Phosphorylation of the RNA-binding protein tristetraprolin (TTP) by p38 MAPK/MK2 does not prevent its RNA interaction and switches the mode of TTP action from destabilization to stabilization of the HIF-1α mRNA and subsequent activation of HIF-1 signaling.
Abstract
Hypoxia-inducible factor-1 (HIF-1) is a well-studied transcription factor mediating cellular adaptation to hypoxia. It also plays a crucial role under normoxic conditions, such as in inflammation, where its regulation is less well understood. The 3′-untranslated region (UTR) of HIF-1α mRNA is among the most conserved UTRs in the genome, hinting toward posttranscriptional regulation. To identify potential trans factors, we analyzed a large compilation of expression data. In contrast to its known function of being a negative regulator, we found that tristetraprolin (TTP) positively correlates with HIF-1 target genes. Mathematical modeling predicts that an additional level of posttranslational regulation of TTP can explain the observed positive correlation between TTP and HIF-1 signaling. Mechanistic studies revealed that TTP indeed changes its mode of regulation from destabilizing to stabilizing HIF-1α mRNA upon phosphorylation by p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein kinase 2. Using a model of monocyte-to-macrophage differentiation, we show that TTP-driven HIF-1α mRNA stabilization is crucial for cell migration. This demonstrates the physiological importance of a hitherto-unknown mechanism for multilevel regulation of HIF-1α in normoxia.
INTRODUCTION
The transcription factor hypoxia-inducible factor (HIF) is regarded as a key factor in the cellular adaptation of gene expression to oxygen deprivation. HIF consists of a regulatory α subunit and a constitutively expressed β subunit. There are three closely related isoforms of the α subunit: HIF-1, -2, and -3. Of these, HIF-1α is ubiquitously expressed, whereas HIF-2α is cell specific, and both are known to be critical for the hypoxia response. The role of HIF-3α, however, is less well understood (Castrop and Kurtz, 2010). Regulation of HIF activity occurs mainly by posttranslational hydroxylation of the regulatory α subunits by HIF-prolyl hydroxylases (PHD1, 2, 3; also known as HPH2, 1, 3 or EGLN2, 1, 3, respectively), as well as by factor-inhibiting HIF (FIH; Semenza, 2003). Prolyl hydroxylation of HIF-α causes ubiquitination and subsequent proteolysis by the 26S proteasome. HIF target genes are involved in a broad range of cellular processes, such as proliferation and apoptosis, glycolysis, pH regulation, erythropoiesis, iron metabolism, extracellular matrix remodeling, inflammation, transcription, and angiogenesis (Semenza, 2003). Although regulation of HIF by hydroxylation of its α subunit seems to be crucial for cellular oxygen sensing and adaptation of gene expression, recent studies indicate that HIF-1α is also regulated transcriptionally, especially in the context of inflammation (Frede et al., 2007). Moreover, HIF-1 can be activated under normoxic conditions, which allows the initiation of an inflammatory response before tissues become hypoxic (Eltzschig and Carmeliet, 2011). This suggests that alterations in HIF-1α mRNA levels by either a change in the transcription rate or in mRNA turnover should also have a considerable impact on HIF-1 function under normoxic conditions. Consistently, a number of RNA-binding proteins (RBPs) that interact and thus may influence HIF-1α mRNA stability have been described (Gorospe et al., 2011).
The control of mRNA turnover is mainly attributed to the mRNA 3′-untranslated region (3′ UTR), which contains regulatory elements that govern the spatial and temporal expression of mRNAs (Kuersten and Goodwin, 2003). One important example of specific cis-acting elements controlling the half-life of mRNAs is the group of adenylate- and uridylate-rich elements (AREs) found in the 3′ UTRs of many unstable mammalian mRNAs (Bevilacqua et al., 2003). AREs influence mRNA stability through different mechanisms, some of which comprise mRNA deadenylation (Zhang et al., 2002). Examples of mRNAs that are regulated by alterations in mRNA stability include granulocyte-monocyte colony–stimulating factor, interleukins, interferons, tumor necrosis factor α (TNF-α), vascular endothelial growth factor (VEGF), and some proto-oncogenes (e.g., c-fos, k-ras, and pim-1; Bakheet et al., 2001).
It has been reported that the 3′ UTR contains many AREs, which may function in the control of HIF-1α mRNA turnover (Sheflin et al., 2004). Recently, two independent groups identified the ARE-interacting RBP tristetraprolin (TTP; ZFP36) as a trans factor mediating HIF-1α destabilization under hypoxic conditions (Kim et al., 2010; Chamboredon et al., 2011). To understand the role of HIF-1α mRNA stability and its regulation by trans factors under normoxic conditions, we decided to perform an in silico screen for potential regulators of the HIF-1α mRNA turnover. Contrary to our expectation, we found that the mRNA of the known regulator TTP positively correlates with HIF-1 target gene expression. Using mathematical modeling and detailed molecular experimentation, we were able to resolve this paradox and identify a novel level of regulation by TTP: phosphorylation of TTP by the p38 mitogen-activated protein kinase (MAPK) pathway leads to stabilization of HIF-1α mRNA and subsequent enhanced HIF-1 signaling under normoxic conditions. Using a model of macrophage differentiation, we further demonstrate that this hitherto-unknown multilevel regulation of HIF-1α function by TTP is essential for inflammatory response–triggered migration in normoxia.
RESULTS
The 3′ UTR of HIF-1α mRNA includes highly conserved information content
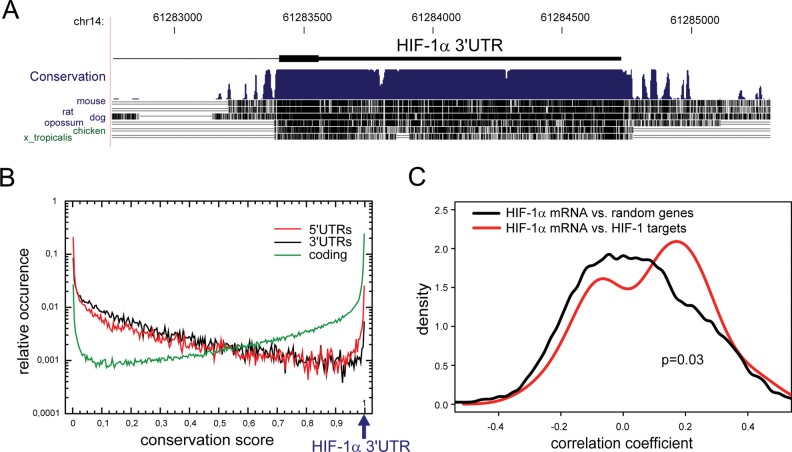
In a multispecies alignment using the University of California, Santa Cruz, human genome BLAT search (http://genome.ucsc.edu), we observed an unexpectedly high conservation of the HIF-1α mRNA 3′ UTR (size: 1320 nucleotides; Figure 1A). Of interest, this extreme conservation was noted for >95% of the length of the UTR and does not correlate with base composition as previously described for most other mammalian 3′ UTRs (Shabalina et al., 2003). To determine whether the conservation of HIF-1α mRNA 3′ UTR is significantly above average, we estimated a conservation score for all human 5′ UTRs, coding regions, and 3′ UTRs (Figure 1B). Coding regions were found to display the highest conservation level, whereas only ∼1.5% of all 3′ UTRs were strongly conserved (conservation score, ≥0.6). Indeed, the HIF-1α mRNA 3′ UTR belongs to the top 20 most-conserved UTRs out of 7953 genes with UTR lengths >1000 nucleotides, which corresponds to the 0.213 percentile (Supplemental Table S1).
FIGURE 1:
HIF-1α mRNA 3′ UTR conservation and correlation study of HIF-1α mRNA and HIF-1 target genes. (A) Depiction of sequence conservation as obtained by BLAT search multispecies alignment (http://genome.ucsc.edu) for 3′ UTR of HIF-1α mRNA (B) Conservation score for all human 5′ UTRs (red), coding regions (green), and 3′ UTRs (black). The position of the HIF-1α conservation score is indicated by an arrow. (C) Histogram of pairwise correlation of HIF-1α mRNA with HIF-1 target genes. The correlation was performed by using data sets from the Stanford Microarray Database with >1200 microarray-based expression profiles. In comparison with randomly chosen genes (black), correlation of HIF-1α mRNA with HIF-1 target genes (red) causes a shift to a significant positive correlation. HIF-1 target genes were chosen as described (Semenza, 2003).
Moreover, according to the classification of AREs (Barreau et al., 2005), the HIF-1α mRNA 3′ UTR sequence contains eight AUUUA motifs in a U-rich context (class I ARE), as well as two overlapping UUAUUUA(U/A)(U/A) nonamers (class II ARE; Rossignol et al., 2002). Because most AREs are functionally important for RNA–protein interactions involving the control of mRNA stability, we conclude that the gene expression rate of HIF-1α is likely to be regulated through its mRNA stability.
Conservation of regulatory elements suggests a crucial function in the control of gene expression. The extreme conservation grade that is maintained throughout the entire length of the HIF-1α mRNA 3′ UTR indicated that this 3′ UTR plays an important role in the control of HIF-1α expression. HIF-1 activity is regulated at the posttranslational level through oxygen-dependent degradation of HIF-1α (Semenza, 2007), whereas transcriptional regulation of HIF-1 activity has only recently been described, mainly in the context of nonhypoxic HIF-1 activation (Kuschel et al., 2012). Our hypothesis, therefore, was that an alteration of HIF-1α mRNA level affects HIF-1 activity.
To investigate whether HIF-1α mRNA level and HIF-1 activity are correlated, we used a compilation of large-scale gene expression studies (Stuart et al., 2003) originating from the Stanford Microarray Database with >1200 experiments. The data contain a collection of different independent investigations, including expression profiles from tumor samples, cancer cell lines, and different organ-specific tissue samples, as well as expression data from studies of diverse biological processes such as cell cycle, stress, signaling, and apoptosis. We correlated HIF-1α mRNA levels with randomly chosen genes or with verified HIF-1 target genes (Semenza, 2003), as described earlier (Mrowka et al., 2008). Of note, we found a significant positive correlation between HIF-1α mRNA levels and HIF-1 target gene expression (Figure 1C), supporting the view that HIF-1α mRNA level, and thus mRNA stability, is important for HIF-1 function.
The RNA-binding protein tristetraprolin is positively correlated with HIF-1 target genes but acts as their negative regulator
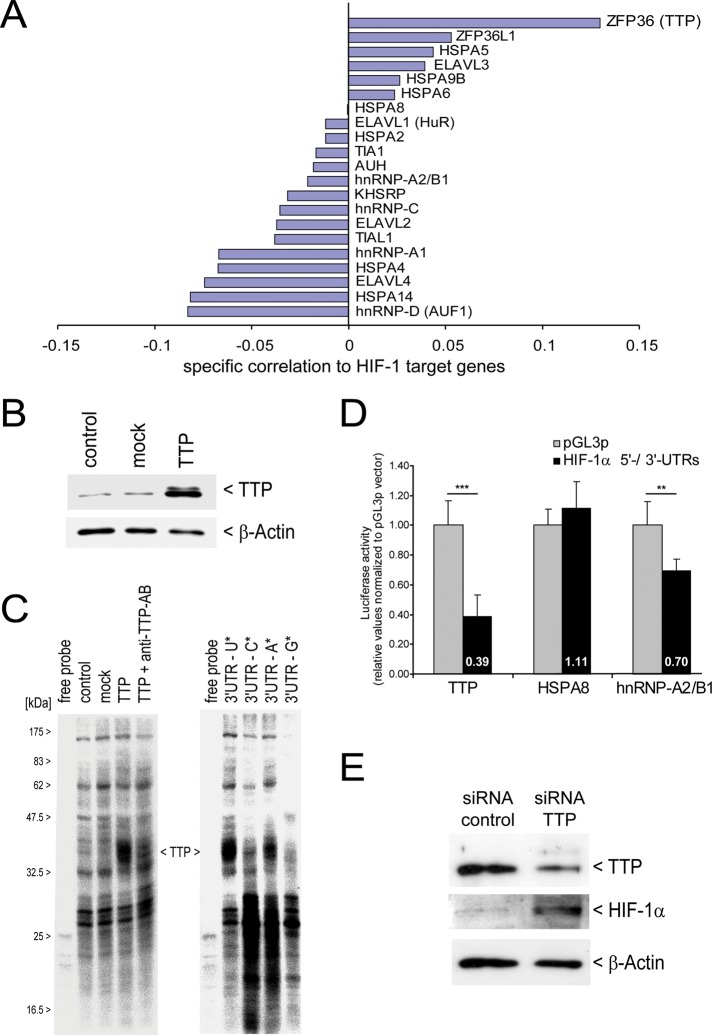
To identify trans factors that influence HIF-1α mRNA level, we used the same expression profile database and correlated the mRNA levels of known ARE-binding proteins (ARE-BPs) with the mRNA levels of verified HIF-1 target genes. We expected that factors stabilizing HIF-1α mRNA would show a positive correlation with HIF-1 target gene expression and factors that destabilize HIF-1α mRNA would show a negative correlation. As shown in Figure 2A, several ARE-BPs were not correlated with HIF-1 target gene expression (e.g., HSPA8), were only weakly correlated, or showed a clearly negative correlation, such as hnRNP-D (alias, AUF-1). In contrast, the factor with the strongest correlation, the ARE-BP TTP (ZFP36), is positively correlated with HIF-1 target genes. Because TTP has been described as an ARE-binding protein that mediates HIF-1α mRNA destabilization (Chamboredon et al., 2011; Kim et al., 2010), the strong positive correlation of TTP with HIF-1 target genes is unexpected and contradicts the current model of how TTP regulates HIF-1α expression.
FIGURE 2:
Influence of ARE-BPs on HIF-1. (A) Specific correlation of individual ARE-BP mRNAs with HIF-1 targets. A set of known AU-rich element–binding proteins was correlated with HIF-1 target genes (Figure 1C). Strongest positive correlation is observed for TTP. (B) HEK293 cells were nontransfected (control) or transiently transfected with empty vector (mock) or using a human TTP expression vector (TTP). Western blot for TTP; β-actin served as loading control. (C) UV cross-linking was performed using 32P-labeled transcripts representing the HIF-1α mRNA 3′ UTR. Left, radioactive signals represent RNA-BPs that directly bind to HIF-1α mRNA 3′ UTR. The signal representing TTP is indicated. The TTP signal can be suppressed by preincubation of the cytosolic extract that result from forced TTP expression (lane TTP in 2B) using a specific anti-TTP antibody (TTP + anti–TTP-AB). Right, HIF-1α mRNA 3′ UTR transcripts were separately labeled by U, C, A, or G and incubated with cytosolic extracts from cells overexpressing TTP (lane TTP in 2B). Effective label transfer is observed by A- and U-labeled transcripts. (D) HIF-1α mRNA UTR–dependent reporter gene assays following forced expression of select cotransfected ARE-BPs in HEK293 cells. (E) siRNA-mediated knockdown of TTP in human monocytic THP-1 cells. THP-1 cells were transfected with either control siRNA or siRNA targeting TTP mRNA. TTP and HIF-1α protein levels were assessed by Western blotting. Detection of β-actin served as loading control.
To confirm whether TTP regulates HIF-1α expression and subsequent HIF-1 activity in normoxia, we investigated binding and regulation of HIF-1α mRNA by TTP. After TTP overexpression in human embryonic kidney (HEK) 293 cells (Figure 2B), we applied an ultraviolet (UV) cross-linking method and compared in vitro binding of proteins to HIF-1α mRNA in cytosolic extracts from control cells (nontransfected), mock (empty vector) transfected cells, and cells overexpressing TTP. We observed a strong labeling of TTP after the UV cross-linking procedure as evidenced by an increase in signal intensity when TTP is overexpressed and followed by its decrease in the presence of a specific anti-TTP antibody. This confirms that TTP interacts directly with the HIF-1α mRNA 3′ UTR also at normoxic conditions (Figure 2C, left). Moreover, nucleotide-specific labeling of transcripts of the 3′ UTR of HIF-1α mRNA revealed that label transfer takes place preferentially with A- and U-labeled transcripts (Figure 2C, right). The latter finding provides evidence that TTP binds to AREs in the HIF-1α mRNA 3′ UTR.
Furthermore, we established a luciferase reporter gene assay to study the effect of TTP on HIF-1α mRNA UTR function in HEK293 cells that show weak basal TTP expression levels (Cao et al., 2006). To test the specific UTR-mediated response, we replaced the 5′ and 3′ UTRs of the firefly luciferase by the specific HIF-1α mRNA UTRs. In the correlation study (Figure 2A), heat-shock 70-kDa protein 8 (HSP-A8) showed no correlation with HIF-1 target genes, whereas heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP-A2/B1) showed a negative correlation. These data were confirmed in vitro in HIF-1α mRNA UTR-dependent reporter gene assays following forced expression of ARE-BPs HSP-A8 and hnRNP-A2/B1 in HEK293 cells (Figure 2D). However, forced expression of human TTP, as shown in Figure 2B, negatively affects the reporter activity (Figure 2D), which is in line with previous reports (Chamboredon et al., 2011; Kim et al., 2010) but is contrary to the positive correlation of TTP with HIF-1 target genes. We further confirmed the negative influence of TTP on HIF-1α gene expression by small interfering RNA (siRNA)–mediated down-regulation of TTP in THP-1 cells, a monocytic cell line that has a higher basal TTP expression level. TTP knockdown resulted in an increase in HIF-1α protein level (Figure 2E), again supporting the view of TTP being a negative regulator of HIF-1α synthesis. Taken together, these data indicate that TTP binds to the UTR of HIF-1α and negatively regulates HIF1α mRNA and protein levels in normoxia but is positively correlated with HIF-1 targets.
Mathematical modeling of TTP action predicts a phosphorylation-dependent positive effect on HIF-1 activity
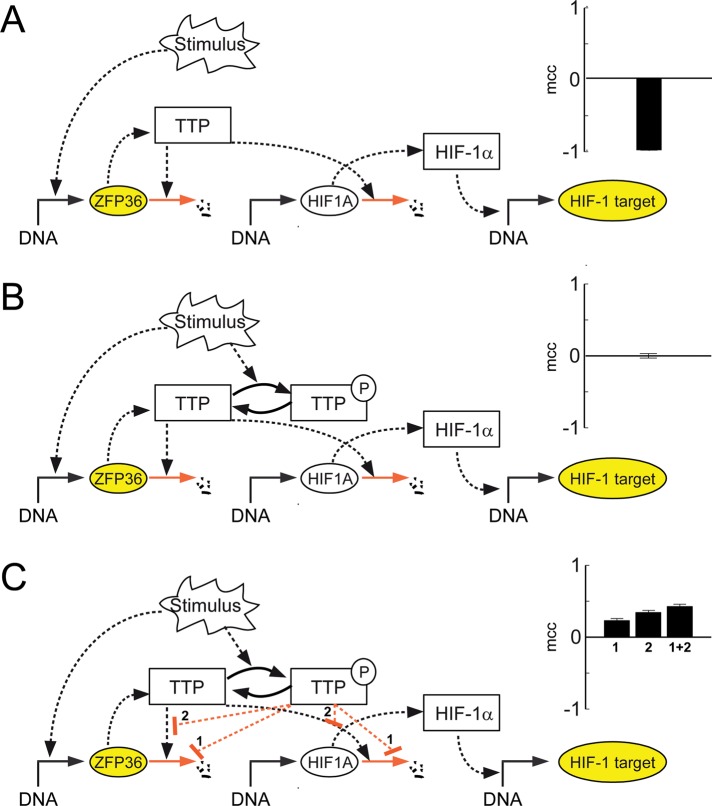
Because TTP was verified to be a negative regulator of HIF-1α expression, it is difficult to understand why TTP mRNA is strongly positively correlated with HIF-1 targets. To test different hypotheses, we decided to use mathematical modeling to test candidate mechanisms that could explain the observed correlation. We modeled TTP action on HIF-1 activity by applying a Monte Carlo simulation (as described in Materials and Methods). We tested the following three putative models of TTP action on HIF-1 signaling that would yield a negative, none, or a positive correlation.
The negative feedback model (Figure 3A) assumes that TTP acts as an mRNA-degrading (destabilizing) factor only (in agreement with previous reports and results shown in Figure 2, D and E). According to Brooks et al. (2004), it has to be kept in mind that TTP interacts with its own 3′ UTR. Thus the model assumes that TTP destabilizes the mRNA of HIF-1α, as well as its own mRNA. As a consequence, the predicted correlation of TTP mRNA and HIF-1 target gene expression is strongly negative and is not supported by the experimental findings obtained from the large-scale gene expression profiles (see Figure 2A).
FIGURE 3:
Mathematical model of HIF-1α regulation under normoxic condition predicts the dual role of TTP. For each potential structure of the networks depicted in A–C, a model based on modular response analysis was constructed. Parameters were chosen randomly in a Monte Carlo approach, and the frequency of positive and negative coregulation of ZFP36 and HIF-1 targets (both marked as yellow filled ovals) upon stimulation was calculated and evaluated using the Matthews correlation coefficient (mcc). (A) Negative feedback model: TTP mRNA (ZFP36) is constitutively expressed, and TTP protein destabilizes both its own and mRNA of HIF-1α (HIF1A). Model simulation predicts that TTP mRNA is anticorrelated with HIF-1 target mRNAs. (B) TTP protein sequestration model: If in addition TTP is phosphorylated, and the phosphorylated form does not destabilize 3′ UTR–containing mRNA, model simulation predicts no correlation between mRNA levels of TTP and HIF-1 targets. (C) Competition model: If either binding of phosphorylated TTP actively stabilizes the bound mRNA (scenario1) or phosphorylated TTP competes with TTP for limited binding sites (scenario 2), model simulation predicts positive correlation of the two mRNAs. Both mechanisms in combination would further increase the positive correlation (scenarios 1 + 2).
The protein sequestration model (Figure 3B) assumes that TTP can be phosphorylated and that phosphorylation of TTP causes release of the bound mRNAs, thus enhancing gene expression rate by abolishing its destabilizing influence. This model is based on findings indicating that phosphorylated TTP does not interact with target mRNAs (Carballo et al., 2001; Hitti et al., 2006). Prediction by this simulation indicated no correlation of TTP mRNA and HIF-1 targets and thus also is not supported by the experimental findings obtained from the correlation studies.
The competition model (Figure 3C) assumes that phosphorylated TTP is able to recognize and interact with target mRNAs and that binding of phosphorylated TTP further stabilizes bound mRNAs. Thereby the stabilization effect could be perceived in two scenarios. One way is the direct stabilization of the mRNA (scenario 1), which is based on findings published earlier showing that TTP phosphorylation by p38 MAPK/MAPK-activated protein kinase 2 (MK2) causes transcript stabilization and recruitment of bound transcripts into polysomes, leading to an elevated gene expression rate of TTP targets (Sandler and Stoecklin, 2008; Stoecklin et al., 2008). Accordingly, it was recently shown that phosphorylation of TTP by MK2 primarily prevents recruitment of the deadenylation machinery downstream of RNA binding (Clement et al., 2011). In the second scenario, stabilization was modeled by assuming that phosphorylated and unphosphorylated TTP bind to the same sequence of 3′ UTR but with only unphosphorylated TTP being able to induce degradation (scenario 2). In both cases, model simulation predicts a positive correlation of TTP mRNA and HIF-1 target genes, which is further enhanced if one assumes that scenarios 1 and 2 occur simultaneously. Therefore we conclude that either one or both scenarios of the competition model would provide an explanation for the observed positive correlation of TTP mRNA and HIF-1 target genes.
Phosphorylation of TTP is critical for its influence on HIF-1α synthesis
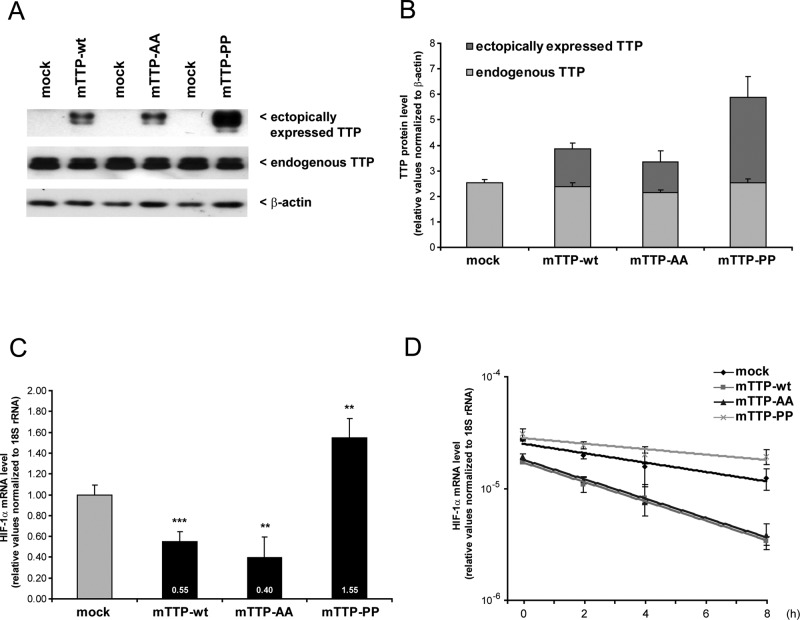
According to the competition model, posttranslational modification of TTP by, for example, phosphorylation may be crucial for understanding how HIF-1α is regulated. Multiple pathways are known to phosphorylate TTP, among them the p38 MAPK/MK2 pathway. Therefore we hypothesized that p38 MAPK may regulate TTP function with regard to the control of posttranscriptional HIF-1α gene expression. To test this hypothesis, we expressed murine TTP mutants as described in Stoecklin et al. (2004) in HEK293 cells (Figure 4A). The S52A/S178A double mutant mTTP-AA cannot be phosphorylated by p38 MAPK/MK2 and effectively prevents the assembly of TTP:14-3-3 complexes. In contrast, mutation of V54 and S180 to proline (mTTP-PP) confers strong binding to 14-3-3 and thus mimics phosphorylated TTP (Stoecklin et al., 2004). We observed differences in mTTP expression levels between murine wild-type TTP (mTTP-wt), mTTP-AA, and mTTP-PP that are consistent with a previous report indicating that phosphorylation of TTP leads to decreased TTP degradation (Brook et al., 2006). Consistently, we found highest accumulation of mTTP-PP; however, ectopically expressed mTTP variants ranged between ratios of ∼0.5:1 and ∼1:1 compared with endogenous TTP (Figure 4, A and B). In support of our findings that TTP causes HIF-1α mRNA degradation, we observed down-regulation of HIF-1α mRNA level (Figure 4C) as well as stability (Figure 4D) after forced expression of mTTP-wt and mTTP-AA, with a stronger tendency mediated by the nonphosphorylatable mTTP-AA variant. However, the opposite influence was found in overexpression of mTTP-PP, which causes a significant up-regulation of HIF-1α mRNA level, which is in line with a weak mRNA stabilization compared with mock transfection (Figure 4, C and D). In Figure 4B we observe a >50% proportion of ectopically expressed mTTP-PP in relation to endogenous TTP protein levels. Because mTTP-PP increases HIF-1α mRNA levels significantly (Figure 4C), this suggests an actively stabilizing role of TTP, depending on its phosphorylation state.
FIGURE 4:
Influence of ectopically expressed TTP on HIF-1α mRNA. HEK293 cells were transfected for 24 h with backbone vector (mock) or murine wild-type TTP (mTTP-wt). Mutated variants that either cannot phosphorylate (mTTP-AA) or mimic phosphorylated TTP by enhanced binding of 14-3-3 were transfected accordingly as described earlier (Stoecklin et al., 2004.). (A) Western blotting to detect endogenous and ectopically expressed TTP. A representative blot is shown. (B) Quantification of endogenous and ectopically expressed TTP expression levels. n = 3. (C) HIF-1α mRNA quantification after forced expression of murine TTP, as well as mutated TTP variants. Wild-type TTP and nonphosphorylatable TTP caused down-regulation of HIF-1α mRNA. In contrast, overexpression of mTTP-PP, which mimics phosphorylated TTP, leads to increased HIF-1α mRNA level. (D) mRNA stability assays indicate that the alterations in HIF-1α mRNA levels are attributed to changes in its half-life (relative to mock: mTTP-wt, 0.44 fold**; mTTP-AA, 0.44 fold**; mTTP-PP, 1.39-fold*). n = 6; *p < 0.05, **p < 0.01, ***p < 0.001.
The data support the notion that TTP acts directly via the HIF-1α mRNA 3′ UTR in its nonphosphorylated as well as in phosphorylated form. Because ectopically expressed mTTP can further lead to lowering of HIF-1α mRNA stability, which is prevented by the mTTP-PP mutant that mimics phosphorylated TTP, the enhanced HIF-1α mRNA level may be explained either by competition of nonphosphorylated and phosphorylated TTP or by active stabilization as outlined in the competition model (see Figure 3C).
These data support mathematical modeling indicating that TTP can function as inhibitor as well as enhancer of HIF-1α synthesis, depending on the activation of the p38 MAPK pathway.
p38 MAPK and TTP regulate HIF-1α expression in a model of monocyte-to-macrophage differentiation
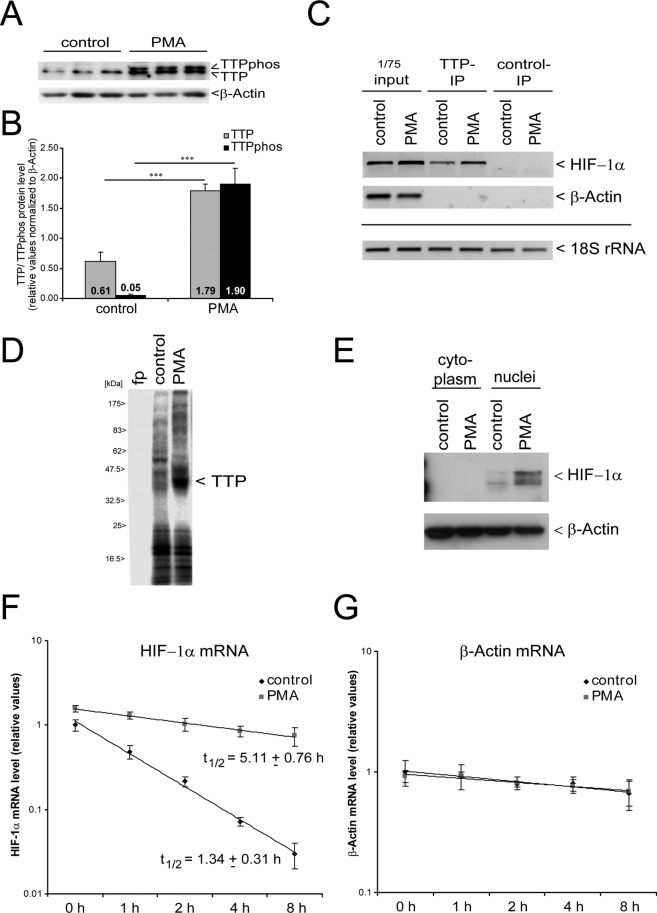
To generalize and validate our data in a physiological model, we screened for conditions in which TTP is phosphorylated in monocytic THP-1 cells. We observed that treatment of THP-1 cells with phorbol 12-myristate 13-acetate (PMA) leads to phosphorylation of TTP (TTPphos) (Figure 5, A and B), as seen by a doublet on SDS–PAGE gel (Cao et al., 2006). PMA administration induces the differentiation of monocytic THP-1 cells to adherent growing macrophages (Auwerx, 1991). Although we observed a general increase in TTP protein level after PMA treatment, the TTP:TTPphos ratio markedly shifted in favor of the phosphorylated form (increased TTP level is ∼3-fold and that of TTPphos is ∼26-fold).
FIGURE 5:
HIF-1 activation during monocytic cell (THP-1) differentiation induced by PMA treatment. (A) Western blot analysis of TTP after differentiation of THP-1 cells by PMA. Detection of relative β-actin levels served as a loading control. Representative samples of three of six independent experiments are shown. (B) Statistical analysis of PMA-induced TTP synthesis and phosphorylation. n = 6. **p < 0.01, ***p < 0.001. (C) Ribonucleoprotein immunoprecipitation using a specific anti-TTP antibody or a control antibody and detection of HIF-1α mRNA via real-time PCR in controls or PMA-treated THP-1 cells. Detection of β-actin mRNA served as negative control. 18S rRNA was used to facilitate RNA precipitation after immunoprecipitation and thus served as positive control. (D) UV cross-linking assay using the HIF-1α mRNA 3′ UTR and cytosolic extracts from undifferentiated (control) and differentiated (PMA) cells. fp, free probe (negative control, labeled transcript only). The signal representing TTP is indicated. (E) Western blot analysis of HIF-1α protein level after PMA-mediated differentiation. β-Actin served as a loading control. A representative blot out of six independent experiments is shown. (F, G) mRNA stability assays were performed in order to estimate the half-life of HIF-1α mRNA in undifferentiated (control) or differentiated (PMA) THP-1 cells. PMA-mediated differentiation leads to increased HIF-1α mRNA stability (F), whereas mRNA stability of β-actin mRNA is stable (G).
Ribonucleoprotein immunoprecipitation using a specific anti-TTP antibody revealed binding of endogenous TTP to HIF-1α mRNA under conditions of nonphosphorylated TTP (control), as well as favorably phosphorylated TTP after PMA treatment (Figure 5C).
Moreover, UV cross-linking assays revealed a marked label transfer from an in vitro transcript representing the HIF-1α mRNA 3′ UTR to TTP after PMA treatment of THP-1 cells. This supports the view that TTP is a crucial trans factor interacting with the HIF-1α mRNA during macrophagocytic differentiation (Figure 5D). Both data sets—endogenous TTP protein:HIF-1α mRNA interaction and in vitro label transfer—crucially support the competition model (see Figure 3C). TTP phosphorylation and binding to the HIF-1α mRNA 3′ UTR are associated with elevated HIF-1α protein and mRNA levels (Figures 5E and 6C), as well as a 3.8-fold elevated HIF-1α mRNA stability (Figure 5F). For comparison, as seen in Figure 5G, PMA treatment had no effect on β-actin mRNA stability, which was used as a control transcript. Our findings suggest that TTP-mediated posttranscriptional control of HIF-1α expression in THP-1 cells after PMA-induced macrophagocytic differentiation participates in activated HIF-1 signaling.
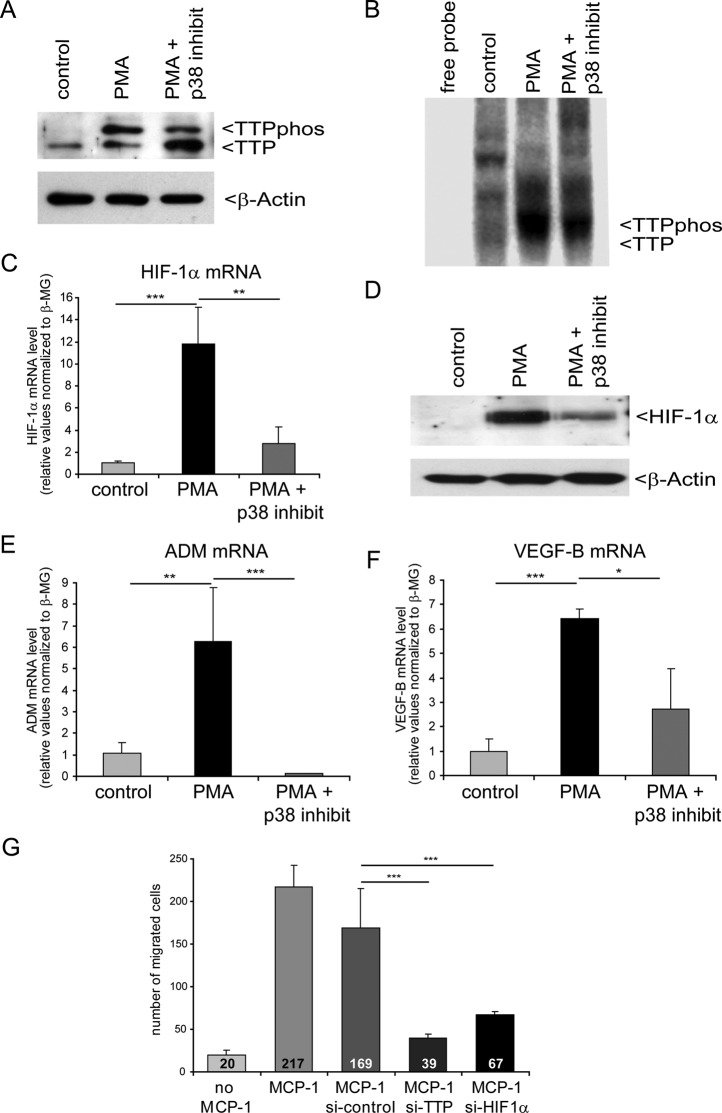
FIGURE 6:
Inhibition of p38 MAPK suppresses TTP-mediated HIF-1 activation during macrophage differentiation. For Western blot analyses and UV cross-linking assays, pools of six independent samples for each condition are shown. (A) Western blot analysis from undifferentiated (control), differentiated (PMA), and differentiated THP-1 cells with p38 MAPK inhibition (PMA + p38 inhibit). The phosphorylated TTP (TTPphos) and nonphosphorylated TTP forms are indicated. Detection of relative β-actin levels served as a loading control. (B) UV cross-linking assay using U-labeled HIF-1α mRNA 3′ UTR and cytosolic extracts as shown in A. (C) Estimation of relative HIF-1α mRNA level and (D) protein level after THP-1 cell differentiation in comparison to differentiation while p38 MAPK was inhibited. (E, F) Determination of HIF-1 target gene mRNA levels via real-time PCR. mRNA levels were normalized to levels of β2-macroglobulin. n = 12; *p < 0.05, **p < 0.01, ***p < 0.001. (G) Cell migration assay. Macrophage migration was assessed using THP-1 cells in a modified Boyden chamber assay. MCP-1 was used to induce chemotactic movement of THP-1 cells. The number of macrophages that had migrated to the underside of the membranes was counted and is indicated in the figure. Untreated cells without (no MCP-1) or with MCP-1 served as negative or positive control, respectively. Knockdown of either TTP or HIF-1α was performed by siRNA transfection. Pools of nontargeting siRNAs were used for control transfection (si-control). **p < 0.01, ***p < 0.001.
To provide further supporting evidence that p38 MAPK-dependent phosphorylation of TTP is involved in elevated HIF-1α activity, we sought to inhibit the phosphorylation of TTP during monocyte-to-macrophage differentiation by simultaneously inducing differentiation of THP-1 cells and inhibiting p38 MAPK with SB 220025, a routinely used p38 MAPK inhibitor. Of interest, p38 MAPK inhibition did not prevent the increase in TTP protein levels but caused an alteration of the TTP phosphorylation state in favor of the nonphosphorylated form (Figure 6A). UV cross-linking assays indicated that this change in TTP:TTPphos-ratio does not inhibit binding of TTP to the HIF-1α mRNA 3′ UTR, although there was a tendency toward a decreased affinity (Figure 6B). The decreased phosphorylation of TTP by inhibition of p38 MAPK correlated well with decreased HIF-1α mRNA and protein levels (Figure 6, C and D). In addition, mRNA levels of HIF-1 target genes adrenomedullin (ADM) and VEGF-B were reduced upon p38 MAPK inhibition (Figure 6, E and F), indicating decreased HIF-1 activity. It is important to note that both target genes showed an increase upon PMA treatment alone, reflecting increased HIF-1 activity that might be at least in part a consequence of the enhanced HIF-1α mRNA stability that was observed. The decrease in ADM mRNA level to a nearly undetectable level after p38 MAPK inhibition is surprising and cannot be explained by HIF-1 activity alone and warrants further investigation.
An interesting experiment would be to test whether knockdown of TTP leads to reduced levels of HIF1α in PMA-stimulated THP1 macrophages. However, there is a drastically reduced viability of THP1 cells after transfection in a maturation setting. This observation is in line with data form other groups (Schnoor et al., 2009). Therefore the study is limited regarding this issue.
HIF-1α is crucial for cellular aggregation, motility, and invasiveness of myeloid cells (Cramer et al., 2003). To correlate our data with HIF-1–dependent cellular response under normoxic conditions, we quantified cell migration in the presence or absence of TTP. Cell migration was measured using a modified Boyden chamber assay toward a monocyte chemoattractant protein-1 (MCP-1) gradient. MCP-1 alone does not induce monocytic differentiation to macrophages, and thus does not induce HIF-1 activity. Under these conditions, cellular differentiation and HIF-1 activation take place when the cells attach to the porous membrane and start to migrate. Under control conditions, MCP-1 induced monocytic cell migration, which was not affected by transfection with a scrambled siRNA (Figure 6G). However, down-regulation of TTP by siRNA treatment significantly impaired monocyte migration to an extent similar to that seen after siRNA-mediated HIF-1α knockdown (Figure 6G). These data strongly support the pivotal function of the multilevel control of HIF-1 by TTP during myeloid cell differentiation.
DISCUSSION
Our results highlight the crucial role of HIF-1α mRNA turnover in HIF-1 signaling independent of the oxygen tension. By a multispecies sequence alignment that included more than 15 species, we demonstrate that the 3′ UTR of HIF-1α mRNA is strongly conserved among multiple vertebrate species. By ranking all genes regarding the conservation score of its 3′ UTR, we find that the HIF-1α mRNA 3′ UTR is among the top 20 of 7953 genes, with a UTR length of ≥1000 nucleotides. These observations, in combination with the AU-rich context of the HIF-1α mRNA 3′ UTR, indicate a regulatory role of mRNA turnover as a modulator of HIF-1α function. By evaluating large-scale microarray-based gene expression profiles, we identified a novel role for the ARE-BP TTP. Contrary to its known role in destabilizing mRNAs, we show that TTP binds to the HIF-1α mRNA 3′ UTR in its phosphorylated form, causing HIF-1α mRNA stabilization by competition with nonphosphorylated TTP, active stabilization, or both. We provide several lines of evidence for this function, including mathematical modeling, ribonucleoprotein immunoprecipitation, in vitro binding assays, and a monocyte-to-macrophage differentiation model. The stabilized transcript is functional, as evidenced by an increase in HIF-1α protein level and HIF-1 activity. We also show that, when dephosphorylated, TTP performs its known destabilizing function, as previously reported (Chamboredon et al., 2011; Kim et al., 2010). The correspondence of our findings with data that we analyzed from large-scale microarray-based gene expression profiles from a variety of different experimental conditions further supports our conclusion. A number of trans factors have been reported to interact with the HIF-1α mRNA, including RNA-binding proteins like Hu antigen R (HuR, ELAVL1) and polypyrimidine tract binding protein, both of them with known function for mRNA turnover (Galban and Gorospe, 2009; Gorospe et al., 2011). However, the only confirmed trans factor that is able to modulate HIF-1α mRNA stability is the RNA-BP TTP. In two independent studies TTP has been implicated in down-regulation of HIF-1α under hypoxic conditions (Kim et al., 2010; Chamboredon et al., 2011). Of note, here we provide evidence that TTP is able to modulate the HIF-1 pathway under normoxic conditions. The ARE-binding zinc-finger protein TTP is known to destabilize bound mRNAs (Bermudez et al., 2011). It was first demonstrated for the proinflammatory cytokine TNF-α (Carballo et al., 1998), which is strongly up-regulated in TTP knockout mice. As a consequence, TTP knockout mice develop generalized inflammation (Taylor et al., 1996). Between 100 and 250 mRNA targets have been described for TTP (Lai et al., 2006; Stoecklin et al., 2008), many of them being cytokine mRNAs and participating in the inflammatory response. Of interest, it was only by applying large-scale mRNA correlation studies, based on microarray data from 1202 different database entries and thus experimental conditions, that we found the ARE-BP TTP with highest positive correlation to HIF-1 target genes. At first glance, this might contradict the notion of TTP being a destabilizing factor. However, we provide evidence that TTP-mediated HIF-1 activation is dependent on TTP phosphorylation state. In its phosphorylated form, TTP has a stabilizing effect on HIF-1α mRNA. In different mathematical models based on a Monte Carlo simulation, only one model, namely the competition model, was able to predict the experimental findings of a positive correlation of TTP mRNA with HIF-1 target genes. This model assumed that phosphorylated TTP binds to target mRNAs and mediates mRNA stabilization either by competition with nonphosphorylated TTP or by active stabilization, which is consistent with previous reports showing that phosphorylation of TTP by MK2, itself a target of p38 MAPK, allows binding of 14-3-3 adaptor proteins, causing TTP-mediated mRNA stabilization and recruitment into polysomes (Johnson et al., 2002; Stoecklin et al., 2004; Sandler and Stoecklin, 2008). Phosphorylation of TTP by MK2 has also been shown to stabilize the transcript by its inability to recruit the deadenylation machinery to the target mRNA (Clement et al., 2011).
Activated p38 MAPK inhibits TTP-mediated repression of cytokine mRNAs. One study implicated p38 MAPK in stabilizing proinflammatory mRNAs by inhibiting the ARE-mediated decay by TTP (Tudor et al., 2009), whereas another showed that an increase in interleukin 6 (IL6) mRNA transcript stability is regulated by TTP through an alteration in the affinity for the transcript that occurs in a p38 MAPK-dependent manner (Zhao et al., 2011). However, although the underlying molecular mechanisms of TTP function are not yet completely understood, it is clear that p38 MAPK/MK2 phosphorylation of TTP plays a definite role. In an effort to understand more about the phosphorylation-dependent function of TTP, several phosphorylation-dependent TTP interacting partners have been identified (Tiedje et al., 2010). Of interest, these include the RBPs HuR and AUF1, both of which are known HIF-1α mRNA–binding proteins. However, our UV cross-linking experiments revealed that TTP, as well as phosphorylated TTP, binds directly at the HIF-1α mRNA 3′ UTR via AU-rich elements. To our knowledge, this is the first study that links phosphorylation of TTP to HIF-1α mRNA stability and therefore HIF-1 activity under normoxic conditions.
Although inhibition of p38 MAPK during PMA-mediated THP-1 cell differentiation suppresses TTP phosphorylation and subsequent HIF-1 activity, it does not abolish TTP phosphorylation, indicating alternate routes to TTP phosphorylation than by p38 MAPK alone. This is in line with findings indicating that HIF-1α mRNA induction in response to PMA-induced differentiation can be suppressed by protein kinase C (PKC) inhibition (Oda et al., 2006). Of interest, PKC has also been implicated in TTP phosphorylation (Cao et al., 2006). These results suggest that PMA-induced differentiation of THP-1 cells activates TTP at multiple levels. Moreover, among p38 MAPK, MK2, and PKC, it has been predicted that TTP bears at least 10 high-probability in vivo sites for phosphorylation, which are potential sites for further kinases such as protein kinase A and B, glycogen synthase kinase-3, c-Jun N-terminal kinase, and extracellular signal-regulated kinases 1 and 2 (Cao et al., 2006). How these kinases (alone or in combination) influence TTP function regarding mRNA binding and modulation of mRNA fate remains to be elucidated. In the case of macrophagocytic differentiation one may hypothesize that the combined action of both pathways, p38 MAPK/MK2 and PKC, synergistically causes TTP-mediated HIF-1 activation.
The finding that the anti-inflammatory factor TTP is crucial for HIF-1 activation during macrophagocytic differentiation is important for the understanding of macrophage function. Several studies have shown that activation of HIF-1 by inflammatory mediators is crucial for survival, differentiation, and functionality of immune cells (Frede et al., 2007). Furthermore, in myeloid-specific HIF-1α knockout mice, HIF-1 was identified as an indispensable transcription factor that coordinates gene expression in myeloid cells during their extravasation (Cramer et al., 2003). Macrophages typically produce most of their ATP through glycolysis, a process known to be activated by HIF-1. Enhanced expression of glycolytic enzymes might be necessary for the elevated energy demand of activated macrophages. Consistently, in our experiments TTP, as well as HIF-1α knockdown, in THP-1 cells showed a loss in their ability to migrate.
Increasing evidence suggests a role of macrophages in a different stress response mechanism, namely the adaptive growth of blood vessels in response to vascular stenosis or occlusion (Carmeliet, 2005). Inflammatory angiogenesis is promoted by macrophages, which infiltrate tissues in response to inflammatory stimuli and produce angiogenesis-promoting factors such as VEGF, interleukins, and matrix metalloproteinases (Ono, 2008)—all of them known HIF targets. The pivotal role of HIF-1 in tumor development is undisputable. Recently it was demonstrated that TTP regulates an important subset of cancer-related genes that are involved in cellular growth, invasion, and metastasis (Al Souhibani et al., 2010). Angiogenesis and inflammation share common pathways that are closely coupled to cancer (Ono, 2008). Therefore TTP-mediated HIF-1 activation in macrophages may be relevant for tumor growth and vascularization.
In summary, this study provides a new perspective on the function of TTP on HIF-1α mRNA stability independent of hypoxia. By combining different approaches, we consistently showed that the influence of TTP on HIF-1α expression depends on its phosphorylation status, which is modulated by p38 MAPK/MK2 signaling. The role of TTP in the activation of HIF-1–dependent pathways provides a new link between inflammation and the HIF-1 response, as well as nonhypoxic HIF-1 activation that to date has not been well defined.
MATERIALS AND METHODS
Estimation of conservation score of UTRs and coding sequence
We obtained genome-wide conservation scores based on the phastCons method (Siepel et al., 2005). The data contain conservation scores for multiple alignments of the following assemblies to the human genome (Mar. 2006 (NCBI36/hg18), Human Genome Browser–hg18 assembly): chimp, mouse, rat, rabbit, macaque, dog, cow, armadillo, elephant, tenrec, opossum, chicken, frog, zebrafish, tetraodon, and fugu. The data originate from ftp://hgdownload.cse.ucsc.edu, downloaded in October 2008. A c++ program was devised that both maps data sets and calculates a conservation score for each specific region of the human genome.
mRNA correlation studies
Human microarray data were obtained from the Stanford Microarray Database (Demeter et al., 2007) as used in Stuart et al. (2003). All preprocessing, such as normalization of the microarray data, has been previously described (Stuart et al., 2003). This set contains 13,555 gene entries and 12,435 genes out of those matched to ENSEMBL genes. The data set used contains 1202 experiments. The data contain a collection of different independent investigations, including expression profiles from tumor samples, cancer cell lines, and different organ-specific tissue samples, as well as expression data from studies of diverse biological processes such as cell cycle, stress, signaling, and apoptosis. For correlating HIF-1α mRNA levels with verified HIF-1 target genes we chose an HIF-1 target gene compilation by Semenza (2003). The same HIF-1 target set was used for calculating the expression correlation with correlation of ARE-BP mRNAs.
Modeling
To assess whether a model structure results in positive, no, or negative correlation, we applied mathematical modeling based on the concepts of modular response analysis (Bruggeman et al., 2002; Kholodenko et al., 2002). This concept relates the so-called global response matrix R (response of the entire network to a perturbation) to the local response matrix r (direct regulatory strength between two adjacent nodes in a network):
|
|
Here u is a vector containing the perturbation. For the biological network considered here, the regulatory strength between nodes is unknown, but the sign (i.e., inhibition or activation) is given by the network structure. Hence, we applied a Monte Carlo method, by which we randomly generated instances of local response matrices that are compatible with the reaction network. As a realization of r as an adjacency matrix representing the network structure, each entry of r was either set to 0, if no direct interaction is present in the model, or given a random number drawn from the standard uniform distribution on the open interval (0, 1) multiplied with 1 or −1 if the edge is activatory or inhibitory, respectively. Next r and an arbitrary perturbation strength (drawn from the standard normal distribution) were applied to Equation 1, yielding the steady state matrix (R) from which the entries corresponding to stimulus-to-TTP mRNA and stimulus-to-HIF-1 target mRNA were extracted and converted into binaries (up or down). This procedure was repeated 1 million times, collecting the binary data for both mRNAs into vectors. Then, as a measure of correlation, the Matthews r was calculated from the two binary vectors.
Cell culture
HEK293 cells (ATCC 305) were obtained from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). HEK293 cells were maintained in DMEM (high glucose; PAA Laboratories, Pasching, Austria), supplemented with 10% fetal calf serum (Biochrom KG, Berlin, Germany), 50 U/ml penicillin, 50 μg/ml streptomycin, 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and 2 mmol/l glutamine at 37°C and 5% CO2.
Human monocytic THP-1 cells (ATCC TIB-202) were obtained from the American Type Culture Collection (Manassas, VA). THP-1 cells were cultivated in RPMI 1640 medium (PAA Laboratories) supplemented with 10% fetal calf serum, 100 U/ml penicillin, 1% glutamate, and 100 μg/ml streptomycin (all from Invitrogen, Carlsbad, CA). For experiments with undifferentiated cells, before all experiments, undifferentiated cells were expanded to a concentration of 106 cells/ml at 37°C in a humidified atmosphere of 5% CO2 in air. Phagocytic differentiation of THP-1 cells was induced by treating 106 cells/ml with phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, Munich, Germany) at a final concentration of 1 μmol/l for 24 h. p38 MAPK inhibition was achieved using SB 220025 (10 μmol/l; Sigma-Aldrich) for 24 h during THP-1 cell differentiation.
Cell transfection and HIF-1α mRNA UTR–dependent reporter gene assays
For reporter gene assays the firefly luciferase vector pGL3-promotor (Promega, Madison, WI) was used. The vector-specific 5′ and 3′ UTRs of firefly luciferase mRNA were replaced by the human HIF-1α UTRs. The quality of processed vectors was confirmed by sequencing. The resulting vector construct expressed a constitutively transcribed firefly luciferase transcript (SV40 promoter) bearing the specific HIF-1α mRNA 5′ and 3′ UTRs.
Cells were cultured in 96-well plates (μClear Platte 96K; Greiner Bio-One, Frickenhausen, Germany) and cotransfected with the modified pGL3-promotor vector variant bearing the HIF-1α mRNA UTRs and the Renilla luciferase phRL-TK vector using the FuGENE 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's protocol. Cell transfection using the original pGL3-promotor vector served as control. Forced expression of ARE-BPs was performed by cotransfection using a fivefold excess of ARE-BP expression vectors compared with pGL3-promoter vector. Full-length human cDNA expression vectors were purchased from the Deutsches Ressourcenzentrum für Genomforschung GmbH (Heidelberg, Germany). TTP: clone, IRATp970E0610D6; hnRNP-A2/B1: clone, IOH-3534; HSPA8: clone, IOH-11329). The myc-tagged murine wild-type TTP (mTTP-wt); the S52A/S178A double mutant TTP (mTTP-AA), which is not phosphorylated by p38 MAPK/MK2 and cannot bind 14-3-3; and the TTP variant with V54 and S180 to proline mutation, which binds to 14-3-3 constitutively (mTTP-PP), were generous gifts from Georg Stoecklin (Deutsches Krebsforschungszentrum DKFZ-ZMBH Allianz, Heidelberg, Germany) and have been described (Stoecklin et al., 2004).
After 6 h, the transfection medium was removed and fresh medium was added. Cells were harvested 24 h after transfection for mRNA and protein detection. The luciferase activity was detected as described earlier (Fähling et al., 2006a) using a luminometer (Luminoscan RS, Labsystems, Helsinki, Finland) programmed with individual software (Luminoscan RII, Ralf Mrowka).
RNA interference–mediated TTP knockdown
For siRNA transfection, THP-1 cells were used at densities of 4 × 105 cells/ml. siRNAs against human TTP, human HIF-1α, or a pool of nontargeting siRNAs (all from Dharmacon, Lafayette, CO) without homology to any vertebrate transcriptome as a negative control were transfected using DharmaFECT (Perbio Science, Helsingborg, Sweden) according to the manufacturer's instructions. For maximal efficiency, a mix of four targeting siRNAs (50 pmol/well) was used. At 48 h after siRNA transfection, differentiation was induced using PMA for another 24 h as described.
Cell migration assay
Macrophage migration was assessed in a modified Boyden chamber assay (Kappert et al., 2009). In brief, THP-1 monocytes were seeded into 8-μm polycarbonate membrane Transwell inserts (250,000/ml). Chambers were inserted into 24-well culture dishes containing MCP-1 (50 ng/ml). After 3 h at culture conditions, migrated cells were fixed and stained, and migration was quantified by determining the number of macrophages that had migrated to the underside of membranes per high-power field, counting three randomly chosen high-power fields per filter. Results are means ± SD from three independent experiments performed in triplicate.
RNA isolation, real-time PCR, and mRNA stability
Total RNA was isolated from cells using the TRIzol system (Invitrogen) according to the manufacturer's protocol. First-strand cDNA synthesis was performed by using random hexamers (Superscript II Reverse Transcriptase Kit; Invitrogen). Primers were designed using the VectorNTI Software (Invitrogen) bridging at least one intron. Real-time PCR amplification was performed in triplicate with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on a GeneAmp5700 thermocycler (PerkinElmer Life Sciences, Waltham, MA). A list of the quantitative PCR primers used in this study is provided in Supplemental Table S2. The expression levels of HIF-1α mRNA were normalized to the housekeeping genes β2-macroglobulin and β-actin, respectively, using the ΔCt method. Parallelism of standard curves of the test and control was confirmed.
Estimation of mRNA stability was performed as described earlier (Fähling et al., 2006a).
Protein isolation and immunodetection
Isolation of protein fractions for HIF-1α quantification was performed as described in Schreiber et al. (1989). Cytosolic protein extracts for Western blotting and UV cross-linking assays were prepared from cultured cells as described previously (Fähling et al., 2006b). The following primary antibodies were used: 1) polyclonal mouse anti–human HIF-1α (H72320; BD Biosciences, Heidelberg, Germany), 2) polyclonal rabbit anti–human ZFP36 (tristetraprolin) antibody (ARP34385; Aviva Systems Biology, San Diego, CA), and 3) polyclonal goat anti–human β-actin (sc-47778; Santa Cruz Biotechnology). Primary antibodies were detected using peroxidase-coupled goat anti–mouse immunoglobulin G (IgG; sc-2031; Santa Cruz Biotechnology) or donkey anti–rabbit IgG (sc-2317; Santa Cruz Biotechnology), respectively. Reaction products were visualized with an enhanced chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ). The anti–β-actin antibody was applied after stripping of the membranes with 0.2 M NaOH at room temperature for 2–5 min to detect differences in protein loading.
Immunoprecipitation assay
The method was adapted from Niranjanakumari et al. (2002). HEK293 cells were harvested as described and centrifuged at 2000 rpm for 2 min in a 15-ml conical tube. To cross-link RNA–protein interactions, 0.2% formaldehyde/PBS was added to the cell pellet and incubated at room temperature with rotation. After 10 min, 1 ml of 1.5 M glycine, pH 7.0, was added and incubated for 5–10 min. Cell pellets were washed twice with PBS and sedimented. The cross-linked cell pellet was resuspended in 1.5 ml of Empigen buffer (20 mM Tris, pH 7.5, 500 mM NaCl, 2.5 mM MgCl2, 1% Empigen, 200 U/ml RNase inhibitor), and cells were lysed by sonication. The supernatant of a 10,000 × g centrifugation for 10 min at 4°C was used for immunoprecipitation. Before use in experiments, 20 μl of protein G–Sepharose (Protein G Sepharose 4 Fast Flow, 17-0618-01; GE Healthcare, Piscataway, NJ) per reaction was washed and equilibrated in washing buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.01% Triton X-100). The polyclonal rabbit anti–human ZFP36 (tristetraprolin) antibody or a control antibody (IgG anti-rabbit) were prebound to protein G–Sepharose for 1 h. After three washing steps, cellular extracts were transferred to the protein G–Sepharose beads and incubated for 2 h at 4°C with rotation. Beads were gently washed five times at 4°C for 10 min with rotation. Reverse cross-linking was performed by addition of buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol) and incubation at 70°C for 2 h. Finally, SDS was supplemented to a final concentration of 2%. Rabbit rRNA was added as RNA precipitation facilitator and positive control. RNA extraction using TRIzol and HIF-1α detection was performed as described.
UV cross-linking assay
Transcript labeling by in vitro transcription and subsequent UV cross-linking was performed as described (Fähling et al., 2006b). Briefly, transcripts representing the 3′ UTR of HIF-1α mRNA were radioactively labeled using [α-32P]uridine‑, [α-32P]cytosine-, [α-32P]adenine-, or [α-32P]guanosine-5′-triphosphate (800 Ci/mmol; MP Biomedicals Germany, Eschwege, Germany) nucleotides. In vitro transcripts were purified by BD Chroma Spin-100 (DEPC) columns (BD Biosciences). RNA/protein assembling was performed by incubating labeled transcripts (1–2 ng) with cytosolic protein extracts (35 μg) for 30 min at room temperature in 10 mM HEPES, pH 7.2, 3 mM MgCl2, 5% glycerol, 1 mM dithiothreitol, 150 mM KCl, and 2 U/μl RNaseOUT (Invitrogen) in the presence of rabbit rRNA (0.5 μg/μl) as competitor. After RNA–protein assembling, samples were exposed to UV light (255 nm, 1.6 J, UV Stratalinker; Stratagene, Santa Clara, CA) on ice and subsequently treated with RNase A (30 μg/ml final concentration) and RNase T1 (750 U/ml final concentration) for 15 min at 37°C. Protein extracts were separated in 12% SDS–PAGE, followed by autoradiography using the FLA-3000 phosphoimager system (Fujifilm, Tokyo, Japan).
Statistical analysis
If not indicated otherwise, all values are presented as mean ± SD. Student's paired t test was applied to reveal statistical significances. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Regine Stöbe, Angelika Richter, and Christine Reinhold for excellent technical assistance. M.F. acknowledges financial support from the Deutsche Forschungsgemeinschaft (FA 845/2-1, FA 845/2-2), R.M. from the Sonnenfeld-Siftung and Bundesministerium für Bildung und Forschung (FKZ 0315398A), and N.B. from the Bundesministerium für Bildung und Forschung (FKZ 0315261).
Abbreviations used:
- ARE
adenylate- and uridylate-rich elements
- HIF
hypoxia inducible factor
- hnRNP
heterogeneous nuclear ribonucleoprotein
- MAPK
mitogen-activated protein kinase
- MCP-1
monocyte chemoattractant protein-1
- MK2
MAPK-activated protein kinase 2
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- RBP
RNA-binding protein
- TTP
tristetraprolin
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0949) on August 23, 2012.
REFERENCES
- Al Souhibani N, Al Ahmadi W, Hesketh JE, Blackshear PJ, Khabar KS. The RNA-binding zinc-finger protein tristetraprolin regulates AU-rich mRNAs involved in breast cancer-related processes. Oncogene. 2010;29:4205–4215. doi: 10.1038/onc.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez O, Jouandin P, Rottier J, Bourcier C, Pagès G, Gimond C. Post-transcriptional regulation of the DUSP6/MKP-3 phosphatase by MEK/ERK signaling and hypoxia. J Cell Physiol. 2011;226:276–84. doi: 10.1002/jcp.22339. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, Clark AR. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol. 2006;26:2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA, Connolly JE, Rigby WF. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J Immunol. 2004;172:7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV, Hoek JB, Kholodenko BN. Modular response analysis of cellular regulatory networks. J Theor Biol. 2002;218:507–520. [PubMed] [Google Scholar]
- Cao H, Deterding LJ, Venable JD, Kennington EA, Yates JR, III, Tomer KB, Blackshear PJ. Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochem J. 2006;394:285–297. doi: 10.1042/BJ20051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Cao H, Lai WS, Kennington EA, Campbell D, Blackshear PJ. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J Biol Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- Castrop H, Kurtz A. Functional evidence confirmed by histological localization: overlapping expression of erythropoietin and HIF-2alpha in interstitial fibroblasts of the renal cortex. Kidney Int. 2010;77:269–271. doi: 10.1038/ki.2009.470. [DOI] [PubMed] [Google Scholar]
- Chamboredon S, Ciais D, Desroches-Castan A, Savi P, Bono F, Feige JJ, Cherradi N. Hypoxia-inducible factor-1{alpha} mRNA: a new target for destabilization by tristetraprolin in endothelial cells. Mol Biol Cell. 2011;22:3366–3378. doi: 10.1091/mbc.E10-07-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fähling M, Mrowka R, Steege A, Martinka P, Persson PB, Thiele BJ. Heterogeneous nuclear ribonucleoprotein-A2/B1 modulate collagen prolyl 4-hydroxylase, {alpha} (I) mRNA stability. J Biol Chem. 2006a;281:9279–9286. doi: 10.1074/jbc.M510925200. [DOI] [PubMed] [Google Scholar]
- Fähling M, Mrowka R, Steege A, Nebrich G, Perlewitz A, Persson PB, Thiele BJ. Translational control of collagen prolyl 4-hydroxylase-alpha(I) gene expression under hypoxia. J Biol Chem. 2006b;281:26089–26101. doi: 10.1074/jbc.M604939200. [DOI] [PubMed] [Google Scholar]
- Frede S, Berchner-Pfannschmidt U, Fandrey J. Regulation of hypoxia-inducible factors during inflammation. Methods Enzymol. 2007;435:405–419. doi: 10.1016/S0076-6879(07)35021-0. [DOI] [PubMed] [Google Scholar]
- Galban S, Gorospe M. Factors interacting with HIF-1alpha mRNA: novel therapeutic targets. Curr Pharm Des. 2009;15:3853–3860. doi: 10.2174/138161209789649376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, Tominaga K, Wu X, Fähling M, Ivan M. Post-transcriptional control of the hypoxic response by RNA-binding proteins and microRNAs. Front Mol Neurosci. 2011;4:7. doi: 10.3389/fnmol.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, binding to adenine/uridine-rich element. Mol Cell Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J Biol Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- Kappert K, et al. Chronic treatment with losartan results in sufficient serum levels of the metabolite EXP3179 for PPARgamma activation. Hypertension. 2009;54:738–743. doi: 10.1161/HYPERTENSIONAHA.109.132886. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Kiyatkin A, Bruggeman FJ, Sontag E, Westerhoff HV, Hoek JB. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc Natl Acad Sci USA. 2002;99:12841–12846. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Yim S, Choi BJ, Jang Y, Lee JJ, Sohn BH, Yoo HS, Yeom YI, Park KC. Tristetraprolin regulates the stability of HIF-1alpha mRNA during prolonged hypoxia. Biochem Biophys Res Commun. 2010;391:963–968. doi: 10.1016/j.bbrc.2009.11.174. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Kuschel A, Simon P, Tug S. Functional regulation of HIF-1α under normoxia—is there more than post-translational regulation? J Cell Physiol. 2012;227:514–24. doi: 10.1002/jcp.22798. [DOI] [PubMed] [Google Scholar]
- Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrowka R, Bluthgen N, Fähling M. Seed-based systematic discovery of specific transcription factor target genes. FEBS J. 2008;275:3178–3192. doi: 10.1111/j.1742-4658.2008.06471.x. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Oda T, et al. Activation of hypoxia-inducible factor 1 during macrophage differentiation. Am J Physiol Cell Physiol. 2006;291:C104–C113. doi: 10.1152/ajpcell.00614.2005. [DOI] [PubMed] [Google Scholar]
- Ono M. Molecular links between tumor angiogenesis and inflammation: inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy. Cancer Sci. 2008;99:1501–1506. doi: 10.1111/j.1349-7006.2008.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/s0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoor M, Buers I, Sietmann A, Brodde MF, Hofnagel O, Robenek H, Lorkowski S. Efficient non-viral transfection of THP-1 cells. J Immunol Methods. 2009;344:109–115. doi: 10.1016/j.jim.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Lipman DJ, Kondrashov AS. Patterns in interspecies similarity correlate with nucleotide composition in mammalian 3′ UTRs. Nucleic Acids Res. 2003;31:5433–5439. doi: 10.1093/nar/gkg751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin LG, Zou AP, Spaulding SW. Androgens regulate the binding of endogenous HuR to the AU-rich 3′ UTRs of HIF-1alpha and EGF mRNA. Biochem Biophys Res Commun. 2004;322:644–651. doi: 10.1016/j.bbrc.2004.07.173. [DOI] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Tiedje C, Kotlyarov A, Gaestel M. Molecular mechanisms of phosphorylation-regulated TTP (tristetraprolin) action and screening for further TTP-interacting proteins. Biochem Soc Trans. 2010;38:1632–1637. doi: 10.1042/BST0381632. [DOI] [PubMed] [Google Scholar]
- Tudor C, Marchese FP, Hitti E, Aubareda A, Rawlinson L, Gaestel M, Blackshear PJ, Clark AR, Saklatvala J, Dean JL. The p38 MAPK pathway inhibits tristetraprolin-directed decay of interleukin-10 and pro-inflammatory mediator mRNAs in murine macrophages. FEBS Lett. 2009;583:1933–1938. doi: 10.1016/j.febslet.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- Zhao W, Liu M, D'Silva NJ, Kirkwood KL. Tristetraprolin regulates interleukin-6 expression through p38 MAPK-dependent affinity changes with mRNA 3′ untranslated region. J Interferon Cytokine Res. 2011;31:629–637. doi: 10.1089/jir.2010.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.