Abstract
Transcription factor GATA4 is a key regulator of cardiomyocyte growth, and differentiation and 50% reduction in GATA4 levels results in hypoplastic hearts. Search for GATA4 targets/effectors revealed cyclin D2 (CD2), a member of the D-type cyclins (D1, D2, and D3) that play a vital role in cell growth and differentiation as a direct transcriptional target and a mediator of GATA4 growth in postnatal cardiomyocytes. GATA4 associates with the CD2 promoter in cardiomyocytes and is sufficient to induce endogenous CD2 transcription and to dose-dependently activate the CD2 promoter in heterologous cells. Cardiomyocyte-specific overexpression of CD2 results in enhanced postnatal cardiac growth because of increased cardiomyocyte proliferation. When these transgenic mice are crossed with Gata4 heterozygote mice, they rescue the hypoplastic cardiac phenotype of Gata4+/− mice and enhance cardiomyocyte survival and heart function. The data uncover a role for CD2 in the postnatal heart as an effector of GATA4 in myocyte growth and survival. The finding that postnatal upregulation of a cell-cycle gene in GATA4 haplo-insufficient hearts may be protective opens new avenues for maintaining or restoring cardiac function in GATA4-dependent cardiac disease.
Keywords: hypertrophy, apoptosis, cardiomyocyte proliferation, transgenic mice
d-type cyclins (cyclins D1, D2, and D3) are G1 cyclins that play a vital role in differentiation and cell-cycle regulation (30). In response to mitogenic signals, cyclin Ds were shown to associate with the cyclin-dependant kinases 4 and 6 (CDK4 and CDK6), phosphorylate the tumor suppressor retinoblastoma protein along with the family members p107 and p130, and control cell-cycle progression (12, 30). However, and despite compelling evidence from in vitro studies on the role of D-type cyclins in the cell cycle, in vivo work indicated that during embryonic development, CD-CDK4 and CD-CDK6 complexes are not required for proliferation of the majority of cell types (14). Mice embryos lacking all three D-type cyclins are viable and relatively normal at embryonic day 13.5 but later develop cardiac abnormalities including severely thinned walls and ventricular septal defects as well as severe anemia leading to embryonic death (14, 15). These observations together with the data from the CDK4/CDK6 double-knockout mice led to the conclusion that in certain compartments such as the myocardium and the hematopoietic system, D-type cyclins are indispensible for cell proliferation (15). Consistent with a cell-specific function, D-type cyclins have distinct tissue- and cell-specific expression patterns (3, 15, 33), the transcription mechanisms underlying this spatial specificity remain incompletely understood.
During embryogenesis, heart development involves coordinated cardiomyocyte differentiation and proliferation. Myocyte proliferation drastically decreases soon after birth, and postnatal heart growth occurs mostly through the enlargement of cardiomyocytes size, a process termed cardiac hypertrophy (19). Additionally, evidence suggests that the adult mammalian heart can be induced to regenerate (19). For example, inhibition of p38 MAPK enables proliferation of adult cardiomyocytes (7). Similarly, combined deletion of retinoblastoma protein plus p130 genes enhances myocytes proliferation in part through upregulations of G1-dependent kinase activities (18). A direct role of G1 cyclins in promoting postnatal cardiomyocyte proliferation has been reported (28). Nuclear import of cyclin D1/CDK4 enables postmitotic cardiomyocytes to enter the cell cycle and divide (31), whereas cardiac-specific overexpression of cyclin D2 (CD2) enhances postischemic heart repair (9, 24). More recently, it was shown that neonate mammalian cardiomyocytes could be induced to proliferate following partial surgical resection leading to transient regenerative potential of neonate hearts (25). Interestingly, in zebrafish, which can regenerate their heart after amputation of up to 20% of the ventricle, the regenerated cardiac cells were shown to arise from differentiated cardiomyocytes that undergo limited dedifferentiation followed by proliferation (11); a population of cardiomyocytes in the zebrafish ventricular wall appear to contribute to cardiac muscle regeneration by inducing expression of transcription factor GATA4, a critical regulator of cardiogenesis (13).
Initially identified as an upstream regulator of the cardiac natriuretic peptide precursors A (NPPA) and B (NPPB), GATA4 regulates a plethora of cardiac genes involved in several cellular processes, including differentiation, proliferation, and survival (20, 21). GATA4 deletion from embryonic cardiomyocytes consistently leads to myocardial thinning, supporting a role for GATA4 in myocyte proliferation (22, 26, 34). Although many direct transcription targets have been identified, the effectors of GATA4 actions are incompletely understood. Interestingly, CD2 was shown to be regulated by GATA4 in the anterior heart field of developing embryos (27); GATA4 was also shown to cooperate with KLF13 in activating CD1 (16). Whether CD2 (or CD1) activation mediates GATA4-dependent cardiomyocyte proliferation has not yet been established. In addition to its critical role for embryonic heart development, GATA4 plays an important role in the postnatal heart where it is required for cardiomyocyte survival and adaptive response. A number of stimuli that induce cardiac hypertrophy were shown to increase GATA4 levels, transcriptional activity, and/or DNA binding, and upregulation of GATA4 is sufficient to induce hypertrophic growth of neonate cardiomyocytes (4, 17). Loss of one Gata4 allele results in hypoplastic hearts, increased cardiomyocyte apoptosis, and impaired adaptive response (2). Surprisingly, targeted upregulation of GATA4 specifically in adult hearts either through adenovirus-mediated delivery to rat hearts or inducible transgenesis in mice is cardioprotective but not associated with myocyte proliferation, suggesting that these terminally differentiated cells may lack critical GATA4 cofactors and/or effectors for proliferative growth (10, 29).
A search for GATA4 targets/effectors in neonatal cardiomyocytes revealed that CD2 levels were exquisitely sensitive to GATA4 and chromatin immunoprecipitation confirmed in vivo GATA4 occupancy of the CD2 promoters in these cells. We therefore tested whether CD2 may mediate GATA4 growth effects in cardiomyocytes. We report that when crossed with Gata4+/− mice, transgenic mice with myocardial-specific expression of CD2 are able to rescue Gata4+/− hypoplastic hearts and restore cardiac function to control wild-type (WT) level. In Gata4+/− mice, CD2 rescued basal as well as doxorubicin (Dox)-induced cardiomyocyte apoptosis and promoted cardiomyocyte proliferation as evidenced by the increased number of Ki67+ +ve cells. Together, the data support a role for CD2 as an effector of GATA4 in cardiomyocyte proliferation. They also unravel potential new protective function for CD2 in adult hearts.
MATERIALS AND METHODS
Plasmids.
CD2-luciferase was generated by subcloning the rat CD2 promoter in the PxP1 vector. GATA4 expression vectors were previously described (2, 6). All constructs were confirmed by sequencing.
Cell cultures and transfections.
Primary cardiomyocytes, NIH3T3, and 293T cells were maintained in culture and transfected as previously described (2, 8). Atrial and ventricular cardiomyocyte primary cultures were prepared from 4- to 5-day-old Sprague-Dawley rats (Charles River). Neonatal rats were euthanized by decapitation. Atrial and ventricular tissues were aseptically removed and washed with Joklik's modified Eagle's medium (GIBCO). The tissues were then minced and subjected to three sequential digestions of 30, 20, and 10 min each in 0.1% collagenase (Cooper Biomedicals). To stop the enzymatic digestion, we added cold fetal calf serum to a final concentration of 28.5%. Undigested tissue remnants were removed by filtration through a nylon mesh (pore size, 100 pm). The cell-containing filtrate was then centrifuged, and the resulting cell pellet was resuspended in Dulbecco's modified Eagle's medium, supplemented with 15% fetal calf serum. To eliminate fibroblasts, we preplated the cells for 20- to 30-min periods, after which the unattached cardiomyocyte-enriched cells were collected. The cardiac cells were plated in Primaria (Falcon) plates. The experiments were done at least twice in duplicate with different DNA preparations.
RT-PCR.
The oligonucleotides used for amplification of CD2 were 5′-CGT CTA GAA TGG AGC TGCT GTG CTG CGA GGT GG-3′ (forward) and 5′-GGG GTA CCT CAC AGG TCA ACA TCC CGC ACG TC-3′ (reverse). RT-PCR was performed as previously described (5).
Chromatin immunoprecipitation assays and quantitative PCR analysis.
Chromatin immunoprecipitation assay was performed as previously described (32). The primers used were 5′-AAAGTTTCCGCACGAGGGTCAT-3′ (sense) and 5′-CCCTGAGGCTTAGACTCCTGATAACT-3′ (antisense) for the GATA site, and 5′-TTTGAAGTTTGGTCAGGCCAGC-3′ (sense) and 5′-GCAAGCTGGAAGGGCAGTTAGAT-3′ (antisense) for the GATA-like site.
Real-time PCR analysis.
Total RNA was isolated from mouse tissues using TRIzol reagent (Invitrogen). Transcript levels of the various markers were determined by real-time PCR as previously described (5).
Electrophoretic mobility shift assay.
Nuclear extracts were prepared from cardiomyocytes in primary cultures. Binding reactions were done at room temperature using 1 μg of poly(dI-dC). The probes used for binding are GATA-like, 5′-GAG GGA AAG ATT GAA AGG AG-3′ (sense) and 5′-CTC CTT TCA ATC TTT CCC TC-3′ (antisense); mutated GATA-like, 5′-GAG GGA AAG gTT GAA AGG AG-3′ (sense) and 5′-CTC CTT TCA AcC TTT CCC TC-3′ (antisense); GATA-site, 5′-TCA GAA AGG ATA ATC AAT AG-3′ (sense) and 5′-CTA TTG ATT ATC CTT TCT GA-3′ (antisense); and mutated GATA-site, 5′-TCA GAA AGG gTg ATC AAT AG-3′ (sense) and 5′-CTA TTG ATc AcC CTT TCT GA-3′ (antisense). The atrial natriuretic factor (ANF) probe was previously described (2).
Transgenic mice.
All animal experimentations were carried out in accordance with institutional guidelines for animal care. Experiments were approved by Institutional Animal Care and Use Committee of the University of Ottawa, which conforms to that of the National Institute of Health (assurance number, A5043-01; University of Ottawa). At end points, mice were anesthetized with 2.2 μl/g ip of a cocktail consisting of ketamine (42.86 mg/ml), xylazine (8.57 mg/ml), and acepromazine (1.43 mg/ml) and were euthanized. For euthanasia, CO2 and cervical dislocation were used. Cervical dislocation without prior anesthesia is required for embryo collection in mice. The Gata4+/− mice were previously described (2). To overexpress CD2 in the heart, mouse CD2 cDNA under the control of the α-myosin heavy chain (α-MHC) promoter was subcloned in the SV40 expression vector (23). Cardiac-specific hemagglutinin-CD2 expression in transgenic mice was validated using Western blot analysis. For Masson trichrome staining, hearts were processed and stained as previously described (2, 8). For echocardiography analysis, mice were anesthetized (2.0% isoflurane, 80 ml/min 100% O2) and two-dimensional guided M-mode echocardiography was performed using a Visual-Sonics VEVO 700 and a 30-MHz linear array transducer as described by Aries et al. (2). Dox treatment was done as previously described (2). Briefly, mice were injected with 15 mg/kg Dox intraperitoneally and euthanized at 7-days postinjection.
Immunohistochemistry.
Immunohistochemistry on heart sections from each mouse group was performed as previously described (2). The antibodies used were GATA4 (2), ANF (rabbit anti-atrial natriuretic factor, Peninsula Laboratories T-4014), CD2 (mouse monoclonal, ab3087, Abcam), Ki67 (Clone SP6, rabbit monoclonal, Thermo Scientific RM-9106-S0), phosphohistone H3 (Ser10, rabbit polyclonal, Millipore 06-570), sarcomeric α-actinin (A7811, mouse monoclonal, Sigma), and 4,6-diamidino-2-phenylindole for nuclei staining (molecular probe D3571).
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay was carried out using apoptag kit (S7100, Chemicon) according to the apoptag protocol.
Cell number and surface area.
Total cell number was determined by calculating the number of cells within a squared micrometer area and multiplying by the surface area of the left ventricle wall and the interventricular septum. Surface area was determined using ImageJ and AxioVision LE software.
Statistics.
The data are presented as means ± SE. P < 0.05 by Student's t-test is considered statistically significant.
RESULTS
CD2 is a direct GATA4 transcriptional target.
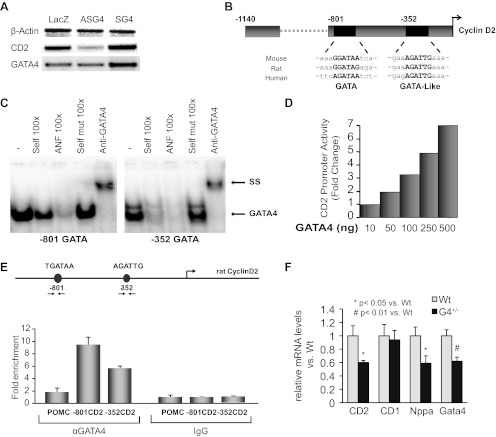
DNA chip analysis using RNA from neonate cardiomyocytes infected with adenoviruses expressing LacZ, or sense or antisense Gata4 transcripts, identified CD2 as a GATA4-inducible transcript. PCR analysis confirmed that CD2 mRNA levels were increased in cells overexpressing GATA4 and decreased in cells with antisense-mediated knockdown of GATA4 (Fig. 1A). Bioinformatics examination of the CD2 promoter revealed two evolutionary-conserved GATA binding sites, a proximal GATA-like site at −352 bp and a distal (−801 bp) GATA consensus site (Fig. 1B). Electromobility shift assays using cardiomyocytes nuclear extracts and probes designed to correspond to the distal and proximal GATA sites of the CD2 promoter confirmed that both sites bound GATA4 with high affinity. The addition of anti-GATA4 antibody confirmed that the bound protein is GATA4 and not any other member of the GATA family (upper arrow in Fig. 1C). We next tested whether GATA4 was able to directly activate the CD2 promoter; we cotransfected the CD2-luc reporter with various doses of a GATA4 expression vector. As shown in Fig. 1D, GATA4 was able to robustly activate the CD2 promoter in a dose-dependent manner. To determine whether GATA4 associates with the CD2 promoter in vivo, we performed chromatin immunoprecipitation assays using neonate primary cardiomyocyte cultures. As shown in Fig. 1E, GATA4 was enriched nine- and fivefold at the distal and proximal domains of the CD2 promoter, respectively. Consistent with a role for GATA4 in CD2 regulation, quantitative PCR (qPCR) analysis revealed a significant decrease in CD2 transcript levels in Gata4 heterozygote embryonic hearts (Fig. 1F). This decrease was not observed for CD1. As positive control, Nppa and Gata4 transcript levels were measured and found to be similarly decreased in Gata4 heterozygote hearts relative to age matched WT littermates. Immunostaining on heart sections of embryonic day 15.5 WT and Gata4+/− embryos showed that CD2 protein levels were greatly reduced in the ventricles of Gata4+/− embryos in line with the qPCR data (data not shown). Together, these findings confirm that CD2 is a direct target of transcription factor GATA4 in postnatal cardiomyocytes.
Fig. 1.
Cyclin D2 (CD2) is a direct GATA4 target. A: RT-PCR analysis of changes in CD2 mRNA levels in cardiomyocytes infected with adeno-lacZ or sense- or antisense GATA4. GATA4 mRNA levels are shown and β-actin is used as an internal control. B: schematic representation of the CD2 promoter showing the location and evolutionary conservation of proximal GATA-like and the distal GATA consensus sites. C: electromobility shift assays were performed using nuclear extracts from rat cardiomyocytes and primers corresponding to the two putative GATA binding sites on the CD2 promoter. Supershifts (SS) were performed using anti-GATA4 antibody. Note how the addition of a cold self probe or the consensus atrial natriuretic factor (ANF) GATA site competes with the binding but not the addition of a self-mutant (mut). D: dose-dependent activation of CD2 promoter by GATA4. Transient cotransfections were carried out in NIH3T3 cells using increasing doses of rat GATA4 expression vectors and the CD2-luc reporter. The data are the mean of n = 4. E: enrichment of GATA4 on the endogenous CD2 promoter as revealed by chromatin immunoprecipitation. The results are from one representative experiment carried out in duplicates on primary cardiomyocytes. POMC, proopiomelanocortin. F: quantitative PCR (qPCR) analysis of transcript levels in total RNA from embryonic day 15.5 wild-type (WT) and Gata4+/− (G4+/−) embryos. Nppa, natriuretic peptide precursor A.
Enhanced cardiac growth in myocardial CD2 transgenic mice.
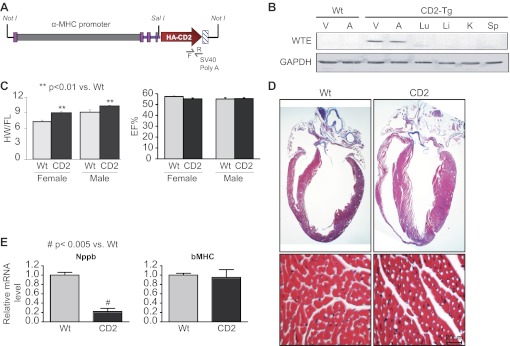
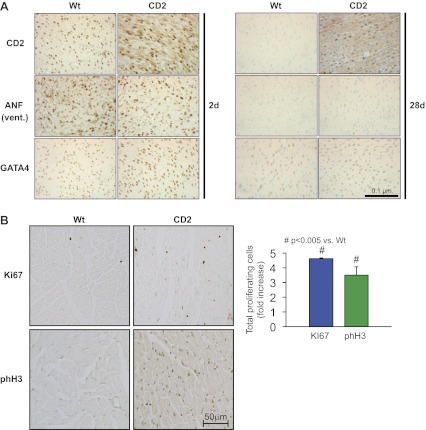
To determine which action of GATA4 is mediated by CD2 in cardiomyocytes, we developed a mouse line with myocardial-specific CD2 overexpression using the mouse α-MHC promoter (Fig. 2A). This promoter drives the expression of transgenes specifically to the heart and can deliver genes that promote various cell fates in postnatal ventricles including cardiomyocyte hypertrophy (8, 23). Tissue-specific transgene expression was monitored at the protein level using Western blot analysis. As expected, CD2 expression was restricted to the ventricles and atria. No expression of the transgene was detected in the lungs, liver, kidney, or spleen (Fig. 2B). Western blot analysis as well as immunohistochemistry showed that exogenous CD2 was upregulated in both nuclear and cytoplasmic compartments (Fig. 3A, and data not shown). The transgenic mice were viable and phenotypically appeared normal. Two independent transgenic lines out of six obtained were used to analyze the effect of CD2 overexpression on the heart, and both produced identical results. Analyses of hearts of CD2 transgenic mice showed a statistically significant increase in heart weight corrected by femur length compared with the WT littermate hearts (Fig. 2C, left). Echocardiography on WT and transgenic mice showed normal ejection fraction and other cardiac functions in CD2 transgenics compared with their WT littermates (Fig. 2C, right, and data not shown). Masson trichrome staining of adult heart sections confirmed a thicker left ventricular wall in CD2 transgenic mice compared with WT littermates. Consistent with the functional data, higher magnification of trichrome-stained section revealed no pathological cardiac remodeling; there was no sign of fibrosis or cardiomyocyte hypertrophy (Figs. 2D and 4A). A time-course analysis of cardiac changes revealed that heart mass was similar in neonate WT and transgenic mice and that increased left ventricular mass-to-body weight ratio was detectable in 30 days and older CD2 transgenics. These findings, which are consistent with postnatal activation of the α-MHC promoter, suggest enhanced postnatal cardiac growth in CD2 transgenics. qPCR analysis using mRNA from hearts of CD2 transgenic mice and their WT littermates revealed no upregulation of genes associated with pathological hypertrophy such as Nppa and Nppb (the genes encoding the cardiac hormones ANF and BNP) as well as skeletal actin (ACTA1) and β-MHC. In fact, a consistent significant decrease in Nppb was detected (Fig. 2E). Immunostaining on ventricular sections from mice confirmed that ANF was not increased in the ventricles of CD2 transgenics (Fig. 3A). Given the above, we tested whether increased cardiomyocyte proliferation accounts for enhanced postnatal cardiac growth. Immunostaining for two proliferation markers, Ki67 and the mitosis marker phosphohistone H3, showed a three- to fourfold increase in the percentage of proliferation in the hearts of adult CD2 transgenic mice compared with their WT littermates (Fig. 3B). Consistent with this, the number of cardiomyocytes per surface area was significantly increased in adult CD2 transgenic ventricles (Fig. 4D). These results suggest that CD2 overexpression promotes postnatal cardiac growth with maintained function, mainly through enhanced cardiomyocyte proliferation.
Fig. 2.
Myocardial overexpression of CD2 in transgenic mice. A: schematic representation of the α-myosin heavy chain (α-MHC)-hemagglutinin (HA)-CD2 transgene. Forward (F) and reverse (R) primers were used for screening. B: cardiac-specific expression of the CD2 transgene (Tg) as determined by Western blot analysis using HA antibody. WTE, whole tissue extract. C: measurements of the heart weight corrected by the femur length (HW/FL) and the ejection fraction (EF%) in young (∼80 days old) WT and transgenic female and male mice; n = 16–23 for each group. The results shown are means ± SD. D: Masson trichrome staining of whole heart sections from adult (170 days old) WT and CD2 transgenic mice. Note the thicker left ventricular (LV) wall of the transgenic heart. E: measurement of natriuretic peptide precursors B (Nppb) and β-MHC mRNA levels by qPCR analysis using RNA from heart tissues of CD2 transgenic mice and their WT littermates.
Fig. 3.
A: immunohistochemical analysis on ventricular (Vent) heart sections from neonate [2 day (2d), left; and 28 day (28d), right] WT and CD2 transgenics. Note how ANF staining is similar in WT and CD2 hearts. B: immunohistochemical staining for proliferation markers Ki67 and phosphohistone H3 (phH3) on adult heart sections (∼170 days old) from WT and CD2 transgenics. The histogram represents quantification of 24 fields from 2 different hearts for each group. The results are expressed as the average percentage of positive cells.
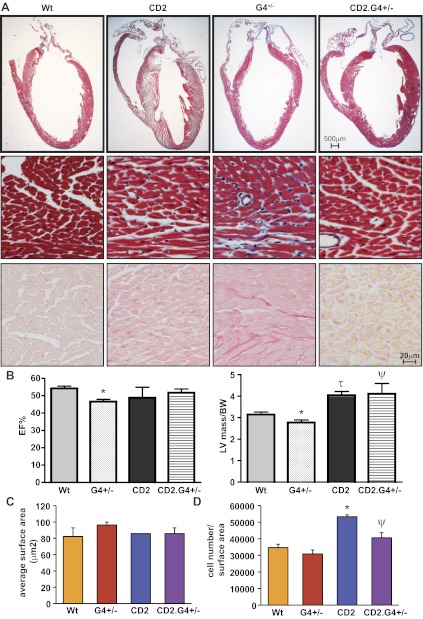
Fig. 4.
CD2 rescues G4+/− haplo-insufficient hearts. A: trichrome staining (top and middle) and Sirius red staining (bottom) of whole heart sections from adult WT, HA-CD2 transgenic, G4+/−, and CD2.G4+/− mice (170 days old). Note the hypoplastic heart of G4+/− mice and the larger LV of the CD2 mice. Note as well the collagen deposition (blue in trichrome and red in Sirius red staining) in the G4+/− heart as evidence of fibrosis. B: echocardiography analysis showing the LV mass measurement corrected by the body weight (LV/BW) and the EF% of the different mouse genotypes; N = 5–6 for each group. C: average cell surface area calculated from ventricular sections of adult WT, CD2 transgenic, G4+/−, and CD2.G4+/− mice (170 days old). The histogram represents quantification of 5 fields from 2 different hearts for each group. The results presented are means ± SE. D: total cell number in ventricular and interventricular septum sections of adult WT, CD2 transgenic, G4+/−, and CD2.G4+/− mice (170 days old). The results presented are means ± SE; n = 3 for each group. *P < 0.05 vs. WT; τP < 0.005 vs. WT; ΨP < 0.05 vs. G4+/−.
CD2 rescues the hypoplastic cardiac phenotype of Gata4+/− mice.
As stated earlier, GATA4 haplo insufficiency results in hypoplastic hearts. To determine whether CD2 may be a GATA4 effector in cardiomyocyte proliferation, we tested whether myocardial-specific overexpression could rescue cardiac growth in Gata4+/− mice. For this, CD2 transgenic mice were crossed with Gata4 heterozygote mice. Masson trichrome staining of heart sections showed that the CD2.Gata4+/− (CD2.G4+/−) mice had normal hearts in contrast to the hypoplastic hearts of Gata4+/− mice; higher magnification of the Trichrome-stained sections also showed increased interstitial collagen deposition in Gata4+/− but not in CD2.Gata4+/− hearts. Staining of heart sections with Sirius red confirmed these observations. (Fig. 4A, middle and bottom). Echocardiography revealed that the ejection fraction and left ventricular mass-to-body weight ratio of the CD2.G4+/− mice were almost restored to normal levels (Fig. 4B). Cell surface area measurements revealed no significant difference among the four genotypes (Fig. 4C). Interestingly, the number of cardiomyocytes per surface area was significantly increased in adult CD2 transgenic as well as in CD2.G4+/− ventricles compared with WT and Gata4+/− hearts, respectively (Fig. 4D).
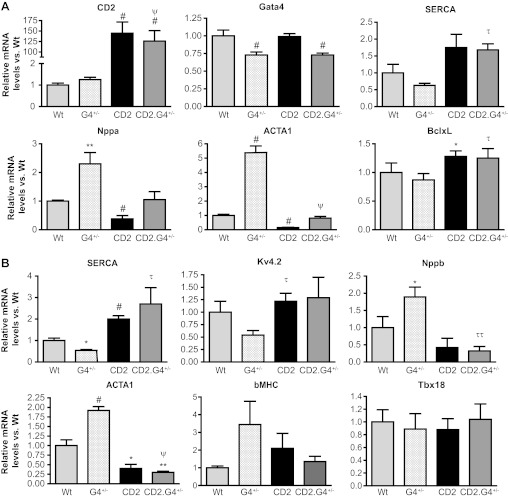
The ability of CD2 to rescue GATA4 deficiency was further assessed at the gene expression level by qPCR analysis on mRNA from postnatal (30–40 days) and adult (170–190 days) hearts. Two sets of genes were measured: on the one hand, those that are biomarkers of pathological cardiac stress and hypertrophy, including natriuretic peptide genes and skeletal actin, which are upregulated in Gata4+/− mice, and on the other, those that are essential for normal heart function such as the sarco(endo)plasmic reticulum Ca2+-ATPase pump and the potassium channel kv4.2. As shown in Fig. 5, CD2 upregulation largely normalized gene expression pattern of postnatal Gata4+/− hearts (Fig. 5A). This normalization was maintained in adult hearts (Fig. 5B).
Fig. 5.
Gene expression changes in CD2 expressing hearts. A: qPCR analysis using RNA from heart tissues of 30- to 40-day-old mice (n = 4 to 5 for each group). Note how Nppa and ACTA1 levels are normalized in the CD2.G4+/− mice vs. G4+/−. B: qPCR analysis using RNA from adult heart tissues (170 days) (n = 3 for each group). Note how most markers are restored back to normal in the CD2.G4+/− mice. SERCA, sarco(endo)plasmic reticulum Ca2+-ATPase. *P < 0.05 vs. WT; **P < 0.01 vs. WT; #P ≤ 0.005 vs. WT; τP < 0.05 vs. G4+/−; ττP ≤ 0.01 vs. G4+/−; ψP < 0.005 vs. G4+/−.
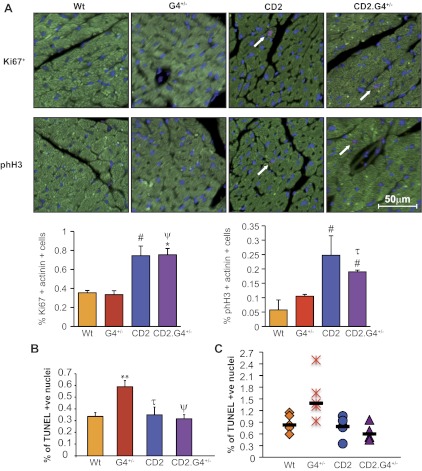
We next analyzed cardiomyocyte proliferation and survival in CD2.G4+/− mice and their control groups. Immunofluorescence was carried out using Ki67 or phosphohistone H3 as a marker for proliferation (red) and actinin costaining (green) to label cardiomyocytes (Fig. 6A, top). Quantification of the fluorescent signals showed increased proliferation in the hearts of CD2 and CD2.G4+/− mice (Fig. 6A, bottom). Gata4+/− mice have increased cardiomyocyte apoptosis (2); terminal transferase-mediated dUTP nick end labeling assays unexpectedly revealed that CD2 overexpression was able to restore the percentage of apoptosis in Gata4+/− mice to the level observed in WT hearts (Fig. 6B). Given this result, we tested whether CD2 overexpression was able to rescue the exaggerated Dox-induced apoptosis in Gata4 haplo-insufficient mice. As seen in Fig. 6C, CD2 restored drug-induced apoptosis of Gata4+/− mice to WT levels.
Fig. 6.
Analysis of cardiomyocyte proliferation and survival. A, top: immunofluorescent analysis of Ki67 and phH3 expression on adult heart sections. Green is actinin staining to mark cardiomyocytes, red is for Ki67 or phH3 staining as indicated, and blue marks nuclei (4,6-diamidino-2-phenylindole). White arrows indicate positive nuclei. A, bottom: percentage of Ki67/actinin and phH3/actinin-positive nuclei vs. WT. Note the significant increase in the level of proliferation in the CD2 and CD2.G4+/− mice. The results presented are means ± SE (n = 4 for each mouse group). B: quantification of terminal transferase-mediated dUTP nick end labeling (TUNEL) assays on adult heart sections (170 days) showing the percentage of TUNEL-positive (+ve) nuclei. Ten regions from each heart section were counted. The results shown are means ± SE (n = 3 for each group). Note the statistically significant increase in the percentage of apoptotic nuclei in the G4+/− mice, which was completely restored back to WT level in the CD2/G4+/− mice. C: scatter plot showing the percentage of TUNEL-positive cardiomyocytes in heart sections from adult (180 days) doxorubicin-treated mice. Sixteen regions from each heart section were taken (n = 4–7 for each group). #P < 0.05 vs. WT; **P < 0.01 vs. WT; τP < 0.05 vs. G4+/−; ψP < 0.005 vs. G4+/−.
Together, these results show that myocardial CD2 upregulation is able to restore, at least in part, the genetic and growth alterations caused by GATA4 haplo insufficiency and suggest that CD2 is a GATA4 effector in cardiomyocyte growth.
DISCUSSION
It is now well established that the zinc finger transcription factor GATA4 is crucial for proper heart development and maturation. Mutations in GATA4 cause various congenital heart problems reflecting its pleitropic role in cardiac cell proliferation and differentiation. However, the effectors of GATA4 actions are incompletely understood. In the presented work, we provide evidence that CD2 is not only a direct target of GATA4 but is also an effector of GATA4 in postnatal cardiomyocytes growth. Remarkably, upregulation of CD2 in cardiomyocytes rescues growth and adaptive stress response of GATA4 haplo-insufficient hearts. These findings provide a new paradigm that may help protect cardiac function of GATA4 haplo-insufficient hearts.
Cardiac-specific overexpression of CD2 caused an increase in cardiac mass with maintained normal cardiac function as evidenced from echocardiography and biomarker analysis. The fact that these transgenic mice were viable with normal cardiac function throughout adult life (up to 300 days) suggests that enhanced cardiac expression of CD2 may have beneficiary effects on the postnatal heart, possibly through limited regenerative capacity. Indeed, our results show increased cardiomyocyte proliferation in the hearts of adult CD2 transgenic mice. These results are generally consistent with those reported for a similar α-MHC overexpressing mouse line (24).
Crossing the CD2 transgenic mice with the Gata4+/− mice, which have hypoplastic hearts and increased sensitivity to drug-induced cardiotoxicity (2, 22), revealed that CD2 can compensate, at least in part, for GATA4 haplo insufficiency. Indeed, CD2 rescued heart size and adaptive response to drug-induced apoptosis of Gata4 heterozygote mice. Gata4+/− mice develop stress-induced pathological hypertrophy, a likely consequence of a lower number of myocytes and decreased myocyte survival and contractile function (2). We found that CD2 upregulation was sufficient to restore cardiac mass through increased myocyte proliferation not hypertrophy. CD2 also improved heart function as evidenced from biomarker analysis. It is interesting to note that cardiac growth in CD2 transgenics on either WT or Gata4+/− background occurred mostly through postnatal cardiomyocyte proliferation not hypertrophy suggesting that GATA4 effectors in hypertrophy and proliferation may be distinct.
The finding that CD2 rescued basal and drug-induced survival of cardiomyocytes with GATA4 haplo insufficiency was unexpected and noteworthy. While a more direct effect of CD2 on myocyte survival cannot be excluded, it is possible that by normalizing cardiac size and function, CD2-reduced basal oxidative stress level and enhanced adaptive response. CD2 has been suggested to play an important role in biomechanical stress response of the heart based on the attenuated response to transaortic constriction of CD2 null mice (1). Be it as it may, the finding that postnatal upregulation of CD2 is beneficial to GATA4 hypoplastic hearts may open new avenues for cardioprotection for individuals with heterozygous mutations in GATA4 or GATA4-dependent cardiac dysfunction.
GRANTS
This work was supported by Canadian Institutes of Health Research Grant MOP36382.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
A.Y., R.T., W.M., S.C., P.P., and A.A. performed experiments; A.Y., W.M., and M.N. analyzed data; A.Y. interpreted results of experiments; A.Y. and R.T. prepared figures; A.Y. and M.N. drafted manuscript; M.N. conception and design of research; M.N. edited and revised manuscript; M.N. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the Institut de recherches cliniques de Montréal (IRCM) transgenic core facilities and the IRCM and University of Ottawa histology core for technical support. We are indebted to laboratory members for helpful insight and suggestions.
REFERENCES
- 1. Angelis E, Garcia A, Chan SS, Schenke-Layland K, Ren S, Goodfellow SJ, Jordan MC, Roos KP, White RJ, MacLellan WR. A cyclin D2-Rb pathway regulates cardiac myocyte size and RNA polymerase III after biomechanical stress in adult myocardium. Circ Res 102: 1222–1229, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aries A, Paradis P, Lefebvre C, Schwartz RJ, Nemer M. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc Natl Acad Sci USA 101: 6975–6980, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brooks AR, Shiffman D, Chan CS, Brooks EE, Milner PG. Functional analysis of the human cyclin D2 and cyclin D3 promoters. J Biol Chem 271: 9090–9099, 1996 [DOI] [PubMed] [Google Scholar]
- 4. Charron F, Tsimiklis G, Arcand M, Robitaille L, Liang Q, Molkentin JD, Meloche S, Nemer M. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev 15: 2702–2719, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Debrus S, Rahbani L, Marttila M, Delorme B, Paradis P, Nemer M. The zinc finger-only protein Zfp260 is a novel cardiac regulator and a nuclear effector of alpha1-adrenergic signaling. Mol Cell Biol 25: 8669–8682, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2–5 and GATA-4 are mutual cofactors. EMBO J 16: 5687–5696, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev 19: 1175–1187, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Georges R, Nemer G, Morin M, Lefebvre C, Nemer M. Distinct expression and function of alternatively spliced Tbx5 isoforms in cell growth and differentiation. Mol Cell Biol 28: 4052–4067, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hassink RJ, Pasumarthi KB, Nakajima H, Rubart M, Soonpaa MH, de la Riviere AB, Doevendans PA, Field LJ. Cardiomyocyte cell cycle activation improves cardiac function after myocardial infarction. Cardiovasc Res 78: 18–25, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heineke J, Auger-Messier M, Xu J, Oka T, Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ, Robbins J, Crombleholme TM, Molkentin JD. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J Clin Invest 117: 3198–3210, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464: 606–609, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev 7: 331–342, 1993 [DOI] [PubMed] [Google Scholar]
- 13. Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464: 601–605, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, Akashi K, Sicinski P. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118: 477–491, 2004 [DOI] [PubMed] [Google Scholar]
- 15. Kozar K, Sicinski P. Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle 4: 388–391, 2005 [DOI] [PubMed] [Google Scholar]
- 16. Lavallee G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, Horb ME, Nemer M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J 25: 5201–5213, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang Q, De Windt LJ, Witt SA, Kimball TR, Markham BE, Molkentin JD. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem 276: 30245–30253, 2001 [DOI] [PubMed] [Google Scholar]
- 18. MacLellan WR, Garcia A, Oh H, Frenkel P, Jordan MC, Roos KP, Schneider MD. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol Cell Biol 25: 2486–2497, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes Dev 25: 299–309, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nemer G, Nemer M. GATA4 in Heart Development and Disease. In: Heart Development and Regeneration, edited by Rosenthal N, Harvey RP. San Diego, Academic Press, 2010, p. 599–616 [Google Scholar]
- 21. Nemer M. Genetic insights into normal and abnormal heart development. Cardiovasc Pathol 17: 48–54, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Oka T, Maillet M, Watt AJ, Schwartz RJ, Aronow BJ, Duncan SA, Molkentin JD. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res 98: 837–845, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Paradis P, Dali-Youcef N, Paradis FW, Thibault G, Nemer M. Overexpression of angiotensin II type I receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc Natl Acad Sci USA 97: 931–936, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res 96: 110–118, 2005 [DOI] [PubMed] [Google Scholar]
- 25. Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol 275: 235–244, 2004 [DOI] [PubMed] [Google Scholar]
- 27. Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol 28: 5420–5431, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubart M, Field LJ. Cardiac regeneration: repopulating the heart. Annu Rev Physiol 68: 29–49, 2006 [DOI] [PubMed] [Google Scholar]
- 29. Rysa J, Tenhunen O, Serpi R, Soini Y, Nemer M, Leskinen H, Ruskoaho H. GATA-4 is an angiogenic survival factor of the infarcted heart. Circ Heart Fail 3: 440–450, 2010 [DOI] [PubMed] [Google Scholar]
- 30. Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev 18: 2699–2711, 2004 [DOI] [PubMed] [Google Scholar]
- 31. Tamamori-Adachi M, Ito H, Sumrejkanchanakij P, Adachi S, Hiroe M, Shimizu M, Kawauchi J, Sunamori M, Marumo F, Kitajima S, Ikeda MA. Critical role of cyclin D1 nuclear import in cardiomyocyte proliferation. Circ Res 92: e12–e19, 2003 [DOI] [PubMed] [Google Scholar]
- 32. Wang J, Paradis P, Aries A, Komati H, Lefebvre C, Wang H, Nemer M. Convergence of protein kinase C and JAK-STAT signaling on transcription factor GATA-4. Mol Cell Biol 25: 9829–9844, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiong Y, Menninger J, Beach D, Ward DC. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics 13: 575–584, 1992 [DOI] [PubMed] [Google Scholar]
- 34. Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest 115: 1522–1531, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]