlc1-mCherry-natMX6 | Irene Tebbs |
| JW1349 | h+ nmt41-GFP-CHD(rng2)-leu1+ rlc1-tdTomato-natMX6 ade6-M210 leu1-32 ura4-D18 | Jian-Qiu Wu |
| JW1085 | h- sad1-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | Jian-Qiu Wu |
| FY528 | h+ leu1-32 ura4-D18 his3-D1 ade6-M210 | Lab collection |
| JM673 | h- cdr2-mEGFP::kanMX6 leu1-32 | Moseley et al. (2009) |
| JM454 | h- cdr1-3xGFP::kanMX6 | Moseley et al. (2009) |
| JM206 | h+ Δblt1::kanMX6 | Moseley et al. (2009) |
| JM569 | h- Δcdr2::natR | Moseley et al. (2009) |
| MBY310 | h+ cdc12-112 ade6-M210 leu1-32 ura4-D18 | Chang et al. (1997) |
| KGY1187 | h90 arp2-1 mam2::LEU2 ade6-M210 leu1-32 ura4-D18 | Morrell et al. (1999) |
| FC1218 | h- nmt41-GFP-CHD(rng2)-leu1+ ade6-M216 leu1-32 ura4-D18 | Martin and Chang (2006) |
Microscopy
Cells were usually grown in exponential growth phase (OD595 values between 0.05 and 0.5) in YE5S medium for 36–48 h at 25ºC before imaging. Strains expressing gene products from nmt1 promoters were grown at 25ºC for 24 h in EMM5S media containing 5 μg/ml thiamine, followed by growth in EMM5S media for 16–24 h before imaging. An Olympus IX71 inverted microscope (Olympus, Tokyo, Japan) fitted with 488-, 514-, and 568-nm argon ion lasers, a Yokogawa CSU-X1 spinning-disk confocal scanning unit (Yokogawa, Tokyo, Japan), and an Andor iXon electron-multiplying charge-coupled device (EMCCD) camera (model 897; Andor Technology, South Windsor, CT) was used for imaging. Micrographs in Figure 7 were acquired with the same microscope equipped with a PerkinElmer UltraView RS spinning-disk confocal scanning unit (PerkinElmer, Waltham, MA) and a Hamamatsu ORCA-ER CCD camera (Hamamatsu, Hamamatsu, Japan).
Image analysis
Micrographs were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). Three-dimensional reconstructions of cells were made using the Projector 4D plug-in in ImageJ software. These reconstructions are shown in Figures 1 and 5 as projections in two dimensions.
Supplementary Material
Acknowledgments
We thank J. B. Moseley, F. Chang, K. L. Gould, J.-Q. Wu, D. R. Kovar, Q. Chen, R. Arasada, and I. Tebbs for strains; C. D. McCormick and J. Berro for help with image analyses; M. J. Solomon and Y.-L. Wang for helpful suggestions on the text and organization of the manuscript; and members of our lab for helpful discussions. This work was supported by National Institutes of Health Research Grant GM-026338 to T.D.P. and an Edward L. Tatum Fellowship to S.S. S.S. dedicates this work to his parents, Sukumar Saha and Madhuchhanda Saha.
Abbreviations used:
- CCD
charge-coupled device
- CHD
calponin-homology domain
- DIC
differential interference contrast
- EMCCD
electron-multiplying charge-coupled device
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- IQGAP
IQ motif containing GTPase-activating protein
- mEGFP
monomeric enhanced green fluorescent protein
- mYFP
monomeric yellow fluorescent protein
- SD
standard deviation
- SIN
septation initiation network
- SPB
spindle pole body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0535) on August 23, 2012.
REFERENCES
- Almonacid M, Celton-Morizur S, Jakubowski JL, Dingli F, Loew D, Mayeux A, Chen JS, Gould KL, Clifford DM, Paoletti A. Temporal control of contractile ring assembly by Plo1 regulation of myosin II recruitment by Mid1/anillin. Curr Biol. 2011;21:473–479. doi: 10.1016/j.cub.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A. Spatial control of cytokinesis by Cdr2 kinase Mid1/anillin nuclear export. Curr Biol. 2009;19:961–966. doi: 10.1016/j.cub.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Arai R, Mabuchi I. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 2002;115:887–898. doi: 10.1242/jcs.115.5.887. [DOI] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol Cell Biol. 2004;24:10621–10635. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR 3rd, Gould KL. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Chang F. Dynamic positioning of the fission yeast cell division plane. Proc Natl Acad Sci USA. 2005;102:8228–8232. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza VM, Naqvi NI, Wang H, Balasubramanian MK. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct Funct. 2001;26:555–565. doi: 10.1247/csf.26.555. [DOI] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Grallert B, Zilahi E, Szilagyi Z, Sipiczki M. Eleven novel sep genes of Schizosaccharomyces pombe required for efficient cell separation and sexual differentiation. Yeast. 1999;15:669–686. doi: 10.1002/(SICI)1097-0061(19990615)15:8<669::AID-YEA411>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Morrell JL, Morphew M, Gould KL. A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast. Mol Biol Cell. 1999;10:4201–4215. doi: 10.1091/mbc.10.12.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell JL, Nichols CB, Gould KL. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J Cell Sci. 2004;117:5293–5302. doi: 10.1242/jcs.01409. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165:685–695. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Bakka K, Sevugan M, Naqvi NI, D'Souza V, Tang X, Mishra M, Balasubramanian MK. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr Biol. 2011;21:467–472. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Pollard TD. Characterization of structural and functional domains of the anillin-related protein Mid1p that contribute to cytokinesis in fission yeast. Mol Biol Cell. 2012;23:3993–4007. doi: 10.1091/mbc.E12-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M, Grallert B, Miklos I. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J Cell Sci. 1993;104:485–493. doi: 10.1242/jcs.104.2.485. [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Takaine M, Numata O, Nakano K. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 2009;28:3117–3131. doi: 10.1038/emboj.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Lee IJ, Runge KW, Wu JQ. Roles of putative Rho-GEF Gef2 in division-site positioning and contractile-ring function in fission yeast cytokinesis. Mol Biol Cell. 2012;23:1181–1195. doi: 10.1091/mbc.E11-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.