Abstract
Increasing evidence suggests that the circadian clock plays an important role in the control of renal function and blood pressure. We previously showed that the circadian clock protein Period (Per)1, positively regulates the expression of the rate limiting subunit of the renal epithelial sodium channel (αENaC), which contributes to blood pressure regulation. Casein kinases 1δ and 1ε (CK1δ/ε) are critical regulators of clock proteins. CK1δ/ε must phosphorylate the circadian clock protein Per1 in order for the latter to enter the nucleus. We used a commercially available CK1δ/ε inhibitor, PF670462, to test the effect of CK1δ/ε blockade and inhibited Per1 nuclear entry on αENaC in a model of the renal cortical collecting duct (mpkCCDc14 cells). CK1δ/ε blockade prevented Per1 and Clock from interacting with an E-box from the αENaC promoter. CK1δ/ε inhibition reduced αENaC mRNA levels by <60%. A similar decrease in αENaC mRNA was observed following siRNA-mediated CK1δ/ε knock-down. Inhibition of CK1δ/ε effectively prevented the transcriptional response of αENaC to aldosterone, suggesting an interaction between the circadian clock and aldosterone-mediated regulation of αENaC. CK1δ/ε inhibition significantly reduced αENaC but increased Caveolin-1 membrane protein levels; transepithelial current, a measure of ENaC activity, was decreased. Importantly, single channel analysis in amphibian renal cells demonstrated a dramatic decrease in the number of patches with observable ENaC current following CK1δ/ε inhibition. The present study shows for the first time that CK1δ/ε inhibition and impaired Per1 nuclear entry results in decreased αENaC expression and ENaC activity, providing further support for direct control of ENaC by the circadian clock.
Keywords: kidney, transcription, sodium transport, Per1
the circadian clock has a significant impact on the regulation of many physiological functions such as blood pressure, metabolism, immune response, and renal function (1, 10, 31, 35, 40). Of note, sodium excretion, potassium excretion, glomerular filtration rate, and renal blood flow exhibit rhythmic fluctuations (2, 30, 40, 43). The circadian clock consists of multiple core proteins that interact with each other to affect transcription of target genes through promoter E-box response elements (14). There are four canonical clock proteins: Period (Per), Cryptochrome (Cry), Bmal1, and Clock. Period and Cryptochrome each have homologs (Per1–3 and Cry1–2). Clock and Bmal1 form a dimer that interacts with E-boxes to transcriptionally regulate clock controlled genes. Per and Cry likely dimerize and interact with Clock and Bmal1 to repress transcriptional activity. This creates a negative feedback loop mechanism wherein Bmal1/Clock activate clock controlled genes and Per-Cry repress Bmal1/Clock activity (4, 5, 14, 47). It is becoming increasingly apparent that, outside of the core clock mechanism, Per1 may act as an activator of gene expression, perhaps in a tissue and gene-specific manner (11, 19, 20).
Accumulating evidence supports an important role for the core clock proteins in the control of blood pressure and renal function. Zuber et al. (48) linked the loss of Clock with hypotension, dysregulated sodium excretion, and partial diabetes insipidus. Doi et al. (16) showed that Cry1/Cry2 knockout animals exhibit salt-sensitive hypertension. This phenotype was due to greater aldosterone production because of increased expression of 3-beta dehydrogenase-isomerase, an enzyme in the aldosterone synthesis pathway (16). We previously demonstrated that Per1 regulates both basal and aldosterone-mediated regulation of αENaC, the rate-limiting subunit of the epithelial sodium channel (ENaC) (20, 29). We later showed that this regulation is via Per1-mediated transcriptional activation of the αENaC promoter (19).
The circadian clock undergoes posttranslational modifications through the phosphorylation of the Per proteins by Casein kinase 1 isoforms δ/ε (CK1δ/ε) and dephosphorylation by protein phosphatase 1 (PP1). CK1δ/ε phosphorylates the Per proteins, allowing them to enter the nucleus; PP1 removes this phosphorylation, preventing nuclear entry (17, 18, 21, 24). This creates a balance between activation and deactivation of the Per proteins and thus regulates the circadian negative feedback loop via control of Per protein phosphorylation state (17, 18, 21). The availability of CK1δ/ε inhibitors makes it possible to study the mechanisms of the circadian clock via pharmacological manipulation. Indeed, treatment with the CK1δ/ε inhibitor PF670462 caused a phase shift delay in rats under entrained and free-running conditions, and, importantly, PF670462 inhibited Per protein nuclear entry (7). PF670462 has exhibited less broad spectrum effects, such as apoptosis and cell cycle arrest, than other CK1δ/ε inhibitors (12). Thus PF670462 is a unique tool for elucidating circadian clock mechanisms. In the present study we used PF670462 to evaluate the effects of CK1δ/ε inhibition and the subsequent inhibition of Per1 nuclear entry, on αENaC expression and transepithelial current in mpkCCDc14 cells (a model of the renal cortical collecting duct). Importantly, single channel recordings made in amphibian kidney cells demonstrate that CK1δ/ε inhibition also markedly affects ENaC activity.
MATERIAL AND METHODS
Cell culture.
Alain Vandewalle (INSERM, Paris, France) graciously provided the mpkCCDc14 cells (8). Cells were maintained in DMEM-F12 plus 10% FBS (Invitrogen) and 50 μg/ml gentamicin (Sigma). For inhibitor experiments, 6 × 105 cells were plated in each well of a six-well Corning Costar Transwell dish (24 mm 0.4 μm pore polyester membranes insert). Twenty-four hours after cells reached 100% confluence, cells were treated with 10 μM PF670462 (Santa Cruz) or vehicle (DMSO) for 72 h. For time course experiments, cells were treated with 0.1, 1, 10, 100 μM, DMSO, or untreated for 24, 48, and 72 h, respectively. Final DMSO concentration in both vehicle- and inhibitor-treated cells was 0.1%. Aldosterone treatments were performed as described previously (19).
RNA isolation and QPCR.
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. RNA samples (10 μg) were treated with DNA-free DNaseI (Ambion). DNaseI-treated RNA (2 μg) samples were used as template for reverse transcription with Superscript III (Invitrogen). The resulting cDNAs (20 ng) were then used as template in quantitative real-time PCR (QPCR) reactions (Applied Biosystems) to evaluate changes in αENaC, ET-1, and Fxyd5 mRNA levels. Cycle threshold (Ct) values were normalized against β-actin and relative quantification was performed using the ΔΔCt method (26). Fold change values were calculated as the change in mRNA expression levels relative to the vehicle-treated control. TaqMan primer/probe sets were purchased from Applied Biosystems.
DNA affinity purification assay.
Experiment was performed as described previously (41). Nuclear extracts were isolated from mpkCCDc14 cells treated with vehicle or PF670462 for 72 h using the NE-PER kit (Pierce) according to the manufacturer's instructions. Single-stranded, biotinylated probes were ordered from GenoSys. Once annealed, the double-stranded DNA affinity purification assay (DAPA) probe was incubated with 175 μg of nuclear extract in the presence of Streptavidin agarose beads (Sigma). The sequence of the DAPA probe was 5′ tggtgggggccagcaggtgcttcccagttt.
RNA silencing.
SMARTpool siRNA for CK1δ, CK1ε, and Non-Target was purchased from Dharmacon (Cat. #L-044377-00-0005, L-040108-00-0005, and D-001206-14-20) and used according to the manufacturer's instructions. The mpkCCDc14 cells were transfected using Dharmafect 4, as described previously (19, 20).
Protein isolation.
Nuclear and cytosolic extracts were isolated with the NE-PER kit (Pierce) according to the manufacturer's instructions. Membrane proteins were isolated by differential centrifugation with sucrose gradients. We collected mpkCCDc14 cells by scraping in ice-cold PBS and centrifuging at 3,000 g for 10 min. Cells were resuspended in sucrose buffer A (10 mM Tris, 1 mM EDTA, 50 mM sucrose) and homogenized. An equal volume of sucrose buffer B (10 mM Tris, 1 mM EDTA, 250 mM sucrose) was added, followed by additional homogenization. Nuclei were pelleted at 1,000 g for 10 min and then discarded. Organelles were pelleted at 10,000 g for 20 min and then discarded. Supernatants were centrifuged at >60,000 g for 18 h. Membrane protein pellets were resuspended in 50 μl of sucrose buffer B and phosphatase and protease inhibitors. Protein concentrations were then quantified by BCA assay (Pierce).
Western blot analysis.
Proteins were separated on a 4–20% Tris·HCl Ready Gel (Bio-Rad) and transferred to a PVDF membrane. The membrane was blocked in 2% Rodeo Blocker in TBS-S (TBS plus 0.05% Rodeo Saddle Soap) (USB) and incubated overnight at 4°C with anti-αENaC (1:1,000; generous gift of Dr. Carolyn Ecelbarger, Georgetown University, Washington, DC), anti-Caveolin-1 (1:1,000, Santa Cruz), or anti-actin (1:500, Santa Cruz). Actin is often associated with the plasma membrane fraction of cells and was used as a loading control (15). The membrane was washed with 2% Rodeo Blocker in TBS-S for 15 min and then incubated with horseradish peroxidase conjugate anti-rabbit secondary antibody and incubated in 2% Rodeo Blocker in TBS-S for 1 h at 4°C. After incubation, the blot was washed with TBS-S for 15 min. Detection was performed with Rodeo ECL detection reagents. Densitometry was performed using ImageJ (http://rsbweb.nih.gov/ij).
Transepithelial current analysis.
The mpkCCDc14 cells used in these experiments were cultured as described (8). The cells were plated in six-well (24 mm) polyester Transwell permeable supports (Corning) and allowed to grow several days past confluence. Transepithelial voltage (Vte) and resistance (Rte) were measured with chop-stick electrodes (EVOM, World Precision Instruments) preceding treatment and at 1, 24, 48, and 72 h after treatment with vehicle (0.1% DMSO or water) or 10 μM PF670462. In some experiments, these parameters were also measured before and after a 10 min exposure to 10 μM amiloride on the apical side of the inserts. Transepithelial current (Ite) was calculated using Ohm's law, Ite = Vte/Rte, and corrected for surface area of the well.
Single channel recordings.
For cell-attached patch clamp experiments, A6 cells (subclone 2F3; passages 96–106) were grown on rings as previously described (28). The cells were treated with vehicle (0.1% DMSO or water) or 10 μM PF670462 for 72 h prior to single channel recordings. Glass electrodes (TW-150F, World Precision Instruments) were pulled on a two-stage vertical glass puller to achieve resistances of 5–10 MΩ in patch solution (96 mM NaCl, 3.4 mM KCl, 0.8 mM CaCl2, 0.8 mM MgCl2, and 10 mM HEPES titrated to a pH of 7.4 with NaOH). For patch experiments, A6 cell medium was replaced with patch solution, and gigaohm seals were made on individual cells. Single channels recordings were made (5–10 min) with patch clamp amplifiers (Dagan PC-One and Axopatch 200B) with a low-pass 3-pole Bessel filter of 1 kHz and digitized (Digidata 1440a and pCLAMP10, Axon Instruments) at a sampling frequency of 5 kHz and digitally filtered at 30 Hz. The recording period was ∼5–10 min. Channel density (N) was calculated as the number of individual current levels that occurred during the recording period. The pCLAMP 10 software program calculated the NPo using the equation:
where i denotes the number of open channels, ti the time the channels are open, and T the total recording time. Channel open probability (Po) was calculated as the ratio of NPo/N using only those patches with observable channel currents. Only patches with less than or equal to eight observable channels were included in the analysis due to constraints of the analysis software.
Statistical analysis.
Student's unpaired t-test (Microsoft Excel) was used to compare two data sets. A two-way ANOVA (SigmaStat) was used to evaluate the four groups of QPCR in Fig. 6. Data are presented as means ± SE. P values < 0.05 were considered significant. Data analysis was performed, and graphs were generated for transepithelial current experiments and single channel recordings using SigmaPlot 12 (SysStat). Data are presented as means ± SE. Differences in the percentage of patches with channels were assessed with z-test. Since the data were not normally distributed, differences in N, Po, and NPo were determined with a Mann-Whitney rank sum test.
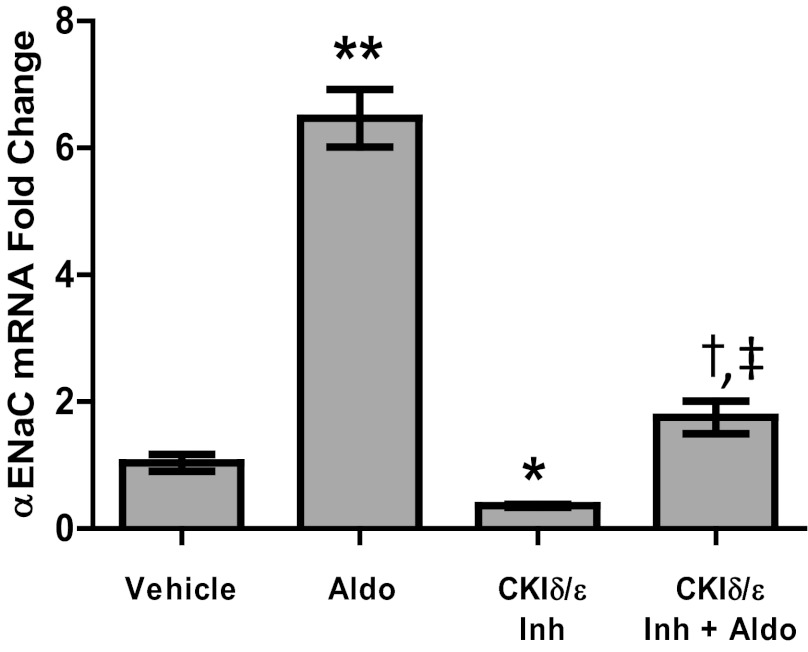
Fig. 6.
Inhibition of CK1δ/ε prevents induction of αENaC by aldosterone (Aldo). mpkCCDc14 cells were treated with DMSO or 10 μM PF670462 once they reached confluence. After 24 h, media were replaced with steroid-free media containing DMSO or PF670462. After an additional 24 h, Aldo or vehicle (ethanol) was added. RNA was isolated 24 h later. Total time of PF670462 or DMSO treatment was 72 h; total time of Aldo or ethanol treatment was 24 h. *P < 0.001 vs. vehicle control, **P < 0.01 vs. vehicle control, †P < 0.001 vs. vehicle control and vs. CK1δ/ε Inh, ‡P < 0.001 interaction via 2-way ANOVA; n = 6. Values are represented as means ± SE.
RESULTS
Inhibition of CK1δ/ε inhibits Per1 nuclear entry.
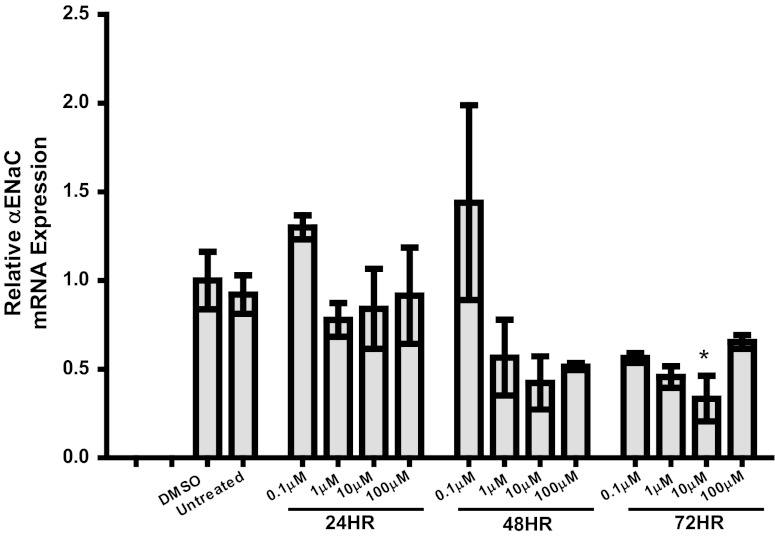
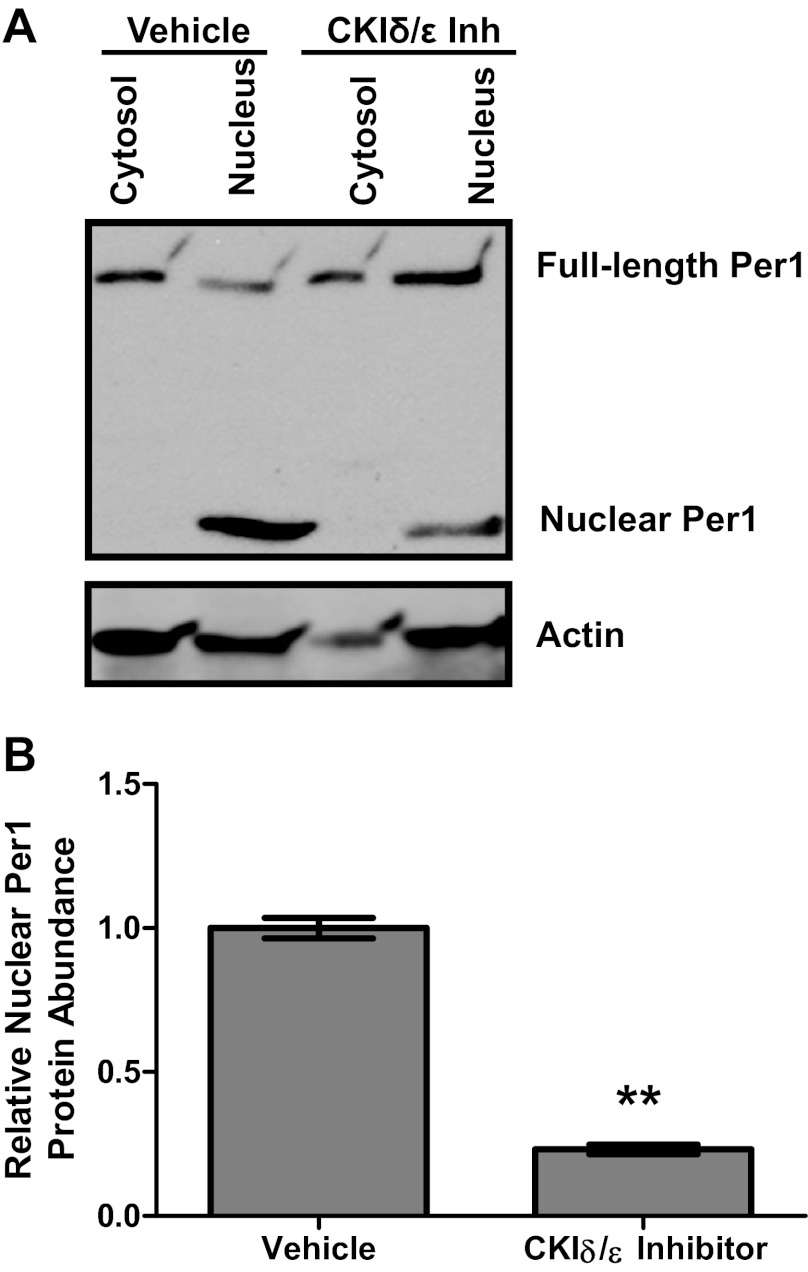
We have previously shown that an ∼50 kDa Per1 fragment interacts with an E-box response element from the Scnn1a promoter (19). Others have observed this lower-molecular-weight Per1 in tissue nuclear extracts (13). Per1 must be phosphorylated by the circadian regulatory casein kinases CK1δ/ε to enter the nucleus (17, 18, 21). PF670462, a pharmacological inhibitor of CK1δ/ε, has been shown to inhibit Per protein nuclear entry with an EC50 of 290 nM (7). For this reason, we evaluated the effect of PF670462 on αENaC expression in mpkCCDc14 cells in a dose response and time course experiment (Fig. 1). As shown, a 72 h treatment with 10 μM PF670462 showed the greatest inhibition of αENaC expression and thus was used for subsequent experiments. This result is consistent with our previous work showing that 72 h of treatment with 10 μM PF670462 was optimal for affecting expression of the Per1 target gene endothelin-1 (42). We tested the effect of this PF670462 treatment on Per1 nuclear entry (Fig. 2). For the first time we show that the ∼50 kDa Per1 fragment was only detected in the nucleus in control cells, compared with the full-length Per1 protein, which was detected in the cytosol and the nucleus (Fig. 2A). PF670462 treatment resulted in a clear decrease in nuclear Per1 levels. Densitometric analysis of these data demonstrated that levels of nuclear Per1 were reduced by >80% (Fig. 2B).
Fig. 1.
Effect of casein kinase (CK)1δ/ε inhibition on α-epithelial sodium channel (ENaC) mRNA expression in mpkCCDc14 cells (a model of the renal cortical collecting duct). Cells were treated with the CK1δ/ε inhibitor PF670462 at 0.1, 1.0, 10, or 100 μM for 24, 48, or 72 h. Quantitative (Q)PCR was used to evaluate changes in αENaC gene expression in PF670462-treated cells vs. DMSO-treated cells. *P < 0.05, n = 2–6.
Fig. 2.
Inhibition of CK1δ/ε inhibits Per1 nuclear entry. Nuclear and cytosolic extracts were collected from mpkCCDc14 cells treated with the CK1δ/ε inhibitor (Inh) PF670462 for 72 h. A: Western blot analysis was performed using anti-Per1 or anti-actin antibodies. Data are representative of 3 independent experiments. B: densitometry analysis was used to quantitate the level of the lower-molecular-weight Per1 labeled “Nuclear Per1” in A. Values are represented as means ± SE. **P < 0.01, n = 3.
Inhibition of CK1δ/ε prevents Per1 interacting with Scnn1a E-box.
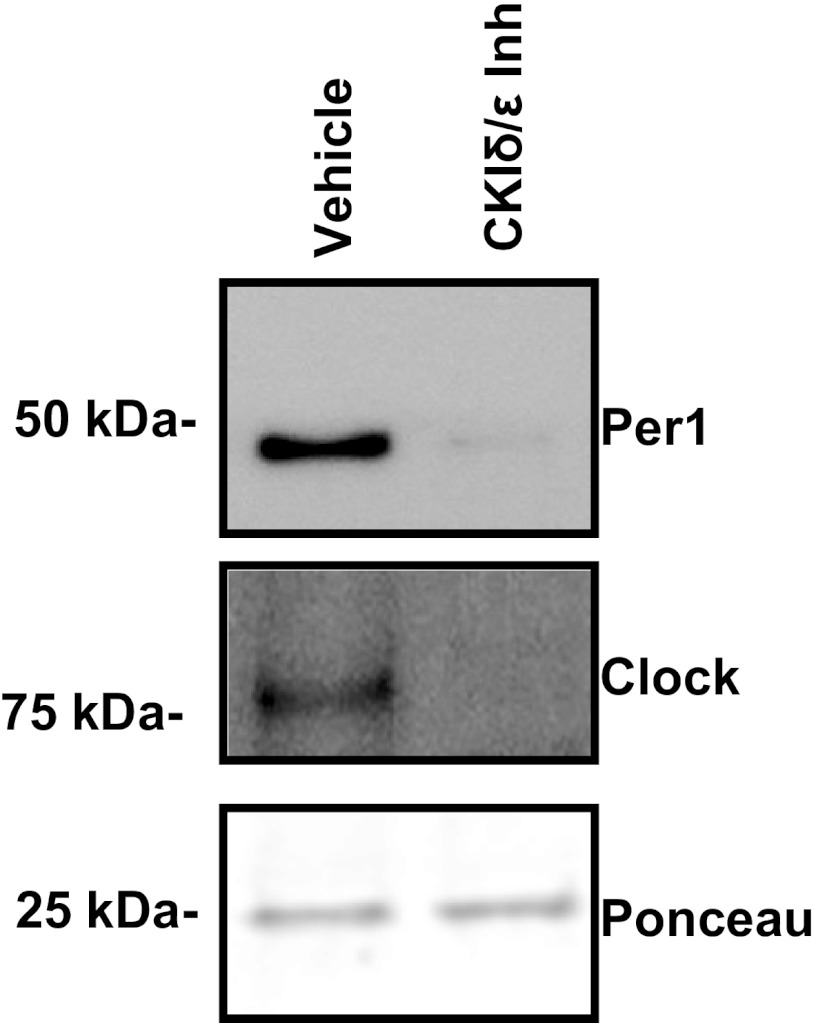
Previously, we detected Per1, presumably in complex with the Clock protein, at an E-box at position −689 relative to the transcription start site in the Scnn1a (αENaC gene) promoter (19). A DAPA was performed with nuclear extracts from mpkCCDc14 cells treated with vehicle (DMSO) or PF670462 for 72 h. Compared with vehicle, CK1δ/ε inhibition greatly diminished both Per1 and Clock protein interaction with the E-box sequence (Fig. 3), providing evidence that CK1δ/ε inhibition prevents Per1 from interacting with the αENaC E-box.
Fig. 3.
Inhibition of CK1δ/ε prevents interaction of Per1 with E-box3 in the Scnn1a promoter. mpkCCDc14 cells were treated with the CK1δ/ε Inh PF670462 for 72 h. Nuclear extracts from treated and control cells were incubated with biotinylated probes containing the sequence 5′ tggtgggggccagcaggtgcttcccagttt, which represents E-box3 from the Scnn1a promoter (see Ref. 19). Western blot analysis was performed using anti-Per1 and anti-Clock antibodies. A nonspecific band at 25 kDa was used as a loading control from the Ponceau S-stained Western blot. Data are representative of 3 independent experiments.
Inhibition of CK1δ/ε reduces basal Scnn1a and Fxyd5 expression.
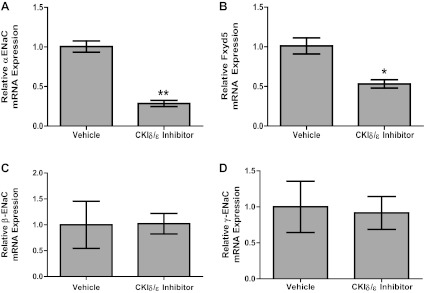
We showed previously that Per1 induces αENaC transcription, the rate-limiting subunit of ENaC (19). To determine the effect of CK1δ/ε inhibition on Scnn1a expression, mRNA levels were measured by QPCR. Following CK1δ/ε inhibition, Scnn1a mRNA levels decreased 60% compared with vehicle (DMSO) (Fig. 4A). We previously identified Fxyd5 as a Per1 target gene (42). Fxyd5 is known to induce Na+,K+-ATPase activity (27). To determine if CK1δ/ε inhibition also affected this independent Per1 target, Fxyd5 expression was analyzed following PF670462 treatment. Fxyd5 mRNA levels decreased 50% compared with vehicle (Fig. 4B). We investigated the effect of CK1δ/ε inhibition on βENaC and γENaC mRNA levels as well. As shown in Fig. 4, C and D, CK1δ/ε inhibition did not appear to alter mRNA expression of the β and γENaC subunits. To rule out cytotoxic effects of PF670462, we examined actin Ct values as an indicator of cell viability with the real-time QPCR data described in Fig. 4. The average Ct value is 18.8 ± 0.1 for vehicle-treated cells and 18.4 ± 0.3 for PF670462-treated cells (P = 0.4, n = 6–12).
Fig. 4.
Inhibition of CK1δ/ε reduces basal Scnn1a and Fxyd5 expression. mpkCCDc14 cells were treated with the CK1δ/ε Inh PF670462 for 72 h. QPCR was used to evaluate changes in αENaC (A), Fxyd5 (B), βENaC (C), or γENaC (D) gene expression in PF670462-treated cells vs. DMSO-treated cells. *P < 0.05, n = 3–6. **P < 0.01, Values are represented as means ± SE.
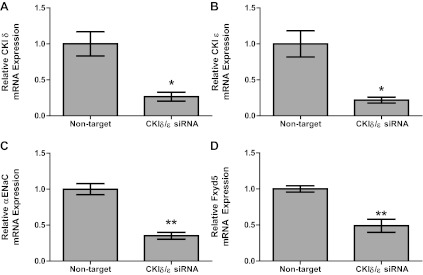
RNA silencing was used to inhibit CK1δ/ε by an alternative method (Fig. 5). The results indicate that knock-down of CK1δ and CK1ε using siRNA caused similar decreases in αENaC and Fxyd5 mRNA levels as what we observed with pharmacological inhibition of CK1δ/ε. Knock-down of CK1δ/ε with siRNA led to a 73% decrease in CK1δ mRNA and a 78% decrease in CK1ε mRNA (Fig. 5, A and B, respectively). Levels of αENaC mRNA were decreased by 70%, and Fxyd5 mRNA levels were decreased by 59% following siRNA-mediated knock-down of CK1δ/ε (Fig. 5, C and D, respectively). To ensure that there was not an effect of DMSO as a vehicle in the pharmacological inhibitor studies shown in Fig. 4, we compared αENaC mRNA expression in DMSO-treated cells to nontarget siRNA transfected cells. The results of this comparison demonstrate that there was not a detectable difference in αENaC mRNA levels in the control conditions for the experiments described in Figs. 4 and 5 [αENaC mRNA 1.0 ± 0.12 (nontarget siRNA) vs. 0.92 ± 0.06 (DMSO), P = 0.6, n = 3].
Fig. 5.
RNA silencing of CK1δ/ε reduces basal αENaC and Fxyd5 expression. A: mpkCCDc14 cells were treated with nontarget or CK1δ and ε siRNA for 48 h. Real-time PCR was used to evaluate changes in CK1δ (A), CK1ε (B), αENaC (C), or Fxyd5 (D) gene expression in CK1δ/ε siRNA-treated cells vs. nontarget. *P < 0.05, **P < 0.01; n = 6–8. Values are represented as means ± SE.
Inhibition of CK1δ/ε prevents aldosterone-mediated induction of Scnn1a.
The steroid hormone aldosterone induces αENaC (Scnn1a) expression, and we previously demonstrated that transient Per1 siRNA knock-down decreases this response to aldosterone (20). To determine if CK1δ/ε inhibition mimics this reduction in aldosterone response, αENaC mRNA levels were analyzed by QPCR in the presence of aldosterone and CK1δ/ε inhibitor (Fig. 6). As expected, αENaC mRNA levels increased sixfold in response to aldosterone alone. Consistent with the results shown in Fig. 4, αENaC mRNA decreased upon treatment with the CK1δ/ε inhibitor. Moreover, CK1δ/ε inhibition reduced the aldosterone response from sixfold to 1.5-fold, showing that, just as with Per1 knock-down by siRNA, CK1δ/ε inhibition inhibits the aldosterone response (Fig. 6). As a control, Fxyd5 mRNA was measured. Fxyd5 mRNA, as shown in Fig. 4B, decreased upon CK1δ/ε inhibition but did not significantly change with aldosterone treatment (data not shown). This result is consistent with what is known about CHIF, another Fxyd family member, that is not aldosterone responsive in the kidney (44).
Inhibition of CK1δ/ε reduces αENaC protein levels and increases Caveolin-1 protein levels in plasma membrane of mpkCCDc14 cells.
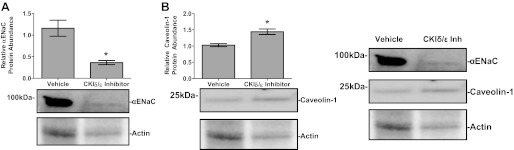
To test whether the effect of CK1δ/ε inhibition on αENaC expression extended to the level of protein, plasma membrane fractions were isolated from mpkCCDc14 cells treated with DMSO (vehicle) or CK1δ/ε inhibitor in the presence of aldosterone. Western blot analysis was performed to measure changes in αENaC protein. In addition, levels of Caveolin-1, a lipid raft protein that inhibits ENaC via changes in membrane recycling (25), were measured. Caveolin-1 was recently identified as a Per1 target gene (42). Cells treated with CK1δ/ε inhibitor showed a 60% decrease in αENaC protein levels (Fig. 7A) and a 40% increase in Caveolin-1 levels (Fig. 7B) compared with control samples.
Fig. 7.
Inhibition of CK1δ/ε reduces αENaC protein levels and increases Caveolin-1 protein levels in membrane of mpkCCDc14 cells. A: mpkCCDc14 cells were treated with the CK1δ/ε Inh PF670462 or DMSO for 72 h; Aldo was present in the media for the last 24 h. Plasma membranes were isolated and Western blot analysis was performed using anti-αENaC, with anti-actin as a loading control. B: same as above but anti-Caveolin-1 antibody was used. Data are representative of 3 independent experiments. *P < 0.05, n = 3. Values are represented as means ± SE.
CK1δ/ε inhibition decreases transepithelial current in mpkCCDc14 cells.
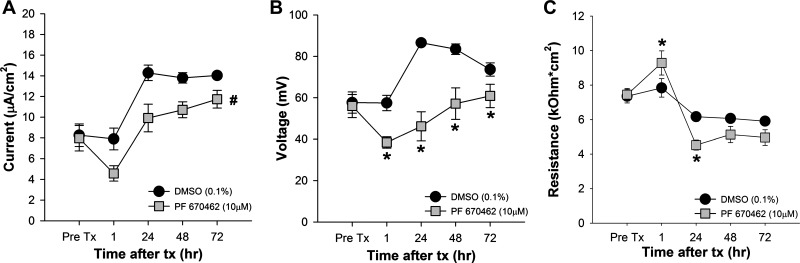
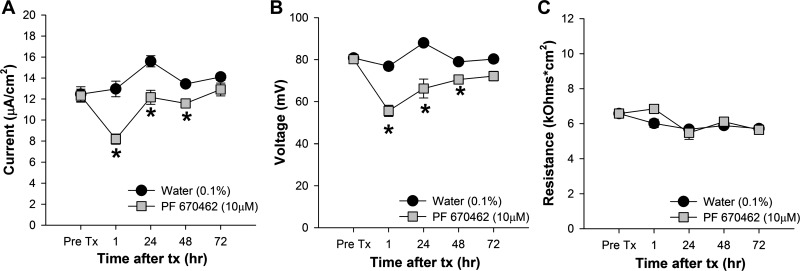
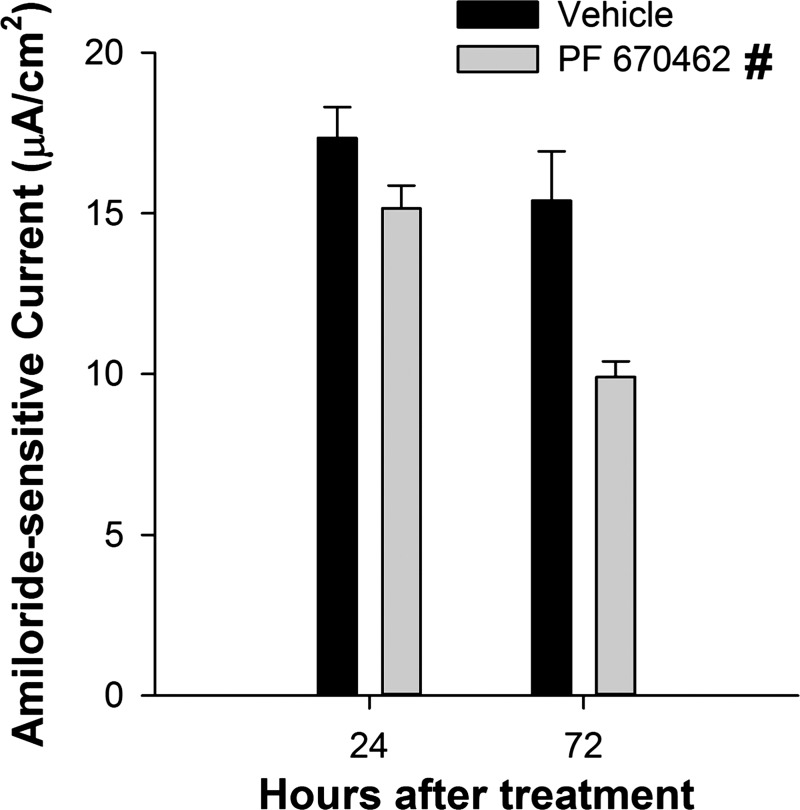
The availability of a pharmacological inhibitor of CK1δ/ε made it possible to test the effect of impaired Per1 nuclear entry on the transepithelial current in mpkCCDc14 cells. Cells were treated with vehicle (DMSO in Fig. 8, water in Fig. 9) or 10 μM PF670462 for 1, 24, 48, or 72 h and the effect on transepithelial (i.e., ENaC) current (Figs. 8A and 9A) was calculated from the differences in transepithelial voltage (Figs. 8B and 9B) and resistance (Figs. 8C and 9C). CK1δ/ε inhibition resulted in an immediate (within 1 h) decrease in transepithelial current, which probably represents an effect of the kinases on the channel itself (see Ref. 46), and this effect was sustained out to 72 h. The vehicle, DMSO, caused an increase in transepithelial voltage by 24 h, an effect not seen when water was used as the vehicle control (Fig. 9B). To determine if the differences in transepithelial current reflected a change in ENaC activity, we next measured transepithelial current in mpkCCDc14 cells before and after treatment with amiloride. As shown in Fig. 10, amiloride-sensitive current was significantly less in PF670462 treated compared with vehicle-treated cells at both 24 and 72 h following treatment.
Fig. 8.
Inhibition of CK1δ/ε decreased transepithelial current in mpkCCDc14 cells. To calculate transepithelial current (A), we measured transepithelial voltage (B) and resistance (C) in mpkCCDc14 cell monolayers before (Pre Tx) and, at specific time intervals, after treatment with vehicle (0.1% DMSO) or 10 μM PF670462 (n = 8–9 in each group). Data shown are means ± SE and were analyzed by 2-way repeated-measures ANOVA with post hoc Student-Newman-Keuls test, when appropriate. For interactions, *P < 0.05 between the treatments groups at a particular time point, #significant effect of treatment across all time points.
Fig. 9.
Inhibition of CK1δ/ε decreased transepithelial current in mpkCCDc14 cells with water as a vehicle. To calculate transepithelial current (A), we measured transepithelial voltage (B) and resistance (C) in mpkCCDc14 cells before (Pre Tx) and, at specific time intervals, after treatment with vehicle (0.1% water) or 10 μM PF670462 (n = 8 in each group). Data shown are means ± SE and were analyzed by 2-way repeated-measures ANOVA with post hoc Student-Newman-Keuls test, when appropriate. For interactions, *P < 0.05 between the treatments groups at a particular time point.
Fig. 10.
Inhibition of CK1δ/ε decreased amiloride-sensitive current in mpkCCDc14 cells. We calculated transepithelial current from transepithelial voltage and resistance measured in mpkCCDc14 cells at 24 and 72 h after treatment with vehicle (0.1% water) or 10 μM PF670462 and following a 10 min exposure to 10 μM amiloride. Data shown are means ± SE and were calculated as the difference in current before and after treatment with amiloride (n = 3 in each group). Statistical differences were determined by 2-way ANOVA with post hoc Holm-Sidak test. #Significant main effect of treatment across all time points.
CK1δ/ε inhibition greatly reduces the number of cell-attached patches with observable ENaC activity in A6 cells.
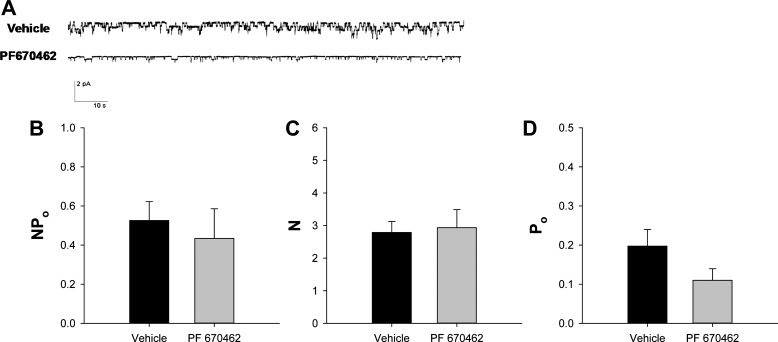
To determine if CK1δ/ε inhibition affects ENaC activity, single channel recordings were made in amphibian kidney A6 cells (clone 2F3). ENaC activity is readily measurable in this well-characterized cell line (45). A representative single channel recording is shown in Fig. 11A. Compared with vehicle, CK1δ/ε inhibition reduced the number of patches with observable channel activity by 55% (Table 1). Despite this reduction in the number of patches with observable activity, the NPo, N, and Po (Fig. 11, B–D, respectively) were not different between the vehicle and PF670462 groups in patches that had observable channel current transitions.
Fig. 11.
CK1δ/ε inhibition did not affect ENaC single channel activity in patches with observable channel openings. A: typical single channel recordings are shown from cell-attached patches of polarized A6 (subclone 2F3) cells treated with vehicle (0.1% DMSO or water) or PF670462 (10 μM) for 72 h. B: ENaC activity (NPo) was calculated from 5–10 min single channel recordings. Vehicle, n = 19; PF670462, n = 16. C: the number of channels (N) in individual patches was determined by determining the number of individual current levels observed during the recording period. D: the open probability (Po) was calculated through the equation: Po = NPo/N. Based on the distribution of the number of channels per patch shown in Fig. 11, we excluded patches without observable channel activity from these data sets. Vehicle, n = 19; PF670462, n = 16. Values are represented as means ± SE.
Table 1.
CK1δ/ε inhibition decreases the number of patches with observable ENaC activity
| Treatment | Total Patches, n | Patches With Channels, n | Proportion |
|---|---|---|---|
| Vehicle (0.1% DMSO) | 29 | 24 | 0.828 |
| PF670462 (10 μM) | 51 | 19 | 0.373* |
CK, casein kinase; ENaC, epithelial sodium channel.
P < 0.001 by z-test with Yates correction.
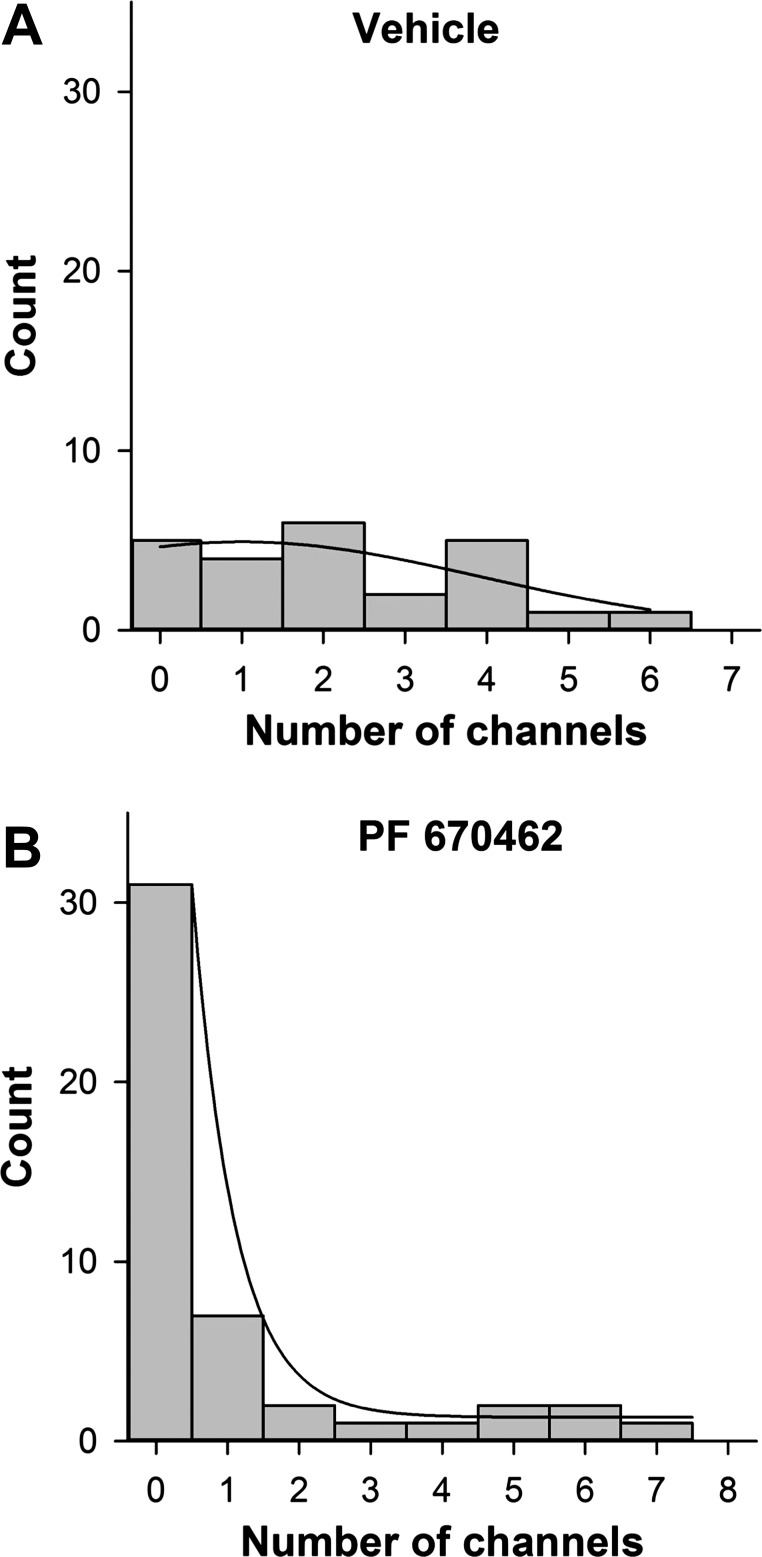
We suspected that the reason we could detect no activity in some patches was because there were no channel proteins in the isolated patches (N equals zero) rather than that there were channel proteins, but that the channels never opened (Po equal to zero). To confirm this, we examined the distribution of the number of channels per patch (Fig. 12). In vehicle-treated cells, channels were distributed independently (χ2-test suggested a probability >0.97 that the channels were independently inserted in the membrane), and the distribution could be fit by a normal distribution with 2.2 ± 0.79 channels per patch. For inhibitor-treated cells, the distribution shifts to strongly favor patches with no measurable channels even though when there are channels in the patches the distribution is similar to untreated cells. Therefore, we conclude that patches with no activity have no channel proteins, and we feel that it is reasonable to calculate NPo, N, and Po only for patches with observable activity as shown in Fig. 11.
Fig. 12.
Inhibition of CK1δ/ε decreased the number of channels present in each patch of A6 cells. These histograms show the distribution of the number of channels observed in each individual patch treated with vehicle (0.1% DMSO or water) or PF670462 (10 μM) for 72 h. The histogram for vehicle data was fitted with a 3-parameter Gaussian nonlinear regression (P = 0.21, R2 = 0.546). The histogram for PF670462 data was fitted with 2 exponential functions using nonlinear regression (P < 0.0001, R2 = 0.998). *P < 0.001 by z-test with Yates correction.
DISCUSSION
The purpose of this study was to utilize pharmacological modulation to explore the role of the circadian clock in the regulation of ENaC. To this end, we determined the effect of inhibition of the circadian regulatory kinases, CK1δ/ε, on αENaC mRNA, protein expression, and ENaC activity. DAPA showed that CK1δ/ε inhibition prevented Per1 and Clock from interacting with the target E-box in the Scnn1a promoter. CK1δ/ε inhibition reduced αENaC mRNA and plasma membrane protein levels significantly. Moreover, CK1δ/ε inhibition repressed the aldosterone-induced response of αENaC expression. Importantly, we show for the first time that inhibition of CK1δ/ε resulted in decreased amiloride-sensitive transepithelial current in mpkCCDc14 cells and significantly fewer patches with observable ENaC activity found in 2F3 A6 cells.
Our results provide evidence that CK1δ/ε promotes transcription and translation of αENaC through regulation of Per1 nuclear translocation. These data are consistent with our previous observations that Per1 appears to positively regulate αENaC expression (19, 20). The same result has been observed for the action of Per1 on prolactin mRNA expression (11). The present study demonstrates a role for Per1 in the positive regulation of αENaC, a result that is also in line with our recent finding that plasma membrane levels of αENaC are reduced following siRNA-mediated knock-down of Per1 in mpkCCDc14 cells (42). We also showed that mice lacking functional Per1 exhibit significantly reduced blood pressure compared with wild-type mice. It remains to be determined if reduced ENaC activity in the absence of Per1 in vivo contributes to the blood pressure phenotype observed in Per1 knockout mice. Together these data support a role for Per1 in transcriptional activation, perhaps in a gene- and tissue-specific manner. It should be noted that CK1 and CK2 directly modulate ENaC already present at the plasma membrane. CK2 directly phosphorylated the COOH terminus of the beta and gamma subunits (36). CK2 phosphorylation of these subunits positively regulates ENaC activity through inhibition of the Nedd4-2 pathway (6). CK1δ also positively regulates human αENaC via enhancing intracellular trafficking and biosynthesis (46). Thus, in addition to increasing synthesis of αENaC via Per1-mediated upregulation, these enzymes phosphorylate membrane-bound ENaC, thereby reducing channel retrieval and degradation. These coordinated actions of CK1 and CK2 on ENaC expression and localization would likely promote sodium reabsorption. In the context of our results, the decrease in ENaC activity produced by inhibition of CK1 δ/ε most likely occurs as a combination of transcriptional repression of αENaC via a Per1-dependent mechanism and enhanced internalization and degradation via less αENaC phosphorylation by CK1δ/ε. Future studies are needed to address the effect of time on these observations. As previously mentioned, CK1δ/ε inhibition impairs the circadian clock through induction of phase delays in rats under free running conditions (7). Cynomolgus monkeys also display a similar phase delay (39), and time of dosing affects the phase shifting (38). In addition to this role in regulation of the circadian clock, CKs (1 and 2 specifically) have been targeted as possible anticancer and antiviral drugs. CK1δ/ε inhibition resulted in apoptosis of cancer cells (12). CK2 is constitutively active, highly expressed in cancerous cells (3, 9), vital for cell proliferation (32), and plays a role in viral infections (34). One CK2 inhibitor used in the treatment of cervical cancer provided positive results (clinical trial CIGB-300) (33, 37). Interestingly, it was recently shown that ENaC and the acid sensing ion channel ASIC1 form hybrid channels in a human cell model of glioblastoma and that knock-down of these channels inhibits cell migration (22, 23). Whether or not this effect involves CK1 or CK2 dysregulation is unknown. Together with the present study, these data suggest that inhibition of CK1δ/ε may be advantageous in disease states.
In summary, the results of the present study provide pharmacological evidence for the regulation of αENaC expression and ENaC activity by the circadian clock. Importantly, we demonstrated for the first time that inhibiting the circadian regulatory kinases CK1δ/ε and subsequent Per1 nuclear entry decreases the number of patches with observable ENaC current. Together with our previous findings (19, 20), these results support a critical role for the circadian clock in the regulation of ENaC. Given the importance of ENaC in sodium homeostasis, these findings may have important implications for blood pressure control.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant DK-085193 and University of Florida, Division of Nephrology funds (to M. L. Gumz) and NIH Grants T32 DK-07518 (to J. Richard), 5R37DK-037963-25 (to D. C. Eaton), and 5K12GM-000680-12 (to D. C. Eaton and M. M. Greenlee).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.R. and M.L.G. conception and design of research; J.R., M.M.G., L.A.J., K.-Y.C., L.G., and M.L.G. performed experiments; J.R., M.M.G., L.G., D.C.E., and M.L.G. analyzed data; J.R., M.M.G., D.C.E., and M.L.G. interpreted results of experiments; J.R., M.M.G., and M.L.G. prepared figures; J.R. drafted manuscript; J.R., M.M.G., L.A.J., D.C.E., and M.L.G. edited and revised manuscript; D.C.E. and M.L.G. approved final version of manuscript.
REFERENCES
- 1. Agarwal R. Regulation of circadian blood pressure: from mice to astronauts. Curr Opin Nephrol Hypertens 19: 51–58, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agarwal R, Light RP. The effect of measuring ambulatory blood pressure on nighttime sleep and daytime activity–implications for dipping. Clin J Am Soc Nephrol 5: 281–285, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmed K. Significance of the casein kinase system in cell growth and proliferation with emphasis on studies of the androgenic regulation of the prostate. Cell Mol Biol Res 40: 1–11, 1994 [PubMed] [Google Scholar]
- 4. Albrecht U. The mammalian circadian clock: a network of gene expression. Front Biosci 9: 48–55, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev 13: 271–277, 2003 [DOI] [PubMed] [Google Scholar]
- 6. Bachhuber T, Almaca J, Aldehni F, Mehta A, Amaral MD, Schreiber R, Kunzelmann K. Regulation of the epithelial Na+ channel by the protein kinase CK2. J Biol Chem 283: 13225–13232, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, Fisher K, Holland J, Kleiman R, Nelson F, Reynolds L, St Germain K, Schaeffer E, Tate B, Sprouse J. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Ther 322: 730–738, 2007 [DOI] [PubMed] [Google Scholar]
- 8. Bens M, Vallet V, Cluzeaud F, Pascual-Letallec L, Kahn A, Rafestin-Oblin ME, Rossier BC, Vandewalle A. Corticosteroid-dependent sodium transport in a novel immortalized mouse collecting duct principal cell line. J Am Soc Nephrol 10: 923–934, 1999 [DOI] [PubMed] [Google Scholar]
- 9. Boldyreff B, Meggio F, Pinna LA, Issinger OG. Casein kinase-2 structure-function relationship: creation of a set of mutants of the beta subunit that variably surrogate the wildtype beta subunit function. Biochem Biophys Res Commun 188: 228–234, 1992 [DOI] [PubMed] [Google Scholar]
- 10. Bollinger T, Bollinger A, Oster H, Solbach W. Sleep, immunity, and circadian clocks: a mechanistic model. Gerontology 56: 574–580, 2010 [DOI] [PubMed] [Google Scholar]
- 11. Bose S, Boockfor FR. Episodes of prolactin gene expression in GH3 cells are dependent on selective promoter binding of multiple circadian elements. Endocrinology 151: 2287–2296, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheong JK, Nguyen TH, Wang H, Tan P, Voorhoeve PM, Lee SH, Virshup DM. IC261 induces cell cycle arrest and apoptosis of human cancer cells via CK1delta/varepsilon and Wnt/beta-catenin independent inhibition of mitotic spindle formation. Oncogene 30: 2558–2569, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chilov D, Hofer T, Bauer C, Wenger RH, Gassmann M. Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. FASEB J 15: 2613–2622, 2001 [DOI] [PubMed] [Google Scholar]
- 14. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549, 2010 [DOI] [PubMed] [Google Scholar]
- 15. Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys 37: 65–95, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, Haraguchi S, Emoto N, Okuno Y, Tsujimoto G, Kanematsu A, Ogawa O, Todo T, Tsutsui K, van der Horst GT, Okamura H. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med 16: 67–74, 2010 [DOI] [PubMed] [Google Scholar]
- 17. Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111: 41–50, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Gumz ML, Cheng KY, Lynch IJ, Stow LR, Greenlee MM, Cain BD, Wingo CS. Regulation of alphaENaC expression by the circadian clock protein Period 1 in mpkCCD(c14) cells. Biochim Biophys Acta 1799: 622–629, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gumz ML, Stow LR, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Weaver DR, Wingo CS. The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice. J Clin Invest 119: 2423–2434, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, Kito R, Nakao K, Kishimoto W, Yoo SH, Shimomura K, Takao T, Takano A, Kojima T, Nagai K, Sakaki Y, Takahashi JS, Ueda HR. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA 106: 15744–15749, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapoor N, Bartoszewski R, Qadri YJ, Bebok Z, Bubien JK, Fuller CM, Benos DJ. Knockdown of ASIC1 and epithelial sodium channel subunits inhibits glioblastoma whole cell current and cell migration. J Biol Chem 284: 24526–24541, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kapoor N, Lee W, Clark E, Bartoszewski R, McNicholas CM, Latham CB, Bebok Z, Parpura V, Fuller CM, Palmer CA, Benos DJ. Interaction of ASIC1 and ENaC subunits in human glioma cells and rat astrocytes. Am J Physiol Cell Physiol 300: C1246–C1259, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA 108: 16451–16456, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee IH, Campbell CR, Song SH, Day ML, Kumar S, Cook DI, Dinudom A. The activity of the epithelial sodium channels is regulated by caveolin-1 via a Nedd4-2-dependent mechanism. J Biol Chem 284: 12663–12669, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta delta C(T) method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 27. Lubarski I, Pihakaski-Maunsbach K, Karlish SJ, Maunsbach AB, Garty H. Interaction with the Na,K-ATPase and tissue distribution of FXYD5 (related to ion channel). J Biol Chem 280: 37717–37724, 2005 [DOI] [PubMed] [Google Scholar]
- 28. Marunaka Y, Eaton DC. Effects of insulin and phosphatase on a Ca2+-dependent Cl− channel in a distal nephron cell line (A6). J Gen Physiol 95: 773–789, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. May A, Puoti A, Gaeggeler HP, Horisberger JD, Rossier BC. Early effect of aldosterone on the rate of synthesis of the epithelial sodium channel alpha subunit in A6 renal cells. J Am Soc Nephrol 8: 1813–1822, 1997 [DOI] [PubMed] [Google Scholar]
- 30. Moore-Ede MC. Physiology of the circadian timing system: predictive versus reactive homeostasis. Am J Physiol Regul Integr Comp Physiol 250: R737–R752, 1986 [DOI] [PubMed] [Google Scholar]
- 31. Naidu KS, Morgan LW, Bailey MJ. Inflammation in the avian spleen: timing is everything. BMC Mol Biol 11: 104, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Padmanabha R, Chen-Wu JL, Hanna DE, Glover CV. Isolation, sequencing, and disruption of the yeast CKA2 gene: casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol Cell Biol 10: 4089–4099, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perea SE, Reyes O, Baladron I, Perera Y, Farina H, Gil J, Rodriguez A, Bacardi D, Marcelo JL, Cosme K, Cruz M, Valenzuela C, Lopez-Saura PA, Puchades Y, Serrano JM, Mendoza O, Castellanos L, Sanchez A, Betancourt L, Besada V, Silva R, Lopez E, Falcon V, Hernandez I, Solares M, Santana A, Diaz A, Ramos T, Lopez C, Ariosa J, Gonzalez LJ, Garay H, Gomez D, Gomez R, Alonso DF, Sigman H, Herrera L, Acevedo B. CIGB-300, a novel proapoptotic peptide that impairs the CK2 phosphorylation and exhibits anticancer properties both in vitro and in vivo. Mol Cell Biochem 316: 163–167, 2008 [DOI] [PubMed] [Google Scholar]
- 34. Pinna LA, Meggio F. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog Cell Cycle Res 3: 77–97, 1997 [DOI] [PubMed] [Google Scholar]
- 35. Schroeder A, Loh DH, Jordan MC, Roos KP, Colwell CS. Circadian regulation of cardiovascular function: a role for vasoactive intestinal peptide. Am J Physiol Heart Circ Physiol 300: H241–H250, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi H, Asher C, Yung Y, Kligman L, Reuveny E, Seger R, Garty H. Casein kinase 2 specifically binds to and phosphorylates the carboxy termini of ENaC subunits. Eur J Biochem 269: 4551–4558, 2002 [DOI] [PubMed] [Google Scholar]
- 37. Solares AM, Santana A, Baladron I, Valenzuela C, Gonzalez CA, Diaz A, Castillo D, Ramos T, Gomez R, Alonso DF, Herrera L, Sigman H, Perea SE, Acevedo BE, Lopez-Saura P. Safety and preliminary efficacy data of a novel casein kinase 2 (CK2) peptide inhibitor administered intralesionally at four dose levels in patients with cervical malignancies. BMC Cancer 9: 146, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sprouse J, Reynolds L, Kleiman R, Tate B, Swanson TA, Pickard GE. Chronic treatment with a selective inhibitor of casein kinase I delta/epsilon yields cumulative phase delays in circadian rhythms. Psychopharmacology (Berl) 210: 569–576, 2010 [DOI] [PubMed] [Google Scholar]
- 39. Sprouse J, Reynolds L, Swanson TA, Engwall M. Inhibition of casein kinase I epsilon/delta produces phase shifts in the circadian rhythms of Cynomolgus monkeys. Psychopharmacology (Berl) 204: 735–742, 2009 [DOI] [PubMed] [Google Scholar]
- 40. Stow LR, Gumz ML. The circadian clock in the kidney. J Am Soc Nephrol 22: 598–604, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stow LR, Gumz ML, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Wingo CS. Aldosterone modulates steroid receptor binding to the endothelin-1 gene (edn1). J Biol Chem 284: 30087–30096, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stow LR, Richards J, Cheng KY, Lynch IJ, Jeffers LA, Greenlee MM, Cain BD, Wingo CS, Gumz ML. The circadian protein period 1 contributes to blood pressure control and coordinately regulates renal sodium transport genes. Hypertension 59: 1151–1156, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vagnucci AH, Shapiro AP, McDonald RH., Jr Effects of upright posture on renal electrolyte cycles. J Appl Physiol 26: 720–731, 1969 [DOI] [PubMed] [Google Scholar]
- 44. Wald H, Goldstein O, Asher C, Yagil Y, Garty H. Aldosterone induction and epithelial distribution of CHIF. Am J Physiol Renal Fluid Electrolyte Physiol 271: F322–F329, 1996 [DOI] [PubMed] [Google Scholar]
- 45. Wills NK, Purcell RK, Clausen C. Na+ transport and impedance properties of cultured renal (A6 and 2F3) epithelia. J Membr Biol 125: 273–285, 1992 [DOI] [PubMed] [Google Scholar]
- 46. Yan W, Spruce L, Rosenblatt MM, Kleyman TR, Rubenstein RC. Intracellular trafficking of a polymorphism in the COOH terminus of the alpha-subunit of the human epithelial sodium channel is modulated by casein kinase 1. Am J Physiol Renal Physiol 293: F868–F876, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Zuber AM, Centeno G, Pradervand S, Nikolaeva S, Maquelin L, Cardinaux L, Bonny O, Firsov D. Molecular clock is involved in predictive circadian adjustment of renal function. Proc Natl Acad Sci USA 106: 16523–16528, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]