Abstract
Understanding the ecology and evolutionary history of symbionts and their hosts requires accurate taxonomic knowledge, including clear species boundaries and phylogenies. Tortoise mites (Mesostigmata: Uropodoidea) are among the most diverse arthropod associates of bark beetles (Curculionidae: Scolytinae), but their taxonomy and host associations are largely unstudied. We tested the hypotheses that (1) morphologically defined species are supported by molecular data, and that (2) bark beetle uropodoids with a broad host range comprise cryptic species. To do so, we assessed the species boundaries of uropodoid mites collected from 51 host species, across 11 countries and 103 sites, using morphometric data as well as partial cytochrome oxidase I (COI) and nuclear large subunit ribosomal DNA (28S). Overall, morphologically defined species were confirmed by molecular datasets, with a few exceptions. Twenty-nine of the 36 uropodoid species (Trichouropoda, Nenteria and Uroobovella) collected in this study had narrow host ranges, while seven species had putative broad host ranges. In all but one species, U. orri, our data supported the existence of these host generalists, which contrasts with the typical finding that widespread generalists are actually complexes of cryptic specialists.
Introduction
Increased access to nucleotide sequencing over the last twenty years has led to exponential growth of molecular-based taxonomy [1]. Modern molecular techniques provide powerful tools to assess species boundaries, and cryptic species (species distinguishable by no or overlooked subtle morphological differences) are being discovered increasingly in a wide range of invertebrate groups [2]–[4]. Species boundaries of symbionts are frequently assessed using molecular markers, and it is often revealed that an apparent widespread host generalist is not a generalist, but rather a complex of cryptic species with narrower host ranges. For instance, Ixodes uriae (Ixodidae) was previously considered to be a host generalist, but microsatellite analysis showed strong genetic divergence across host species, suggesting that I. uriae represents multiple host races with relatively narrower host ranges [5], [6]. Morphological and molecular analyses of Uroobovella nova (Urodinychidae), a single widespread putative generalist uropodoid species collected from silphid beetles worldwide, is actually a complex of cryptic species with varying degrees of host specificity [7].
Bark beetles (Curculionidae: Scolytinae) are a prominent group of wood-borers that feed and mate in the cambium or xylem of numerous tree species worldwide [8]. Mites are one of the most common and diverse associates of scolytines. For instance, 97 species of mites representing 65 genera and 40 families have been collected from under the bark of scolytine infested pine trees [9]. Many or most of these mites reside, feed and reproduce in the galleries of bark beetles, and they attach to dispersing scolytines, hitching a ride to new host trees or coarse woody debris, which would otherwise be difficult to access for most free-living mites.
Uropodoids (Acari: Mesostigmata), or tortoise mites, are among the most frequently collected mite associates of bark beetles, and include three genera Trichouropoda, Nenteria (Trematuridae) and Uroobovella (Urodinychidae). Scolytine-associated uropodoids are often found at a relatively high prevalence (e.g. up to 36% of 8475 beetles had mites in Louisiana; [10]. The superfamily Uropodoidea is represented by over 2,000 described species worldwide, many of which occur in patchy habitats such as nests, woody debris, and dung [11]. Phoresy is therefore a prerequisite for dispersal between such patchy habitats, and deutonymphal uropodoids glue themselves to their host with an anally secreted pedicel. The feeding habits of uropodoids are poorly known but typically they are considered to be omnivorous, feeding on fungal hyphae, slow moving prey, or small particulate matter [12]. The deutonymphs of some species associated with scolytines have been reported as feeding on nematodes and or fungi [13], [14], as well as the eggs and larvae of their bark beetle hosts [15], [16].
Many acarological studies have used mitochondrial cytochrome oxidase I (COI) and nuclear large subunit ribosomal DNA (28S), either alone or combined with other markers, to elucidate species boundaries, uncover cryptic species, and assess phylogenetic relationships of mites [17]–[22]. In this study, we employed morphological and molecular markers (COI and 28S D2–D4) to explore the species boundaries of bark beetle-associated uropodoids and to assess whether morphological species concepts are supported by molecular data. Additionally, we tested whether generalists are truly single species with broad host preferences or instead complexes of cryptic species with narrower host ranges, using quantitative morphological and molecular analyses.
Materials and Methods
Biological Material
Bark beetle specimens were collected across 11 countries and 103 sites, with the majority of sites in Canada and the USA. Canadian specimens were collected in Ontario by W.K. and in various provinces by the Canadian Food Inspection Agency (CFIA) staff as part of the Invasive Alien Species Monitoring program, and examined by W.K. with permission. Specimens from the USA and other countries were collected by A.I.C., and examined by W.K. with permission. All necessary permits and permissions were obtained for the described field studies. Field studies were conducted with a permit to collect in Ontario Provincial Parks issued by Ontario Parks and coordinated by B. Steinberg and B. Crins, as well as permission from private landowners to sample on their property.
In Ontario, bark beetles were collected from mid-April to early August 2009 across four study sites: Algonquin Provincial Park site 1 (45.902, −77.605), Algonquin PP site 2 (45.895, −78.071), one site near Pakenham (45.33, −76.371), and another on Hwy 132 near Dacre (45.369, −76.988). Four Lindgren traps with propylene glycol were placed in each study site. Traps were baited with 95% ethanol and/or α-pinene lures (Synergy Semiochemicals). Traps were emptied every two weeks, trap lures were replaced every eight weeks, and the propylene glycol insecticide was replaced at each visit. Bark beetles were placed individually into 1.5 ml microfuge tubes with 95% ethanol and stored at −20°C. Scolytines were identified to species using keys [8], [23], and tribes were based on the literature [24]. Beetles were examined for uropodoid mites using a dissecting microscope, and all mites found were removed and placed into a 0.5 ml microfuge tube with 95% ethanol and stored at −20°C.
A portion of the bark beetles collected by CFIA staff in 2009 from Canadian provinces, as well as scolytine specimens collected by A.I.C. from USA and several other countries were examined by W.K. for uropodoid mites, and all mites found were removed and stored in 95% ethanol at −80°C. Four species of uropodoids (Uroobovella spp. 1–4) collected from Nicrophorus beetles (Silphidae) in Ontario were used as outgroup specimens. Although the outgroup species are in the same genus as some of the ingroup, the generic position of the outgroup species is contentious, and they are associated with a different family of beetles. Following DNA extraction, mites were recovered from the extraction buffer and slide-mounted in a polyvinyl alcohol medium, and slides were cured on a slide warmer at about 40°C for 3–4 days. Slide-mounted specimens were examined using a compound microscope (Leica DM 5500B or Nikon 80I) and identified to species (or morphospecies) using taxonomically informative morphological characters based on species descriptions from the literature [25]–[30]. Species were identified prior to examining the molecular reconstructions, and in any instances where a conflicting result emerged between the molecular data and morphology-based identifications, both datasets were reexamined. Voucher specimens are deposited in the Canadian National Collection of Insects, Arachnids and Nematodes, in Ottawa, Canada, and the Michigan State University A.J. Cook Arthropod Research Collection, East Lansing, USA.
DNA Extraction, Amplification and Sequencing
Total genomic DNA was extracted from whole specimens for 24 hours using a DNeasy Tissue kit (Qiagen Inc., Santa Clara, CA, USA). Following extraction, mites were removed from the extraction buffer, and genomic DNA was purified following the DNeasy Tissue kit protocol.
PCR amplifications were performed in a total volume of 25 µl, with 13 µl ddH2O, 2.5 µl 10× PCR buffer, 2.5 µl 25 mM MgCl2, 0.5 µl of each 10 µM primer, 0.5 µl 10 mM dNTPs, 0.5 µl Taq DNA polymerase (Promega Corp., Madison, WI, USA), and 5 µl genomic DNA template. In the instances where semi-nested or nested primers were employed, 1 µl of primary PCR product was used as template and the ddH20 was increased to 17 µl. PCR amplification cycles were performed on an Eppendorf ep Gradient S Mastercycler (Eppendorf AG, Hamburg, Germany). Primer pairs LCO1490+ LoDog, and LCO1490+ BB R4 (Table 1), were used to amplify 643 and 603 bp fragments, respectively, of the mitochondrial COI gene. Specimens that did not produce detectable PCR products using either of these primer pairs were reamplified using 1 µl of the primary PCR product and semi-nested, LCO1490+ BB R3Lo, or nested, BB F + BB R3Lo, primer combinations (Table 1), which amplified 592 and 475 bp fragments, respectively. The thermocycler protocol for COI amplification was as follows: initial denaturation cycle at 94°C for 3 min, followed by 40 cycles of 94°C for 45 s, primer annealing at 45°C for 45 s, 72°C for 1 min, and a final extension at 72°C for 5 min. The primer annealing temperature was reduced to 43°C when primer BB R4 was employed.
Table 1. Primer sequences (5′–3′) used to amplify partial COI and 28S D2–D4 sequences from uropodoid mites collected from bark beetles (*primers from this study).
| Gene | Primer | Sequence 5′–3′ | Reference |
| COI | LCO1490 | GGTCAACAAATCATAAAGATATTGG | 51 |
| BB F | TAATTGGWRATGAYCAAATTTTTAA | * | |
| BB R2 | AATHGTDGTAATAAAATTAATTGA | * | |
| BB R3Lo | CCTCCTGCTAADACHGG | * | |
| BB R4 | GTATAGTAATRGCTCCTGC | * | |
| LoDog | GGRTCAAAAAAAGAWGTRTTRAARTTTCG | * | |
| 28S | D23F | GAGAGTTCAAGAGTACGTG | 52 |
| 28S Fb | GAGTACGTGAAACCGCWTWGA | * | |
| 28Sa | GACCCGTCTTGAAACACGG | 53 (modified) | |
| 28S F1 | GGCGHAATGAAATGTGAAGG | * | |
| 28S R3 | GGCTTCRTCTTGCCCAGGC | * | |
| 28S R4 | GGCTTCGTCTTGCCCAGGC | * | |
| 28Sb | CGGAAGGAACCAGCTAC | 53 (modified) | |
| 28S R2 | CCAGTTCTGCTTACCAAAAATGG | * |
Primer pairs D23F +28S R2, and 28S Fb +28S R2 (Table 1), were used to amplify a 990 and 980 bp fragment, respectively, from the 5′ end of the nuclear ribosomal 28S gene, spanning the D2–D4 region. In the instances where neither primer pair produced a detectable PCR product, the specimens were reamplified using 1 µl of the primary PCR product and semi-nested primer pairs, D23F +28Sb or 28S Fb +28Sb, which amplified an 800 and 790 bp fragment of 28S rDNA, respectively (Table 1). The PCR protocol for D23F +28S R2, and D23F +28Sb was as follows: initial denaturation cycle at 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, primer annealing at 44°C for 1.5 min, 72°C for 2 min, and a final extension at 72°C for 10 min. The primer annealing temperature was changed to 56°C for 28S Fb +28S R2, and it was changed to 50°C for 28S Fb +28S R2. Additional primers were designed to amplify COI and 28S from uropodoids; all primers designed or used in this study are shown in the primer map (Table 1, Fig. 1).
Figure 1. Primer map showing the relative location of primers used to amplify.
(A) partial COI, and (B) 28S D2–D4 sequences from uropodoid mites collected from bark beetles.
Amplified products and negative controls were visualized on 1% agarose electrophoresis gels, and purified using pre-cast E-Gel CloneWell 0.8% SYBR Safe agarose gels (Invitrogen, Carlsbad, CA, USA) following the protocol of [31]. Sequencing reactions were performed in a total reaction volume of 10 µl, with 3 µl ddH2O, 1.5 µl of 5× sequencing buffer, 0.5 µl of primer, 1 µl of BigDye Terminator (PE Applied Biosystems, Foster City, CA, USA), and 4 µl of purified PCR product. Sequencing was performed at the Agriculture & Agri-Food Canada, Eastern Cereal and Oilseed Research Centre Core Sequencing Facility (Ottawa, ON, Canada). Purification of sequencing reactions was performed using the ABI ethanol/EDTA/sodium acetate precipitation protocol and reactions were analysed on an ABI 3130×l Genetic Analyzer (PE Applied Biosystems, Foster City, CA, USA).
Sequence Alignment and Phylogenetic Analysis
Sequence chromatograms were edited and contiguous sequences were assembled using Sequencher v4.7 (Gene Codes Corp., Ann Arbor, MI, USA). COI sequences were aligned manually in Mesquite v2.74 [32] according to the translated amino acid sequence. 28S was initially aligned in ClustalX v2.0.12 [33] with the default settings, and subsequently adjusted manually in Mesquite, no regions were excised, and due to the absence of any secondary structure for mites for this gene region, no secondary structure alignment was performed. Sequences have been submitted to GenBank (Table 2).
Table 2. Collection locations and host species records of uropodoid mites collected from scolytines (ingroup) and Nicrophorus beetles (outgroup) with GenBank accession no. for COI and 28S (*Uroob = Uroobovella, Trich = Trichouropoda, Nent = Nenteria).
| Beetle no. | Beetle species | Collection location | Lat | Long | Date | Mite species* | COI | 28S |
| 1 - WKB4051 | Pityokteines sparsus | Can, ON, Hwy 132, Dacre | 45.369 | −76.988 | 16 v 2009 | Uroob. orri | JN992226 | – |
| 2 - WKB4057 | Orthotomicus caelatus | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 16 v 2009 | Uroob. n.sp. 6 | JN992227 | – |
| 3 - WKB4095 | Gnathotrichus materiarius | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 16 v 2009 | Trich. parisiana | JN992184 | – |
| 4 - WKB4109 | Ips grandicollis | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 16 v 2009 | Trich. australis | – | – |
| 5 - WKB4190 | Pityokteines sparsus | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 28 v 2009 | Trich. moseri | JN992171 | – |
| 6 - WKB4232 | Polygraphus rufipennis | Can, ON, Carbine Rd. | 45.330 | −76.371 | 16 v 2009 | Uroob. orri | – | – |
| 7 - WKB4429 | Dendroctonus valens | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 16 v 2009 | Uroob. americana | JN992202 | – |
| 8 - WKB4850 | Polygraphus rufipennis | Can, AB, Fort McMurray | 56.016 | −110.88 | 23 vii 2009 | Trich. moseri | JN992172 | – |
| 9 - WKB4869 | Dryocoetes affaber | Can, AB, Fort McMurray | 56.016 | −110.88 | 29 vi 2009 | Uroob. orri | – | – |
| 10 - WKB4943 | Hylesinus aculeatus | Can, ON, Hwy 132, Dacre | 45.369 | −76.988 | 1 v 2009 | Trich. bipilis | JN992155 | – |
| 11 - WKB4987 | Ips pini | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 1 v 2009 | Trich. australis | JN992139 | – |
| 12 - WKB4995 | Trypodendron retusum | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 1 v 2009 | Trich. parisiana | – | – |
| 13 - WKB5224 | Polygraphus rufipennis | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 28 v 2009 | Uroob. orri | JN992228 | – |
| 14 - WKB5226 | Dryocoetes affaber | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 28 v 2009 | Uroob. orri | JN992229 | – |
| 15 - WKB5261 | Hylastes porculus | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 28 v 2009 | Uroob. dryocoetes | JN992211 | – |
| 16 - WKB5344 | Gnathotrichus materiarius | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 28 v 2009 | Trich. parisiana | JN992185 | – |
| 17 - WKB5351 | Dendroctonus valens | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 28 v 2009 | Uroob. dryocoetes | – | – |
| 18 - WKB5563 | Pityogenes hopkinsi | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 28 v 2009 | Trich. n.sp. 3 | – | – |
| 19 - WKB5564 | Polygraphus rufipennis | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 28 v 2009 | Trich. moseri | – | – |
| 20 - WKB5568 | Ips pini | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 28 v 2009 | Trich. australis | – | – |
| 21 - WKB5639 | Orthotomicus caelatus | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 28 v 2009 | Uroob. n.sp. 6 | JN992230 | – |
| 22 - WKB5682 | Dryocoetes autographus | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 25 vi 2009 | Uroob. dryocoetes | JN992212 | – |
| 23 - WKB5759 | Ips grandicollis | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 25 vi 2009 | Trich. lamellosa | – | – |
| 24 - WKB5759 | Ips grandicollis | Can, ON, Algonquin P.P. 1 | 45.902 | −77.605 | 25 vi 2009 | Uroob. orri | JN992231 | – |
| 25 - WKB5797 | Hylurgops pinifex | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 25 vi 2009 | Trich. hirsuta | – | – |
| 26 - WKB5882 | Hylastes porculus | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 25 vi 2009 | Trich. hirsuta | JN992167 | JN992260 |
| 27 - WKB5970 | Dendroctonus ponderosae | Can, AB, Grande Prairie | 2007 | Trich. lamellosa | JN992170 | JN992261 | ||
| 28 - WKHD001 | Gnathotrichus materiarius | Can, QC, La Patrie, Route 212 | 46.345 | −72.576 | 22 v 2009 | Trich. parisiana | – | – |
| 29 - WKHD004 | Pityokteines sparsus | Can, QC, La Patrie, Route 212 | 46.345 | −72.576 | 22 v 2009 | Uroob. orri | JN992232 | – |
| 30 - WKHD008 | Dendroctonus valens | Can, QC, La Patrie, Route 212 | 46.345 | −72.576 | 22 v 2009 | Uroob. americana | – | – |
| 31 - WKHD009 | Polygraphus rufipennis | Can, QC, East Hereford | 45.029 | −71.505 | 22 v 2009 | Uroob. orri | – | – |
| 32 - WKHD010 | Gnathotrichus materiarius | Can, QC, East Hereford | 45.029 | −71.505 | 22 v 2009 | Trich. parisiana | – | – |
| 34 - WKHD012 | Gnathotrichus materiarius | Can, QC, Pont Rouge | 46.806 | −71.679 | 05 vi 2009 | Trich. parisiana | JN992186 | – |
| 35 - WKHD014 | Polygraphus rufipennis | Can, QC, Pont Rouge | 46.806 | −71.679 | 05 vi 2009 | Uroob. dryocoetes | JN992213 | – |
| 36 - WKHD018 | Dendroctonus rufipennis | Can, NS, West Northfield | 01 vi 2009 | Uroob. orri | – | – | ||
| 37 - WKHD030 | Hylastes porculus | Can, NS, Westfield | 44.403 | −64.975 | 28 v 2009 | Uroob. dryocoetes | JN992214 | – |
| 38 - WKHD037 | Hylastes porculus | Can, NB, Bayside, Route 127 | 45.205 | −67.140 | 15 vi 2009 | Uroob. dryocoetes | – | – |
| 39 - WKHD042 | Xyleborinus saxesenii | Can, BC, Stanley Park, Pipeline Dr. | 06 vi 2008 | Trich. parisiana | JN992187 | – | ||
| 40 - WKHD057 | Gnathotrichus materiarius | Can, QC, Parc des iles de Boucherville | 45.601 | −73.466 | 26 v 2009 | Trich. parisiana | – | – |
| 41 - WKHD062 | Dendroctonus valens | Can, QC, Sorel-Tracy | 46.030 | −73.083 | 09 vi 2009 | Uroob. americana | JN992203 | – |
| 42 - WKHD065 | Gnathotrichus materiarius | Can, QC, Sorel-Tracy | 46.030 | −73.083 | 09 vi 2009 | Uroob. orri | – | – |
| 43 - WKHD066 | Hylastes porculus | Can, QC, Sorel-Tracy | 46.030 | −73.083 | 09 vi 2009 | Uroob. dryocoetes | JN992215 | JN992277 |
| 44 - WKHD067 | Dryocoetes autographus | Can, QC, Sorel-Tracy | 46.030 | −73.083 | 09 vi 2009 | Uroob. dryocoetes | – | – |
| 45 - WKHD070 | Dryocoetes affaber | Can, QC, Sorel-Tracy | 46.030 | −73.083 | 09 vi 2009 | Uroob. dryocoetes | – | – |
| 46 - WKHD075 | Hylastes ruber | Can, BC, McPhee Creek Rd. | 49.323 | −117.61 | 29 iv 2009 | Trich. fallax | JN992166 | JN992259 |
| 47 - WKHD078 | Hylurgops pinifex | Can, NS, Greenfield | 44.335 | −64.915 | 11 vi 2009 | Trich. fallax | – | – |
| 48 - WKHD079 | Dendroctonus rufipennis | Can, NS, Annapolis, Granville ferry | 44.810 | −65.537 | 22 vi 2009 | Trich. alascae | JN992137 | – |
| 49 - WKHD079 | Dendroctonus rufipennis | Can, NS, Annapolis, Granville ferry | 44.810 | −65.537 | 22 vi 2009 | Uroob. orri | JN992233 | – |
| 50 - WKHD080 | Dendroctonus rufipennis | Can, NS, Victoria Beach | 44.703 | −65.747 | 22 vi 2009 | Uroob. orri | JN992234 | – |
| 51 - WKHD085 | Dendroctonus rufipennis | Can, NS, Blomidon, Stewart Mtn. Rd. | 45.227 | −64.397 | 19 vi 2009 | Trich. alascae | JN992138 | JN992253 |
| 52 - WKHD085 | Dendroctonus rufipennis | Can, NS, Blomidon, Stewart Mtn. Rd. | 45.227 | −64.397 | 19 vi 2009 | Uroob. orri | JN992235 | – |
| 53 - WKHD114 | Dendroctonus rufipennis | Can, QC, Degelis | 47.561 | −68.644 | 16 vi 2009 | Uroob. orri | – | – |
| 54 - WKHD116 | Hylastes porculus | Can, QC, Saint Come De liniere | 46.014 | −70.483 | 23 vi 2009 | Uroob. dryocoetes | JN992216 | – |
| 55 - WKHD117 | Gnathotrichus materiarius | Can, QC, Degelis | 47.551 | −68.642 | 26 vi 2009 | Trich. parisiana | – | – |
| 56 - WKHD118 | Hylastes porculus | Can, QC, Degelis | 47.551 | −68.642 | 26 vi 2009 | Uroob. dryocoetes | JN992217 | – |
| 57 - WKHD120 | Dendroctonus valens | Can, QC, Pont Rouge | 46.562 | −71.545 | 08 vi 2009 | Uroob. americana | JN992204 | – |
| 58 - WKHD121 | Dendroctonus valens | Can, QC, Saint Pamphile | 46.943 | −69.764 | 17 vi 2009 | Uroob. dryocoetes | JN992218 | – |
| 59 - WKHD129 | Dendroctonus rufipennis | Can, QC, Saint Pamphile | 46.947 | −69.761 | 17 vi 2009 | Uroob. orri | – | – |
| 60 - WKHD130 | Dryocoetes autographus | Can, NB, Monument | 45.954 | −67.767 | 24 vi 2009 | Uroob. dryocoetes | JN992219 | – |
| 61 - WKHD133 | Dendroctonus rufipennis | Can, NS, Sheet Harbour | 44.907 | −62.491 | 19 vi 2009 | Uroob. orri | JN992236 | – |
| 62 - WKHD136 | Polygraphus rufipennis | Can, NS, Sheet Harbour | 44.907 | −62.491 | 19 vi 2009 | Uroob. orri | JN992237 | – |
| 63 - WKHD140 | Dryocoetes autographus | Can, NS, Sheet Harbour | 44.909 | −62.503 | 19 vi 2009 | Uroob. dryocoetes | JN992220 | – |
| 64 - WKHD142 | Dryocoetes affaber | Can, NS, Sheet Harbour | 44.909 | −62.503 | 19 vi 2009 | Trich. hirsuta | – | – |
| 65 - WKHD142 | Dryocoetes affaber | Can, NS, Sheet Harbour | 44.909 | −62.503 | 19 vi 2009 | Uroob. orri | JN992238 | – |
| 66 - WKHD149 | Polygraphus rufipennis | Can, QC, Cookshire | 45.389 | −71.513 | 02 vii 2009 | Trich. hirsuta | – | – |
| 67 - WKHD158 | Dryocoetes autographus | Can, QC, Cookshire | 45.389 | −71.513 | 02 vii 2009 | Uroob. dryocoetes | JN992221 | – |
| 68 - WKHD169 | Dryocoetes affaber | Can, QC, Cookshire | 45.389 | −71.513 | 02 vii 2009 | Uroob. dryocoetes | JN992222 | – |
| 69 - WKHD172 | Dryocoetes affaber | Can, QC, Saint Malo | 45.197 | −71.527 | 02 vii 2009 | Uroob. dryocoetes | – | – |
| 70 - WKHD175 | Hylastes porculus | Can, QC, La Patrie, Route 212 | 46.345 | −72.576 | 02 vii 2009 | Uroob. dryocoetes | JN992223 | – |
| 71 - WKHD177 | Dryocoetes autographus | Can, QC, La Patrie, Route 212 | 46.345 | −72.576 | 02 vii 2009 | Uroob. dryocoetes | – | – |
| 72 - WKHD178 | Orthotomicus caelatus | Can, NS, Goodwood | 44.603 | −63.677 | 27 v 2009 | Uroob. n.sp. 6 | JN992239 | JN992278 |
| 73 - WKHD179 | Ips pini | Can, NS, Goodwood | 44.603 | −63.677 | 27 v 2009 | Trich. australis | JN992140 | JN992254 |
| 74 - WKHD181 | Polygraphus rufipennis | Can, NS, Purcell’s Cove | 44.624 | −63.575 | 03 vi 2009 | Uroob. orri | JN992240 | – |
| 75 - WKHD182 | Dryocoetes affaber | Can, NS, Purcell’s Cove | 44.624 | −63.575 | 03 vi 2009 | Uroob. orri | JN992241 | – |
| 76 - WKHD183 | Dendroctonus rufipennis | Can, NS, Purcell’s Cove | 44.624 | −63.575 | 13 vii 2009 | Uroob. orri | – | – |
| 77 - WKHD184 | Gnathotrichus materiarius | Can, NS, Debert, Industrial Park | 45.428 | −63.429 | 25 vi 2009 | Trich. parisiana | JN992188 | – |
| 78 - WKHD185 | Ips pini | Can, NS, Debert, Industrial Park | 45.428 | −63.429 | 25 vi 2009 | Trich. australis | JN992141 | – |
| 79 - WKHD189 | Ips borealis | Can, NS, Debert, Industrial Park | 45.428 | −63.429 | 25 vi 2009 | Trich. polytricha | JN992191 | – |
| 80 - WKHD193 | Dryocoetes autographus | Can, NS, Debert, Industrial Park | 45.428 | −63.429 | 25 vi 2009 | Uroob. dryocoetes | JN992224 | – |
| 81 - WKHD194 | Dryocoetes affaber | Can, QC, Saint Roch de Mekinac | 46.792 | −72.748 | 23 vi 2009 | Uroob. orri | – | – |
| 82 - WKHD199 | Hylastes porculus | Can, QC, Saint Severin, Route 159 | 46.686 | −72.525 | 23 vi 2009 | Uroob. dryocoetes | JN992225 | – |
| 83 - WKHD204 | Ips grandicollis | Can, ON, Brampton | 43.708 | −79.728 | 06 vii 2009 | Trich. australis | JN992142 | – |
| 84 - WKHD208 | Ips grandicollis | Can, ON, Argentia Rd. Century Ave | 43.598 | −79.744 | 07 vii 2009 | Trich. australis | JN992143 | JN992255 |
| 85 - WKHD228 | Ips pini | Can, QC, Boucherville | 45.601 | −73.466 | 09 vii 2009 | Trich. australis | JN992144 | – |
| 86 - WKHD230 | Ips pini | Can, ON, Argentia Rd. Century Ave | 43.598 | −79.744 | 20 vii 2009 | Trich. australis | – | – |
| 87 - WKHD232 | Ips grandicollis | Can, ON, New Market, 500 Water St. | 44.047 | −79.456 | 23 vii 2009 | Uroob. orri | JN992242 | – |
| 88 - WKHD234 | Ips pini | Can, ON, New Market, 500 Water St. | 44.047 | −79.456 | 23 vii 2009 | Trich. australis | JN992145 | – |
| 89 - WKHD235 | Polygraphus rufipennis | Can, QC, Saint Zacharie | 46.130 | −70.262 | 21 vii 2009 | Trich. moseri | JN992173 | JN992262 |
| 90 - WKHD236 | Polygraphus rufipennis | Can, QC, Woburn | 45.342 | −70.898 | 21 vii 2009 | Trich. moseri | JN992174 | – |
| 91 - WKHD237 | Polygraphus rufipennis | Can, QC, Saint Benjamin | 46.268 | −70.617 | 21 vii 2009 | Trich. moseri | – | – |
| 92 - WKHD252 | Ips borealis | Can, NS, Hantsport, Cobesquid Bay | 45.099 | −64.184 | 21 vii 2009 | Trich. polytricha | – | |
| 93 - WKHD254 | Ips pini | Can, NS, Hantsport, Cobesquid Bay | 45.099 | −64.184 | 11 viii 2009 | Trich. australis | JN992146 | – |
| 94 - WKHD261 | Hylastes subopacus | USA, NM, Bernalillo | 10 x 2008 | Nent. chiapasa | – | – | ||
| 95 - WKB5929 | Dendroctonus valens | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 25 vi 2009 | Uroob. americana | JN992205 | – |
| 96 - WKB5639 | Orthotomicus caelatus | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 28 v 2009 | Uroob. n.sp. 6 | JN992243 | JN992279 |
| 97 - WKB5929 | Dendroctonus valens | Can, ON, Algonquin P.P. 2 | 45.895 | −78.071 | 25 vi 2009 | Uroob. americana | JN992206 | JN992275 |
| 98 - MSU001 | Pityophthorus sp. | USA, CA, El Dorado N.F. Ice House Res. | 38.5 | −120.22 | 25 v 2007 | Trich. n.sp. 2 | JN992178 | JN992265 |
| 99 - MSU004 | Dendroctonus valens | USA, OH, Secrest Arboretum | 40.782 | −81.916 | v 2007 | Uroob. americana | – | – |
| 100 - MSU006 | Ficicis sp. | China, Yunnan, Xishuangbanna | 22.163 | 100.871 | 30 v 2008 | Uroob. australiensis | JN992210 | – |
| 101 - MSU010 | Dendroctonus valens | USA, PA, Keystone Rd. | 40.739 | −76.308 | 30 iv 2009 | Uroob. americana | – | – |
| 102 - MSU012 | Polygraphus sp. | Thailand, Doi Pui | iv 2005 | Trich. polygraphi | – | – | ||
| 103 - MSU014 | Scolytus ventralis | USA, CA, El Dorado N.F. Ice House Res. | 38.5 | −120.22 | 17 vi 2003 | Trich. n.sp. 10 | JN992175 | JN992263 |
| 104 - MSU016 | Hylurgops rugipennis pinifex | USA, UT, Ashley N.F., Gray Head Peak | 39.54 | −110.45 | 11 vi 2003 | Trich. fallax | – | – |
| 105 - MSU020 | Monarthrum dentigerum | USA, TX, Davis Mt. S.P. | 25 v 2001 | Trich. n.sp. 8 | – | – | ||
| 106 - MSU024 | Monarthrum dentigerum | USA, TX, Big Bend N.P. | iv 2004 | Trich. n.sp. 8 | – | – | ||
| 107 - MSU025 | Hylurgops sp. | Mex, South of Amecameca | 19.016 | −98.741 | 11 v 2004 | Uroob. vinicolora | JN992248 | – |
| 108 - MSU028 | Hylastes sp. | USA, WI, Cobma | 11 iv 2004 | Trich. perissopos | – | – | ||
| 109 - MSU030 | Dendroctonus valens | USA, WI, nr. Madison | v 2005 | Uroob. americana | JN992207 | – | ||
| 110 - MSU032 | Pseudips mexicanus | Mex, Jalisco | 5 xi 2003 | Nent. moseri | JN992136 | JN992252 | ||
| 111 - MSU036 | Pityokteines curvidens | Croatia | 2003 | Uroob. orri | JN992244 | JN992280 | ||
| 112 - MSU038 | Pseudips mexicanus | Mex, Jalisco, nr. Ciudad Guzman | 9 ii 2006 | Trich. n.sp. 9 | JN992181 | – | ||
| 113 - MSU040 | Orthotomicus erosus | Italy, Tuscany, nr. San Gusme | 43.360 | 11.501 | 29 xii 2006 | Trich. n.sp. 4 | JN992179 | JN992266 |
| 114 - MSU045 | Ips hunteri | USA, UT, Ashley N.F., Hwy 191 | 40.43 | −109.29 | 10 vi 2003 | Trich. polytricha | – | – |
| 115 - MSU049 | Ips pilifrons utahensis | USA, CO, San Isabel N.F. Monarch Pass | 38.31 | −106.19 | 9 vi 2003 | Trich. polytricha | – | – |
| 116 - MSU050 | Ips cribricollis | USA, NM, Big Burro Mts | 20 viii 2003 | Trich. australis | JN992147 | – | ||
| 117 - MSU051 | Ips perturbatus | USA, MN, Cascade River Park | 12 vi 2001 | Trich. polytricha | JN992192 | – | ||
| 118 - MSU053 | Ips cribricollis | Mex, South of Amecameca | 19.016 | −98.741 | 11 v 2004 | Trich. tegucigalpae | JN992201 | JN992274 |
| 119 - MSU055 | Ips cribricollis | Mex, Landa de Matamoros | 21.263 | −99.177 | 14 v 2004 | Trich. australis | – | – |
| 120 - MSU056 | Ips nitidus | China, Sichuan | 9 vii 2004 | Nent. eulaelaptis | JN992135 | JN992251 | ||
| 121 - MSU057 | Ips cribricollis | Mex, Jalisco, nr. Ciudad Guzman | 9 ii 2006 | Trich. n.sp. 13 | JN992198 | – | ||
| 122 - MSU060 | Ips pilifrons | USA, CO, White River N.F. Lost Lake | 30 vi 2005 | Trich. polytrichasimilis | – | – | ||
| 123 - MSU066 | Ips calligraphus | USA, FL, Naples, Collier | 26.157 | −81.660 | iii - iv 2007 | Trich. australis | – | – |
| 124 - MSU067 | Ips hoppingi | USA, TX, McDonald Observatory | 12 iv 2002 | Trich. californica | JN992156 | – | ||
| 125 - MSU069 | Ips montanus | USA, WA, Hwy 410, nr. Chinook Pass | 11 v 2001 | Trich. polytrichasimilis | – | – | ||
| 126 - MSU071 | Ips pini | USA, AK, Douglas is. nr. Juneau | 4 v 2001 | Trich. idahoensis | JN992168 | – | ||
| 127 - MSU073 | Ips pini | USA, CA, Lassen N.F. Polesprings Rd. | 3 vii 2001 | Trich. idahoensis | JN992169 | – | ||
| 128 - MSU079 | Ips plastographus | USA, CA, | v 2001 | Trich. n.sp. 11 | JN992197 | JN992272 | ||
| 129 - MSU084 | Ips paraconfusus | USA, CA, Mt. Diablo S.P. Contra Costa | 10 vi 2001 | Trich. n.sp. 7 | – | – | ||
| 130 - MSU085 | Ips lecontei | USA, AZ, Coronado N.F. Ladybug Peak | 18 vii 2001 | Trich. australis | JN992148 | – | ||
| 131 - MSU086 | Ips cembrae | Switzerland | v 2002 | Trich. polytricha | JN992193 | JN992270 | ||
| 132 - MSU090 | Ips montanus | USA, CA, El Dorado, Hwy 50 nr. Meyer | 13 vi 2001 | Trich. polytricha | JN992194 | – | ||
| 133 - MSU091 | Pityogenes chalcographus | Norway | v 2002 | Trich. n.sp. 5 | JN992180 | JN992267 | ||
| 134 - MSU094 | Ips confusus | USA, NV, Mt. Charleston Recreation | 36.16 | −115.32 | 27 vi 2003 | Trich. californica | JN992157 | – |
| 135 - MSU099 | Ips confusus | USA, UT, nr. Baker Dam | 37.23 | −113.39 | 28 vi 2003 | Trich. californica | JN992158 | – |
| 136 - MSU104 | Ips confusus | USA, AZ, Kaibab N.F. Hwy 389 | 36.51 | −112.16 | 30 vi 2003 | Trich. californica | JN992159 | – |
| 137 - MSU108 | Ips confusus | USA, AZ, Kaibab N.F. nr. Flagstaff | 35.24 | −111.35 | 2 vii 2003 | Trich. californica | JN992160 | – |
| 138 - MSU111 | Ips confusus | USA, NM, Carson N.F. nr. Los Pinons | 36.25 | −106.01 | 9 vi 2003 | Trich. californica | JN992161 | JN992257 |
| 139 - MSU114 | Ips confusus | USA, NM, Santa Fe | 17 vi 2003 | Trich. californica | JN992162 | – | ||
| 140 - MSU119 | Ips confusus | USA, NV, Risue Canyon | 4 vi 2003 | Trich. californica | JN992163 | – | ||
| 141 - MSU123 | Ips confusus | USA, AZ, Coconino, nr. Red Mt. | 35.31 | −111.5 | vi 2003 | Trich. californica | JN992164 | – |
| 142 - MSU124 | Ips confusus | USA, CO, F.R. 504 | 37.669 | −108.70 | 9 viii 2004 | Trich. californica | JN992165 | – |
| 143 - MSU125 | Ips perturbatus | Can, ON, Marlborough Forest | 19 v 1995 | Trich. australis | – | – | ||
| 144 - MSU127 | Pseudips mexicanus | USA, CA, San Francisco | 20 viii 1995 | Trich. n.sp. 9 | JN992182 | – | ||
| 145 - MSU131 | Ips emarginatus | USA, CA, Lassen, Black Mt. | 7 vii 1995 | Trich. polytrichasimilis | – | – | ||
| 146 - MSU132 | Ips calligraphus | USA, NY, Smithtown | 11 ix 1994 | Trich. australis | – | – | ||
| 147 - MSU133 | Ips pini | USA, NY | 18 x 1995 | Trich. australis | – | – | ||
| 148 - MSU137 | Ips paraconfusus | USA, CA, Mt. Diablo | 3 ix 1995 | Trich. n.sp. 7 | JN992199 | – | ||
| 149 - MSU139 | Ips woodi | USA, AZ, Coronado N.F. Hospital Flat | 4 ix 1996 | Trich. polytricha | JN992195 | – | ||
| 150 - MSU143 | Dendroctonus valens | USA, PA, 225 Yeager Rd. Woodland | 41.049 | −78.349 | 30 iv 2009 | Uroob. americana | JN992208 | – |
| 151 - MSU144 | Ips woodi | USA, AZ, Apache N.F. Hannagan Meadow | 1 ix 1996 | Trich. polytricha | JN992196 | – | ||
| 152 - MSU147 | Ips pilifrons | USA, AZ, Apache N.F. Hannagan Meadow | 31 viii 1996 | Trich. australis | JN992149 | – | ||
| 153 - MSU148 | Ips cribricollis | USA, NM, Otero | v 1994 | Trich. australis | JN992150 | – | ||
| 154 - MSU150 | Ips hunteri | USA, AZ, Apache N.F. Hannagan Meadow | Trich. australis | JN992151 | – | |||
| 155 - MSU152 | Pseudips mexicanus | USA, CA, Albion River Rd. nr. Rt. 1 | 23 iii 1996 | Trich. n.sp. 9 | JN992183 | JN992268 | ||
| 156 - MSU154 | Ips emarginatus | USA, CA, El Dorado N.F. Ice House Res. | 6 ix 1997 | Uroob. orri | JN992245 | – | ||
| 157 - MSU155 | Dendroctonus valens | USA, CA, University of California Berkeley | 14 x 1996 | Uroob. vinicolora | JN992249 | – | ||
| 158 - MSU157 | Ips cribricollis | USA, NM, Cloudcroft | 11 v 1994 | Trich. australis | JN992152 | – | ||
| 159 - MSU162 | Ips bonanseai | Mex, Nuevo Leon | xii 1993 | Trich. tegucigalpae | – | – | ||
| 160 - MSU163 | Ips hoppingi | Mex, Nuevo Leon | 24.505 | −99.985 | 25 x 1993 | Trich. californica | – | – |
| 161 - MSU167 | Ips plastographus | USA, CA, Santa Cruz | 13 x 1993 | Uroob. orri | JN992246 | – | ||
| 162 - MSU168 | Ips pini | USA, RI, Lincoln S.P. | 19 vii 1997 | Trich. australis | JN992153 | – | ||
| 163 - MSU173 | Ips emarginatus | USA, CA, Lassen, Bogard Bultes | 6 xii 1996 | Uroob. orri | JN992247 | – | ||
| 164 - MSU174 | Ips cembrae | Germany, Dresden | 28 v 1986 | Trich. polytricha | – | – | ||
| 165 - MSU179 | Gnathotrichus materiarius | USA, MI, Mt. Pleasant | 28 v 1998 | Trich. parisiana | JN992189 | – | ||
| 166 - MSU180 | Camptocerus auricomis | Panama | 4 ix 2008 | Trich. n.sp. 6 | – | – | ||
| 167 - MSU185 | Corthylus sp. | Panama | 8.862 | −82.743 | 26 viii 2008 | Trich. n.sp. 1 | JN992176 | – |
| 168 - MSU010 | Dendroctonus valens | USA, PA, Keystone Rd. | 40.739 | −76.308 | 30 iv 2009 | Uroob. americana | – | – |
| 169 - MSU084 | Ips paraconfusus | USA, CA, Mt. Diablo S.P. Contra Costa | 10 vi 2001 | Trich. n.sp. 7 | JN992200 | JN992273 | ||
| 170 - MSU123 | Ips confusus | USA, AZ, Coconino, nr. Red Mt. | 35.31 | −111.5 | vi 2003 | Trich. californica | – | JN992258 |
| 171 - MSU143 | Dendroctonus valens | USA, PA, 225 Yeager Rd. Woodland | 41.049 | −78.349 | 30 iv 2009 | Uroob. americana | JN992209 | JN992276 |
| 172 - MSU148 | Ips cribricollis | USA, NM, Otero | v 1994 | Trich. australis | JN992154 | JN992256 | ||
| 173 - MSU154 | Ips emarginatus | USA, CA, El Dorado N.F. Ice House Res. | 6 ix 1997 | Uroob. orri | – | – | ||
| 174 - MSU185 | Corthylus sp. | Panama | 8.862 | −82.743 | 26 viii 2008 | Trich. n.sp. 1 | JN992177 | JN992264 |
| 175 - MSU025 | Hylurgops sp. | Mex, South of Amecameca | 19.016 | −98.741 | 11 v 2004 | Uroob. vinicolora | JN992250 | JN992281 |
| 176 - MSU049 | Ips pilifrons utahensis | USA, CO, San Isabel N.F. Monarch Pass | 38.31 | −106.19 | 9 vi 2003 | Trich. polytricha | – | JN992271 |
| 177 - MSU179 | Gnathotrichus materiarius | USA, MI, Mt. Pleasant | 28 v 1998 | Trich. parisiana | JN992190 | JN992269 | ||
| 2 - WKN084 | Nicrophorus sayi | Can, QC, Pont-Rouge | 46.806 | −71.679 | 05 vi 2009 | Uroob. sp. 2 | JN992096 | – |
| 7 - WKN165 | Nicrophorus orbicollis | Can, ON, Carbine Rd. | 45.330 | −76.371 | 23 vii 2009 | Uroob. sp. 1 | JN992074 | JQ316464 |
| 8 - WKN184 | Nicrophorus vespilloides | Germany, Mooswald Forest, nr. Freiburg | 48.0 | 7.85 | vi 2009 | Uroob. sp. 3 | JN992102 | JQ316465 |
| 21 - WKN165 | Nicrophorus orbicollis | Can, ON, Carbine Rd. | 45.330 | −76.371 | 23 vii 2009 | Uroob. sp. 1 | JN992075 | – |
| 30 - WKN090 | Nicrophorus nepalensis | Taiwan, nr. Meifeng, 5 km Sungkang | 24.088 | 121.171 | 02 v 2007 | Uroob. sp. 4 | JN992103 | – |
| 65 - WKN350 | Nicrophorus sayi | Can, NS, Portobello | 44.75 | −63.6 | 2009 | Uroob. sp. 2 | JN992097 | – |
Pairwise distances were calculated using neighbour-joining (NJ) analyses with the Kimura-2-parameter (K2P) model in PAUP* v4.0b10 [34]. Phylogenetic reconstructions of COI, 28S, and concatenated datasets were performed using Bayesian inference (BI) in MrBayes v3.1.2 [35], [36], and parsimony analyses in TNT v1.1 [37]. Gaps were treated as missing since gaps scored as a fifth state produced the same topology as that observed for gaps as missing for each of the analytical approaches. Analyses of the COI dataset excluding the third codon positions produced poorly supported reconstructions with similar topology to the analyses including the third codon position; hence analyses were performed including the 3rd codon.
MrModeltest v2.3 [38] was used to determine the best-fit model of molecular evolution for each gene, which was determined to be GTR+I+G. Bayesian analysis was performed in MrBayes with a Markov Chain Monte Carlo (MCMC) method, two independent runs, with nucmodel = 4by4, Nst = 6, rates = invgamma, samplefreq = 1000, four chains = one cold and three heated. The COI dataset ran for 20 million generations, and the 28S and concatenated datasets ran for 10 million generations with a burn-in of 1000. In Mesquite, the remaining trees, excluding the burn-in, were used to generate a majority-rule consensus tree displaying the posterior probability supports for each node. Bayesian analyses were performed using the on-line Computational Biology Service Unit at Cornell University, and at the Cyberinfrastructure for Phylogenetic Research (CIPRES) portal [39].
Parsimony analysis was performed using a heuristic search with tree bisection-reconnection (TBR) branch swapping and 1000 random addition sequence replicates, all characters were treated as unordered, equal weighting, and gaps were treated as missing. Multiple trees were obtained and these were presented in a semistrict consensus tree. Node support was assessed in TNT, using jackknife resampling with 36% of characters removed and 1000 replicates, Bremer supports and partitioned Bremer supports (PBS) were also determined using TNT. Node support for the parsimony analysis of the COI and concatenated datasets were mapped onto the corresponding Bayesian phylogenies.
Morphological Analysis
To assess intraspecific morphological divergence of mites used in the molecular analyses, slide-mounted specimens were examined using a Leica DM5500B compound microscope, and 15 and 14 characters (for Trematuridae and Urodinychidae, respectively) were measured using Leica Application Suite, Live and Interactive Measurements Modules v3.5. Characters from different body regions were selected based on their relative ease of measurement and prominence, as well as previously observed variation across specimens. The 15 characters measured for trematurid species were: maximal length and width of the dorsal shield and ventrianal shield; sternal shield (SS) median length; SS width at five levels (from anterior to posterior): maximal width of the SS anterior margin, maximum width of the two expansions at level with coxae II–III and coxae III–IV, minimum width of the posterior constriction level with coxa IV, and width of the SS posterior margin; length of tarsus I; and the length of the following setae: opisthogastric setae V8 and V4 [25] (JV4 and paranal, sensu [40]), the proximoventral setae of femur I, and the longest of anterodorsal setae in the sensory pit of tarsus I. The same characters were measured for Urodinychidae (Uroobovella) species, except that seta V4 and proximoventral setae of femur I were not measured, but the length of dorsal seta j1 was instead. Morphological divergence was visualized by generating an ordination based on semistrong hybrid multidimensional scaling (SSH MDS) with PATN v2.27 [41]. The ordination was based on a Bray-Curtis distance matrix between mite specimens created using morphometric data standardized for body size to eliminate bias linked to body size, and transformed ((value – minimum)/range) to balance the weight of all measured characters. The ordination was generated based on 1000 iterations and 1000 random starts. Significant differences among groups detected in a given ordination were tested using ANOSIM (analysis of similarity), with 1000 iterations.
To ensure that specimens that underwent DNA extraction could be studied morphologically without any bias, the effect of DNA extraction was tested by comparing the morphology of specimens that underwent DNA extraction with specimens of the same species, and from that same host individual, that did not undergo extraction. Thirteen of the aforementioned morphological characters (standardized for body size) were examined for specimens of two species (Uroobovella orri, Trichouropoda californica) using Wilcoxon signed rank tests performed in SPSS v17 (SPSS Inc., Chicago, United States of America). No significant differences in morphology were observed between U. orri mites that underwent DNA extraction versus mites that did not undergo extraction, based on 13 characters and 15 pairwise comparisons (each pair consisting of two mites from the same host individuals; P = 0.078–0.995). DNA extraction had no significant effect on the morphology of T. californica specimens either (P = 0.139–0.799; 13 characters, 10 pairwise comparisons), except for two characters: median length and width of the sternal shield (P = 0.037, P = 0.009). The variation of these characters was most likely an artefact of slide mounting following DNA extraction, in that extraction weakens sclerotized tissue, which may have encouraged shields to fracture. Slide-mounted T. californica specimens that underwent DNA extraction had small fractures on either side of the sternal shield just posterior to the midpoint, and this may have increased sternal shield medial length and width measured relative to that of mites that did not undergo DNA extraction. With the exception of these two characters, DNA extraction did not significantly alter mite morphology, and as a result specimens that underwent extraction can be compared morphologically without any incurred bias.
Results
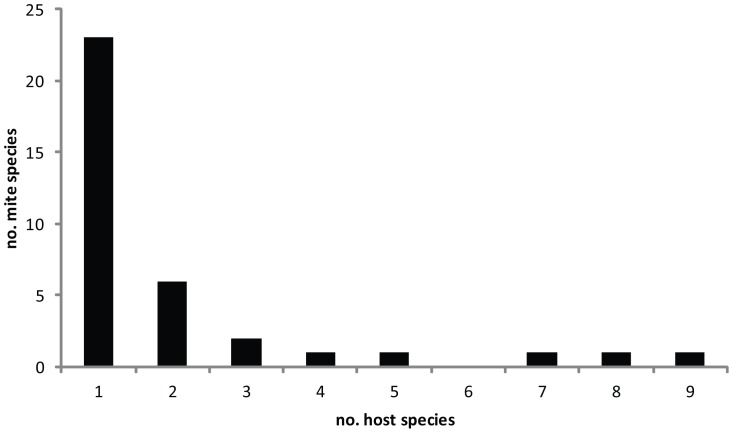
A total of 36 species of uropodoids (from three genera and two families) were found on 51 scolytine species (from 20 genera and 10 tribes), which were collected across 11 countries (Table 2). Of these 36 mite species, 13 are undescribed. The majority of the 36 species were collected from only one (64%) or two (17%) host species; fewer species were collected from three to nine host species (19%) (Fig. 2, Table 2). Most (76%) of the host associations observed in this study represent new records, and 19 of the 23 described species collected in this study had new host records (Table 3). There was little overlap in bark beetle hosts between this study and the literature for many of the common uropodoid species (e.g. T. australis, T. polytricha, and U. orri, each with only 1–3 host species shared; Table 3). The host records of many of the described species collected in this study are novel, when compared with published host records (Table 3). Most bark beetle species were associated with only one or two mite species; four host species had three mite species, and one host species (Polygraphus rufipennis) was associated with four mite species (Table 2).
Figure 2. Distribution of the breadth of host range of uropodoid mites.
Uropodoids collected from 51 species of bark beetles from 11 countries, showing the number of total mite species and the number of scolytine species used by each mite species. Note that these observed host ranges are based on opportunistic sampling from various regions; therefore, the true host ranges are possibly much broader.
Table 3. Comparing observed host records (this study) with published records (publ.) for described mite species collected from scolytines and other families of wood-boring beetles1 (*number of host spp. shared).
| Mite species | No. host spp/genera | Published host species (°spp. shared with present study) | Regions2 | References | |
| This study | Publ. | ||||
| Nenteria chiapasa | 1 | 0 | pine duff (needle litter) | Mexico | 54 |
| N. eulaelaptis | 1 | 0 | no host or habitat provided | Hungary, Mongolia | 25, 54 |
| N. moseri | 1 | 1 | Dendrocontus frontalis | Guatemala | 55 |
| Trichouropoda alascae | 1 | 2*/1 | Dendroctonus obesus, D. rufipennis° | AK | 28,56 |
| T. australis | 8/1 | 12***/3 | Dendroctonus brevicomis, D. frontalis, D. ponderosae, D. terebrans, D. simplex, Ips avulsus, I. bonanseai, I. calligraphus°, I. confusus, I. grandicollis°, I. pini°; CER: Neacanthosinus obsoletus | AZ, LA, MS, TX | 9,57,58 |
| T. bipilis | 1 | 1 | Scolytus pygmaeus | Austria | 29 |
| T. californica | 2/1 | 1* | Ips confusus° | CA | 59 |
| T. fallax | 3/2 | 5*/3 | Dendroctonus adjunctus, Hylastes ater, H. cunicularius, H. interstitialis, Hylurgops pinifex° | LA; Siberia; Belgium | 29,57 |
| T. hirsuta | 4/4 | 15/7 | Dendroctonus approximatus, D. brevicomis, D. frontalis, D. valens, Gnathotrichus materiarius, Ips avulsus, I. calligraphus, I. grandicollis, I. pini, Trypodendron scabricollis; CER: Monochamus carolinensis, M. scutellatus, M. titillator, Neacanthosinus obsoletus, Xyloterus sagittatus | AB, ON; AZ, LA, MS, TX | 9,27,57,58,60 |
| T. idahoensis | 1 | 1* | Ips pini° | ID | 27 |
| T. lamellosa | 2/2 | 10*/6 | Dendroctonus pseudotsugae, Dryocoetes confusus, Ips avulsus, I. calligraphus, I. grandicollis°; CER: Monochamus carolinensis, M. scutellatus, M. titillator, Neacanthosinus obsoletus, Xyloterus sagittatus | AB, ON; AZ, LA, MS | 9,14,57,58,60 |
| T. moseri | 2/2 | 1 | Dendroctonus simplex | AB | 25 |
| T. parisiana | 3/3 | 2/1 | Ips sexdentatus, I. typographus | France | 28 |
| T. perissopos | 1 | 1 | CUR: Perissops sobrinus | Poland | 27 |
| T. polygraphi | 1 | 1 | Polygraphus minor | India | 29 |
| T. polytricha | 7/1 | 7*/4 | Dryocoetes autographus, Hylurgops palliatus, Ips amitinus, I. cembrae°, I. hauseri, I. typographus, Pityogenes chalcographus | Austria, Germany, Poland, Turkey | 29,61 |
| T. polytrichasimilis | 3/1 | 1 | Ips sexdentatus; under bark of Pinus pinaster | France, Portugal | 25,62 |
| T. tegucigalpae | 2/1 | 3**/2 | Dendroctonus frontalis, Ips bonanseai°, I. cribricollis° | Honduras, Mexico | 27 |
| Uroobovella americana | 1 | 7*/3 | Dendroctonus pseudotsugae, D. terebrans, D. valens°, Gnathotrichus materiarius, Ips avulsus, I. calligraphus, I. grandicollis | AZ, LA | 9,57 |
| U. australiensis | 1 | 1 | CER: Pelargoderus arouensis | Australia | 63 |
| U. dryocoetes | 5/4 | 3*/3 | Dryocoetes autographus°, Hylastes cunicularius, Ips sexdentatus | Austria | 29 |
| U. orri | 9/6 | 11**/4 | Dendroctonus brevicomis, D. frontalis, D. obesus, D. pseudotsugae, D. valens, Dryocoetes confusus, Gnathotrichus materiarius°, Ips avulsus, I. calligraphus, I. grandicollis°, I. pini. | AZ, LA, MS, TX | 9,57 |
| U. vinicolora | 2/2 | 1 | Ips typographus | Germany | 61 |
CER = Cerambycidae, CUR = Curculionidae.
Provinces and states of Canada and USA follow accepted abbreviations.
Amplification of COI was attempted with 176 deutonymphal mites, from which only 116 (representing 29 species and three genera) from nine countries and 74 sites yielded sequence data (Table 2). COI was amplified from 122 specimens (116 ingroup and six outgroup specimens), with 608 characters in total, 328 constant, 19 parsimony-uninformative, and 261 parsimony-informative. Mean base pair frequencies (A: 0.294, C: 0.187, G: 0.153, T: 0.366) were found to be heterogeneous across all specimens (χ2 = 504.83, P<0.0001). The 28S D2–D4 region was used to assess the branching patterns observed in the COI reconstructions and to further test species boundaries. Partial 28S was amplified from 31 mites from 25 species (three genera) collected across nine countries and 26 sites, as well as from two outgroup specimens (Table 2), with 1069 characters in total, 446 constant, 114 parsimony-uninformative, and 509 parsimony-informative. Mean base pair frequencies (A: 0.239, C: 0.199, G: 0.283, T: 0.279) were found to be homogeneous across all specimens (χ2 = 92.12, P = 0.59). In each reconstruction, each specimen is labeled with a unique number, followed by the host species and abbreviated state, province or country (Table 2).
Pairwise Divergence
NJ analysis (K2P) of COI was performed on 122 mite specimens including 116 ingroup specimens (29 spp. total: 21 Trichouropoda, 2 Nenteria, and 6 Uroobovella spp.) and six outgroup specimens (four spp.). Average COI intraspecific pairwise distance was lowest among Trichouropoda species (1.5%±1.8) and slightly higher among Uroobovella species (1.9%±2.9) (Table 4). The maximum intraspecific divergence was high for both genera, with a maximum of 10.4% for T. polytricha and 12.5% for U. orri, both of which were between new and old world specimens (Table 4). Mean interspecific divergence within each genus was relatively high for all three genera (16.7–17.3%), and typically greater than intraspecific divergence (Table 4). The maximum divergence between Trichouropoda species was between T. hirsuta and T. moseri (23.4%), and the minimum was between T. n.sp. 11 and T. idahoensis (0.5%). The maximum for Uroobovella was between U. americana and U. orri (20.8%), and the minimum was between U. americana and U. vinicolora (8.4%) (Table 4). Average intergeneric divergence was high (18.6–21.5%), with the maximum divergence between T. hirsuta and U. australiensis (28.1%) (Table 4).
Table 4. Intra- and interspecific nucleotide divergence (%) ±standard deviation (range) of COI and 28S amplified from uropodoid mites associated with bark beetles.
| COI | 28S | |
| mean (range) | mean (range) | |
| Intraspecific | ||
| Trichouropoda | 1.5±1.8 (0–10.4) | 0.3±0.2 (0.1–0.5) |
| Nenteria 1 | – | – |
| Uroobovella | 1.9±2.9 (0–12.5) | 0.0±0.0 (0) |
| Interspecific | ||
| Trichouropoda | 16.7±2.9 (0.5–23.4) | 7.1±5.0 (0–16.6) |
| Nenteria | 16.9±0.0 (16.9) | 10.0±0.0 (10.0) |
| Uroobovella | 17.3±2.7 (8.4–20.8) | 32.7±15.9 (1.5–42.5) |
| Intergeneric | ||
| Trich – Nent | 18.6±1.2 (16.3–23.2) | 16.0±1.1 (13.8–20.0) |
| Trich – Uroob | 21.3±1.4 (17.7–28.1) | 34.9±3.9 (28.5–41.6) |
| Nent – Uroob | 21.5±1.3 (18.6–23.6) | 34.5±3.7 (29.0–41.1) |
Nenteria was represented by only 2 species, and each by a single individual.
NJ analysis of 28S was performed on 33 mite specimens including 31 ingroup specimens (25 spp. total: 18 Trichouropoda, 2 Nenteria, and 5 Uroobovella spp.), and two outgroup species. Average 28S intraspecific pairwise distance was highest among Trichouropoda species (0.3%±0.2), and lowest among Uroobovella species (0%±0) (Table 4). The maximum intraspecific divergence was relatively low for Trichouropoda with a maximum of 0.5% for T. californica, and low for Uroobovella with a maximum of 0% for U. n.sp. 6 and U. americana (Table 4). Mean interspecific divergence within each genus was moderate to very high (7.1–32.7%), and clearly higher than intraspecific divergence (Table 4). The maximum between Trichouropoda species was between T. hirsuta and T. n.sp. 11 (16.6%), and the minimum was between T. lamellosa and T. n.sp. 10 (0%) (Table 4). The maximum for Uroobovella species was between U. dryocoetes and U. orri (42.5%), and the minimum was between U. vinicolora and U. americana (1.5%) (Table 4). Average intergeneric divergence was high (16.0–34.9%), with the maximum pairwise distance between Trichouropoda lamellosa and Uroobovella dryocoetes (41.6%) (Table 4).
Bayesian Inference
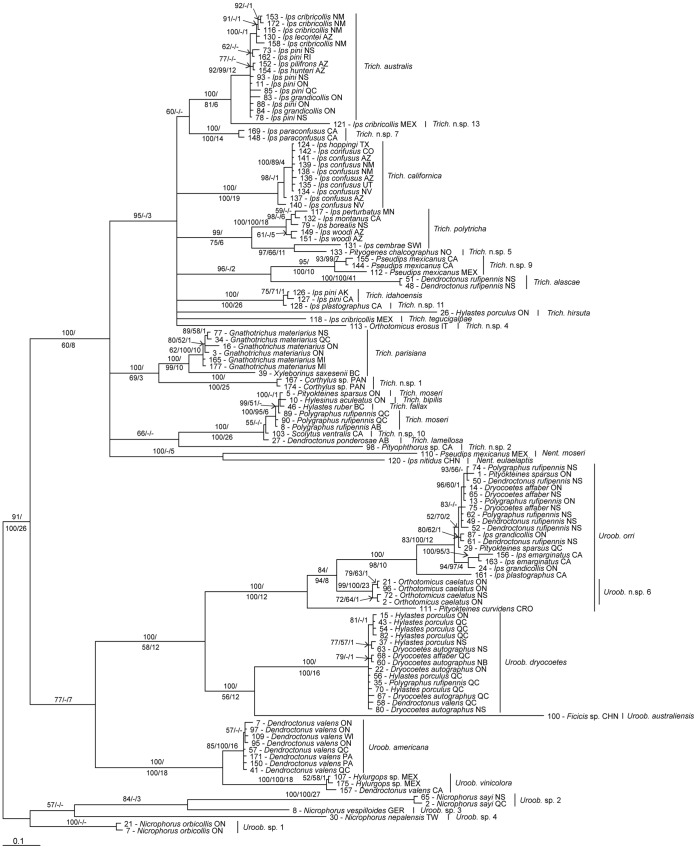
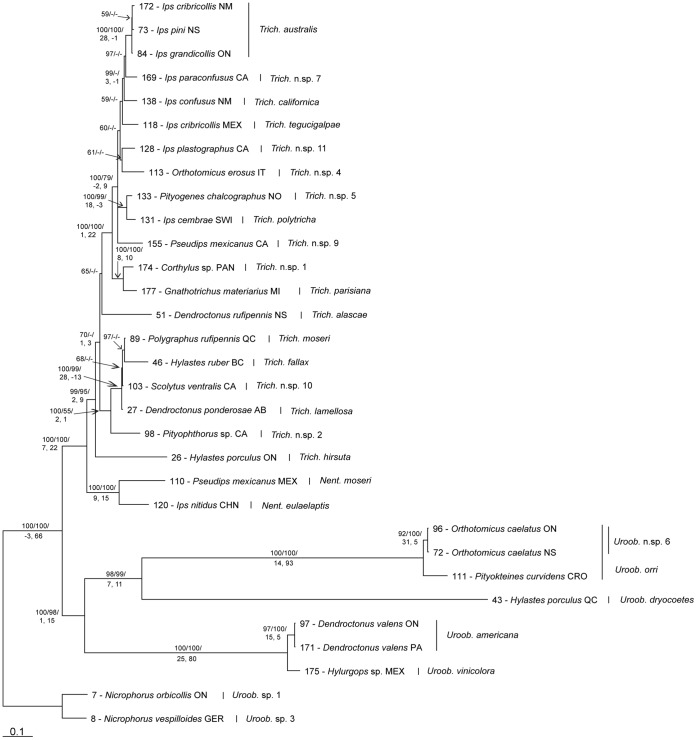
BI of COI was performed for 20 million generations, producing 38002 trees (after burn-in) which were summarized in a majority rule consensus tree (TL = 2021, CI = 0.2459, RI = 0.8277) (Fig. 3). The BI consensus tree was well supported, with most nodes having moderate to high posterior probabilities, with 26 nodes having 100% support, eight of which are basal nodes to ingroup species (Fig. 3). Some species, such as T. australis, T. californica, U. orri, U. dryocoetes, and U. americana, had multiple unresolved nodes collapsing into intraspecific polytomies. BI of 28S was performed for 10 million generations, producing 18002 trees (after burn-in) that were summarized in a majority rule consensus tree (TL = 1465, CI = 0.6881, RI = 0.8204) (tree not shown). The consensus tree was well supported: 12 nodes had 100% support, one of which was the node to the ingroup. BI of the concatenated dataset was performed for 10 million generations, producing 18002 trees (after burn-in) which were summarized in a majority rule consensus tree (TL = 2947, CI = 0.4964, RI = 0.6746) (Fig. 4). The total evidence consensus tree was well supported: 13 nodes had 100% support, including the basal node to the ingroup (Fig. 4).
Figure 3. Bayesian majority rule consensus tree based on COI from bark beetle associated uropodoids.
Majority rule consensus tree of 38002 trees generated by Bayesian MCMC analysis (20 million generations) of 608 bp fragment of COI from 122 uropodoid specimens, 116 ingroup specimens representing 29 species, and six outgroup specimens representing four species (TL = 2021, CI = 0.2459, RI = 0.8277) (Uroob. = Uroobovella, Trich. = Trichouropoda, Nent. = Nenteria). Posterior probability >50%/jackknife support >50%/Bremer support (JKS and BS from parsimony analysis).
Figure 4. Bayesian majority rule consensus tree based on COI and 28S from bark beetle associated uropodoids.
Majority rule consensus tree of 18002 trees generated by Bayesian MCMC analysis (10 million generations) of concatenated dataset of 608 bp fragment of COI and 1069 bp fragment of 28S from 31 specimens, 29 ingroup specimens representing 25 species, and two outgroup species (TL = 2947, CI = 0.4964, RI = 0.6746) (Uroob. = Uroobovella, Trich. = Trichouropoda, Nent. = Nenteria). Posterior probability >50%/jackknife support >50%/partitioned Bremer support (COI, 28S) (JKS and PBS from parsimony analysis).
Parsimony
The parsimony heuristic analysis of COI resulted in 34 most parsimonious trees (TL = 1928, CI = 0.2578, RI = 0.8383) presented in a semistrict consensus tree (tree not shown). Many nodes had moderate to high JKS which were mapped onto the Bayesian analysis of COI (Fig. 3), 18 nodes had 100% jackknife support (JKS). Many nodes had poor Bremer support, with 24 nodes with moderate to strong support (≥10), as shown in the Bayesian phylogeny (Fig. 3). Nine of the nodes with 100% JKS and strong Bremer support are basal nodes to ingroup species. Similar to the BI, T. australis, T. californica, U. orri, U. dryocoetes, and U. americana had multiple unresolved nodes collapsing into intraspecific polytomies. The heuristic analysis of 28S produced 14 most parsimonious trees (TL = 1462, CI = 0.6895, RI = 0.8216) presented in a semistrict consensus tree (tree not shown). Most nodes had moderate to strong Bremer support and nearly every node had JKS, with 12 nodes having 100% JKS, one of which was the basal node to the ingroup. Multiple Trichouropoda species showed little interspecific divergence resulting in a large polytomy. The parsimony analysis of the concatenated dataset resulted in three most parsimonious trees (TL = 2924, CI = 0.5003, RI = 0.6797) presented in a semistrict consensus tree (tree not shown). Most nodes had moderate to strong JKS, with 10 nodes having 100% JKS, including the basal node to the ingroup and to the Trematuridae, and many nodes had moderate to strong PBS, as shown in the Bayesian analysis (Fig. 4).
Summary of Molecular Reconstructions
The parsimony and Bayesian analyses of COI, 28S and concatenated datasets yielded similar results. All COI analyses suggested that each trematurid (Trichouropoda and Nenteria) species was monophyletic, with the exception of T. moseri and T. polytricha. Trichouropoda moseri collected from Pityokteines sparsus consistently grouped separately from those collected from Polygraphus rufipennis. Trichouropoda polytricha collected from Ips cembrae from Switzerland was consistently shown to be more closely related to T. n.sp. 5 from Norway than to other North American T. polytricha specimens.
Overall, the relationships between trematurid species were poorly resolved using 28S, with slightly better resolution in the concatenated dataset, and the best resolution using COI alone. The D2–D4 region of 28S was not effective for examining the relationships between some closely related Trichouropoda species. The 28S and COI analyses were not entirely congruent. In all 28S reconstructions, T. hirsuta was basal to all other species in the genus, whereas T. n.sp. 2 was the basal species in COI reconstructions. COI and 28S also disagreed on the placement of T. fallax and T. alascae. COI provided more insight into the relationships between trematurid species than 28S. The concatenated dataset produced well-supported trees, which were more resolved than those based on 28S alone. The placement of a few Trichouropoda species differed between the 28S and concatenated reconstructions, reflecting the differences in trematurid species relationships independently inferred from COI versus 28S.
Across all reconstructions the monophyly of all Uroobovella species were well supported and the relationships between Uroobovella species were consistent across all analyses. In particular, U. orri, U. n.sp. 6, U. dryocoetes and U. australiensis appear to be most closely related to each other, whereas U. americana and U. vinicolora are most closely related to each other. Across all COI analyses there was a small well-supported clade grouping U. orri specimens from Orthotomicus caelatus beetles, which has been labeled as U. n.sp. 6.
Morphological Analysis
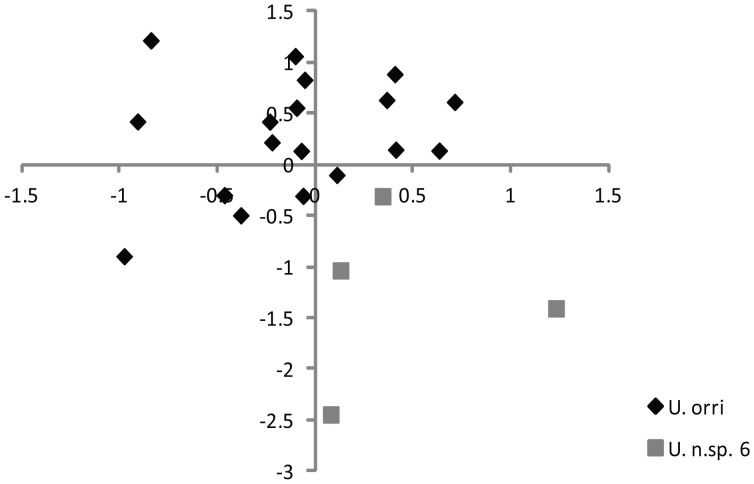
To test whether host generalists displayed cryptic morphological diversity, the level of ‘intraspecific’ morphological divergence was assessed in five species with broad host ranges (T. australis, T. parisiana, T. polytricha, U. orri, U. dryocoetes), and two species with relatively narrow host ranges (T. californica and U. americana). Uroobovella orri was the only species of the seven examined that showed prominent morphological variation, with two apparent groupings in the ordination: mites from Orthotomicus caelatus, labelled as U. n.sp. 6, and mites from hosts (8 host spp.) other than O. caelatus (Fig. 5). The SSH MDS ordination (stress = 0.1571) (Fig. 5) and ANOSIM based on 14 morphological characters measured from 22 U. orri specimens indicate that U. orri and U. n.sp. 6 are significantly distinct morphologically (P = 0.01). Subsequently, slide-mounted specimens were examined closely for variation in discrete morphological characters that could be used to distinguish U. orri and U. n.sp. 6, but this investigation revealed no distinct character states. Mean COI divergence among U. n.sp. 6 specimens was low (0.5% ±0.31), where as the mean divergence between U. n.sp. 6 and other U. orri specimens from North America was 20 times higher (10.5% ±0.4).
Figure 5. SSH MDS ordination showing morphological dissimilarity among Uroobovella species.
Ordination with Bray-Curtis distance performed on measurements ((value – min)/range transformed) of 14 morphological characters from 22 uropodoids representing U. orri and U. n.sp. 6 (stress = 0.1571).
The remaining six generalist and two species with narrow host ranges displayed no significant intraspecific variation in morphometrics or discrete (qualitative) morphological characters; these species also showed low COI intraspecific divergence (<1%), with the exception of T. polytricha and T. parisiana with 4.6% (±3.8) and 2.8% (±2.7) divergence, respectively. The relatively high level of divergence among T. polytricha specimens was largely due to a single specimen from Switzerland; intraspecific divergence among North America specimens was 2% (±0.8).
Discussion
This study indicates that both partial COI and 28S D2–D4 are suitable markers for distinguishing between closely related uropodoid species, with 17% average divergence among species for both markers. 28S appears to be a good marker for separating closely related Uroobovella species, but COI was far more effective at delineating between Trichouropoda species. Most morphologically defined species were well supported in the COI phylogeny, with the exception of T. moseri and T. polytricha. The congruence between morphological and molecular data emphasizes the fact that the best approach is an integrative approach [42], and that morphology-based taxonomy is still relevant and essential [43].
Host Specificity and Cryptic Species
A total of 36 species of uropodoids, including 13 undescribed species, were collected in this study, and these mites exhibited various levels of host specificity. The majority of mite species were collected from one (64%) or two (17%) host species, and seven species (19%) had three or more host species. However, the opportunistic sampling used in this study and the haphazard coverage of hosts and regions may incur a bias towards higher apparent host specificity. Considering published host records, it appears that strict host specificity may be the exception rather than the rule. The observed host associations in this study nearly doubled the number of host records for the described species studied (54% increase from 87 records to 134), and this highlights the lack of knowledge in this group. Considering that only a small proportion of the global bark beetle fauna has been examined for uropodoids, we suspect that many more new and/or cryptic species may be uncovered with further investigations.
Typically, when the species boundaries of symbiotic taxa are assessed using molecular techniques it is revealed that apparent generalists are actually complexes of cryptic specialists (e.g. [5], [7], [44]). To the contrary, in this study molecular and morphological analyses suggested that putative host generalists do not represent complexes of cryptic species with narrower host ranges, but that they are truly single species with a broad host range, with the exception of one species (U. orri). It is possible that some of these apparent generalists comprise rare specialists that remain to be collected, or that additional markers may uncover cryptic specialists, but it is also possible that these species are truly generalists.
Uroobovella orri was the only host generalist that appears to represent at least two distinct species in North America, including a widespread generalist associated with at least eight species and six genera of hosts, and a specialist (U. n.sp. 6) associated with Orthotomicus caelatus (based on COI data). Interestingly, O. caelatus is a host-tree generalist and attacks many species of Pinus, Picea and Larix throughout its range [8]. In addition, the single specimen of U. orri found on Pityokteines curvidens (another conifer generalist) from Croatia may also represent a distinct cryptic species, based upon the level of COI divergence from other U. orri specimens (11.5% ±0.7). Considering that U. orri has been collected from many other bark beetle species that were not included in this study, it is possible that we have only begun to scratch the surface of a diverse complex of cryptic species.
In all COI reconstructions both T. moseri and T. polytricha were paraphyletic, and this may suggest that these two species represent multiple cryptic species associated with different hosts. Trichouropoda moseri collected from Pityokteines sparsus (Ipini) and Polygraphus rufipennis (Polygraphini) were paraphyletic, and these may represent two cryptic host-specific species rather than a single host generalist; however, no morphometric differences were found, and average COI divergence among T. moseri specimens was very low (0.4% ±0.2). Trichouropoda polytricha found on Ips cembrae from Switzerland was more closely related to T. n.sp. 5 from Norway (Pityogenes chalcographus) than to North American T. polytricha. Despite being apparently morphologically identical, it is possible that the North American and European T. polytricha represent two cryptic species. Alternatively, the paraphyly of T. moseri and T. polytricha may be a result of inadequate taxon sampling, or incomplete lineage sorting. More specimens and additional markers are needed to clarify the taxonomic boundaries of these two mites.
The host associations of the closely related uropodoids, T. parisiana and T. n.sp.1, are unique and likely warrant future investigations. Trichouropoda parisiana and T. n.sp. 1 were both associated with ambrosia beetles, an ecological grade of scolytine and platypodine curculionids that carry symbiotic fungi (in complex glandular mycangial structures) which is inoculated into host trees and cultivated as a food source [8]. Trichouropoda parisiana was collected from three distantly related ambrosia beetles, Gnathotrichus materiarius (Corthylini), Xyleborinus saxesenii (Xyleborini) and Trypodendron retusum (Xyloterini), which attack a broad range of unrelated host trees (Pinus and Picea spp.; numerous trees and shrubs; Populus spp., respectively) [8]. Trichouropoda n.sp. 1 is morphologically and genetically similar to T. parisiana, and it was only collected from Corthylus sp. (Corthylini), an ambrosia beetle associated with deciduous trees [8]. It is likely that a common ancestor of T. parisiana and T. n.sp. 1 was originally associated with ambrosia beetles, and that descendant populations tracked some aspect of the mycetophagous life history of their hosts. However, testing this hypothesis further will be difficult given that these two mites are associated with hosts that feed on unrelated host trees in different countries [8]. Trichouropoda n.sp. 6 and T. n.sp. 8 were also collected from ambrosia beetles, Camptocerus auricomis and Monarthrum dentigerum respectively; however, since neither species yielded COI or 28S data, the phylogenetic relationships between these species and T. parisiana and T. n.sp. 1 are not understood.
Coevolution
The evolutionary history of associated symbionts may reflect a long-term coevolutionary relationship, or it may reflect a history of host switching and ecological tracking [45], [46]. Overall, the evolution of scolytine-associated uropodoids shows little evidence of coevolution with their hosts or tracking ecologically similar host species. Phylogenetically related bark beetles [47]–[49] did not necessarily share the same or closely related mite species, and ecologically related host species, which have similar host tree ranges, overlapping geographic ranges or similar phenologies [8], [50] were not necessarily associated with the same or closely related uropodoid species.
An obstacle to the study of coevolution between bark beetles and uropodoids is that phylogenetically related hosts are often ecologically similar (e.g. host tree species, habitat range, feeding ecology, and phenology; [8], [50]), making it difficult to discern the determinants of host associations. For example, T. californica is phoretic on two sister-species, Ips hoppingi and I. confusus [49]. However, I. hoppingi and I. confusus are peripatric and similar ecologically, both feeding on pinyon pine (Pinus) species [8], and therefore it is very difficult to pinpoint the causal factor(s) in the association of T. californica with these two host species. Additionally, the ecology of bark beetle associated uropodoids are poorly understood, which hampers any interpretations of the extent to which mites may be tracking ecologically similar hosts. Future investigations into the extent to which uropodoids may be coevolving with their bark beetle hosts will require much more extensive taxon sampling than that of this study, as well as a more complete and resolved phylogeny of associated mites and their scolytine hosts, and an improved understanding of the ecology of these mites.
Acknowledgments
We thank H. Douglas and the CFIA Invasive Alien Species Monitoring program for providing bark beetles from across Canada. We are grateful to B. Jones, J. Dombroskie, G. Smith, and R.J. Buss for donating specimens. We thank H.W. Knee and T. Knee for sampling for bark beetles in northern Alberta. We also thank H. Klompen for his advice on many of the molecular aspects of the study; J. Gibson and M. Jackson for their input on analyses; and E. Lindquist for his comments on a previous version of the manuscript. T. Hartzenberg and R. Shewchuk for their assistance in the field and the lab, as well as the private land owners who permitted sampling on their property.
Funding Statement
This study was funded by an NSERC Discovery Grant to MRF, and in part by the USDA-FS EDRR program (# 07-DG-11420004-182) and by US-NSF (DEB-0328920) to AIC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, et al. (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22: 148–155. [DOI] [PubMed] [Google Scholar]
- 2. Fontaneto D, Kaya M, Herniou EA, Barraclough TG (2009) Extreme levels of hidden diversity in microscopic animals (Roifera) revealed by DNA taxonomy. Mol Phylogenet Evol 53: 182–189. [DOI] [PubMed] [Google Scholar]
- 3. Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the Neotropical skipper butterfly Astraptes fulgerator . Proc Natl Acad Sci 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilcox TP, Hugg L, Zeh JA, Zeh DW (1997) Mitochondrial DNA sequencing reveals extreme genetic differentiation in a cryptic species complex of Neotropical pseudoscorpions. Mol Phylogenet Evol 7: 208–216. [DOI] [PubMed] [Google Scholar]
- 5. McCoy KD, Boulinier T, Tirard C, Michalakis Y (2001) Host specificity of a generalist parasite: genetic evidence of sympatric host races in the seabird tick Ixodes uriae . J Evol Biol 14: 395–405. [Google Scholar]
- 6. Dietrich M, Kempf F, Gómez-Díaz E, Kitaysky AS, Hipfner M, et al. (2012) Inter-oceanic variation in patterns of host-associated divergence in a seabird ectoparasite. J Biogeogr 39: 545–555. [Google Scholar]
- 7. Knee W, Beaulieu F, Skevington JH, Kelso S, Forbes MR (2012) Cryptic species of mites (Uropodoidea: Uroobovella spp.) associated with burying beetles (Silphidae: Nicrophorus): the collapse of a host generalist revealed by molecular and morphological analyses. Mol Phylogenet Evol 65: 276–286. [DOI] [PubMed] [Google Scholar]
- 8. Wood SL (1982) The bark and ambrosia beetles of North and Central America (Coleoptera: Scolytidae): a taxonomic monograph. Great Basin Nat Mem 6: 1–1359. [Google Scholar]
- 9. Moser JC, Roton LM (1971) Mites associated with southern pine bark beetles in Allen Parish, Louisiana. Can Entomol 103: 1775–1798. [Google Scholar]
- 10. Kinn DN, Witcosky JJ (1978) Variation in southern pine beetle attack height associated with phoretic uropodid mites. Can Entomol 110: 249–251. [Google Scholar]
- 11. Błoszyk J, Bajaczyk R, Markowicz M, Gulvik M (2003) Geographical and ecological variability of mites of the suborder Uropodina (Acari: Mesostigmata) in Europe. Biol Lett 40: 15–35. [Google Scholar]
- 12. Athias-Binche F (1989) General ecological principles which are illustrated by population studies of uropodid mites. Adv Ecol Res 19: 303–344. [Google Scholar]
- 13. Kinn DN (1982) Seasonal distribution of three common mite associates of the southern pine beetle (Coleoptera: Scolytidae) in central Louisiana. Fla Entomol 65: 185–187. [Google Scholar]
- 14. Kinn DN (1987) Incidence of pinewood nematode dauerlarvae and phoretic mites associated with long-horned beetles in central Louisiana. Can J For Res 17: 187–190. [Google Scholar]
- 15. Hofstetter RW, Moser JC, McGuire R (2009) Observations on the mite Schizosthetus lyriformis (Acari: Parasitidae) preying on bark beetle eggs and larvae. Entomol News 120: 397–400. [Google Scholar]
- 16. Moser JC (1975) Mite predators of the southern pine beetle. Ann Entomol Soc Am 68: 1113–1116. [Google Scholar]
- 17. Anderson DL, Trueman JWH (2000) Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp Appl Acarol 24: 165–189. [DOI] [PubMed] [Google Scholar]
- 18. Dowling APG, OConnor BM (2010) Phylogenetic relationships within the suborder Dermanyssina (Acari: Parasitiformes) and a test of dermanyssoid monophyly. Internat J Acarol 36: 299–312. [Google Scholar]
- 19. Kawazoe K, Kawakita A, Kameda Y, Kato M (2008) Redundant species, cryptic host-associated divergence, and secondary shift in Sennertia mites (Acari: Chaetodactylidae) associated with four large carpenter bees (Hymenoptera: Apidae: Xylocopa) in the Japanese island arc. Mol Phylogenet Evol 49: 503–513. [DOI] [PubMed] [Google Scholar]
- 20. Martin P, Dabert M, Dabert J (2010) Molecular evidence for species separation in the water mite Hygrobates nigromaculatus Lebert, 1879 (Acari, Hydrachnidia): evolutionary consequences of the loss of larval parasitism. Aquat Sci 72: 347–360. [Google Scholar]
- 21. Schäffer S, Pfingstl T, Koblmüller S, Winkler KA, Sturmbauer C, et al. (2010) Phylogenetic analysis of European Scutovertex mites (Acari, Oribatida, Scutoverticidae) reveals paraphyly and cryptic diversity: a molecular genetic and morphological approach. Mol Phylogenet Evol 55: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skoracka A, Dabert M (2010) The cereal rust mite Abacarus hystrix (Acari: Eriophyoidea) is a complex of species: evidence from mitochondrial and nuclear DNA sequences. B Entomol Res 100: 263–272. [DOI] [PubMed] [Google Scholar]
- 23.Bright DE (1976) The insects and arachnids of Canada, part 2. The bark beetles of Canada and Alaska Coleoptera: Scolytidae. Canada Department of Agriculture Publication No. 1576. Information Canada, Ottawa.
- 24. Alonso-Zarazaga MA, Lyal CHC (2009) A catalogue of family and genus group names in Scolytinae and Platypodinae with nomenclatural remarks (Coleoptera: Curculionidae). Zootaxa 2258: 1–134. [Google Scholar]
- 25.Hirschmann W (1972) Gangsystematik der Parasitiformes. Acarologie 17: 1–37+1–9 pl.
- 26. Hirschmann W, Wiśniewski J (1985) Gangsystematik der Parasitiformes weltweite revision der gattung Nenteria Oudemans 1915. Acarologie 32: 1–185. [Google Scholar]
- 27. Hirschmann W, Wiśniewski J (1986) Gangsystematik der Parasitiformes weltweite revision der gattung Trichouropoda Berlese 1916 tiel I. Acarologie. 33: 1–181. [Google Scholar]
- 28. Hirschmann W, Wiśniewski J (1987) Gangsystematik der Parasitiformes weltweite revision der gattung Trichouropoda Berlese 1916 tiel II. Acarologie 34: 1–180. [Google Scholar]
- 29. Hirschmann W, Wiśniewski J (1989) Gangsystematik der Parasitiformes weltweite revision der gattung Trichouropoda Berlese 1916 teil IV. Acarologie 36: 1–196. [Google Scholar]
- 30.Hirschmann W, Zirngiebl-Nicol I (1961) Gangsystematik der Parasitiformes teil 4: die gattung Trichouropoda Berlese 1916 nov. comb., die cheliceren und das system der uropodiden. Acarologie 4: 1–41+1–16 pl.
- 31. Gibson JF, Kelso S, Skevington JH (2010) Band-cutting no more: a method for the isolation and purification of target PCR bands from multiplex PCR products using new technology. Mol Phylogenet Evol 56: 1126–1128. [DOI] [PubMed] [Google Scholar]
- 32.Maddison WP, Maddison DR (2010) Mesquite: a modular system for evolutionary analysis v2.74. Available: http://mesquiteproject.org.
- 33. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 34.Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts.
- 35. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 36. Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 37. Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- 38.Nylander JAA (2004) MrModetest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- 39. Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, Louisiana, 14 November 2010: 1–8. [Google Scholar]
- 40. Evans GO, Till WM (1965) Studies on the British Dermanyssidae (Acari: Mesostigmata). Part I. External morphology. Bull Brit Mus Nat Hist Zool 13: 247–294. [Google Scholar]
- 41.Belbin L (2003) PATN. A software for extracting and displaying patterns in any type of complex (multivariate) data, http://www.patn.com.au/.
- 42. Rubinoff D, Cameron S, Will K (2006) A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J Hered 97: 581–594. [DOI] [PubMed] [Google Scholar]
- 43. Hołyński RB (2010) Taxonomy and the mediocrity of DNA barcoding – some remarks on Packer, et al. 2009: DNA barcoding and the mediocrity of morphology. Arthropod Syst Phylogeny 68: 143–150. [Google Scholar]
- 44. Smith MA, Wood DM, Janzen DH, Hallwachs W, Hebert PDN (2007) DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. Proc Natl Acad Sci 104: 4967–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paterson AM, Banks J (2001) Analytical approaches to measuring cospeciation of host and parasites: through a glass, darkly. Int J Parasitol 31: 1012–1022. [DOI] [PubMed] [Google Scholar]
- 46. Kethley JB, Johnston DE (1975) Resource tracking patterns in bird and mammal ectoparasites. Misc Publ Entomol Soc Am 9: 231–236. [Google Scholar]
- 47. Jordal B, Gillespie JJ, Cognato AI (2008) Secondary structure alignment and direct optimization of 28S rDNA sequences provide limited phylogenetic resolution in bark and ambrosia beetles (Curculionidae: Scolytinae). Zool Scr 37: 43–56. [Google Scholar]
- 48. Jordal BH, Sequeira A, Cognato AI (2011) The age and phylogeny of wood boring weevils and the origin of subsociality. Mol Phylogenet Evol 59: 708–724. [DOI] [PubMed] [Google Scholar]
- 49. Cognato AI, Sun JH (2007) DNA based cladograms augment the discovery of a new Ips species from China (Coleoptera: Curculionidae: Scolytinae). Cladistics 23: 539–551. [DOI] [PubMed] [Google Scholar]
- 50. Wood SL, Bright DE (1992) A catalog of Scolytidae and Platypodidae (Coleoptera), part 2: taxonomic index. Great Basin Nat Mem 13: 1–1553. [Google Scholar]
- 51. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–297. [PubMed] [Google Scholar]
- 52. Park JK, Foighil DÓ (2000) Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Mol Phylogenet Evol 14: 75–88. [DOI] [PubMed] [Google Scholar]
- 53. Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC (1997) The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol 46: 1–68. [DOI] [PubMed] [Google Scholar]
- 54.Hirschmann W (1978) Gangsystematik der Parasitiformes. Acarologie 24: 1–129+1–20 pl.
- 55.Hirschmann W (1972) Gangsystematik der Parasitiformes. Acarologie 18: 1–128+1–13 pl.
- 56. Cardoza YJ, Moser JC, Klepzig KD, Raffa KF (2008) Multipartite symbioses among fungi, mites, nematodes, and the spruce beetle, Dendroctonus rufipennis . Environ Entomol 37: 956–963. [DOI] [PubMed] [Google Scholar]
- 57.Hofstetter RW (2008) Information and images of mites associated with bark beetles and their predators. Available: http://oak.ucc.nau.edu/rh245/MiteImages.htm. Accessed 2008 Sept 30.
- 58. Kinn DN, Linit MJ (1989) A key to phoretic mites commonly found on long-horned beetles emerging from southern pines. USDA For Serv Res Note 357: 1–8. [Google Scholar]
- 59. Hirschmann W, Wiśniewski J (1988) Gangsystematik der Parasitiformes weltweite revision der gattung Trichouropoda Berlese 1916 tiel III. Acarologie 35: 1–201. [Google Scholar]
- 60.Knee W, Hartzenberg T, Forbes MR, Beaulieu F (2012) The natural history of mites (Acari: Mesostigmata) associated with the white-spotted sawyer beetle (Coleoptera: Cerambycidae): diversity, phenology, host attachment, and sex bias. Can Entomol. In press.
- 61. Moser JC, Bogenschutz H (1984) A key to the mites associated with flying Ips typographus in south Germany. Z Angew Entomol 97: 437–450. [Google Scholar]
- 62. Levieux J, Lieutier F, Moser JC, Perry TJ (1989) Transportation of phytopathogenic fungi by the bark beetle Ips sexdentatus Boerner and associated mites. J Appl Entomol 108: 1–11. [Google Scholar]
- 63. Wiśniewski J, Hirschmann W (1992) Gangsystematik der Parasitiformes teil 538. Gang und teilgang von 2 neuen Uroobovella-arten der fimicola- und vinicolora-gruppe aus kuba (Dinychini, Uropodinae). Acarologie 39: 100–109. [Google Scholar]