Abstract
Adenosine is a potent modulator of liver fibrosis and inflammation. Adenosine has been shown to regulate such diverse activities as chemotaxis, contraction, and matrix production in hepatic stellate cells (HSC). Ecto-5′-nucleotidase/CD73 [EC 3.1.3.5] is the rate-limiting enzyme in adenosine production. Cd73-deficient mice are resistant to experimental liver fibrosis and have impaired adenosine generation. However, cell-specific expression and regulation of CD73 within the fibrotic liver have not been defined. In particular, prior evidence demonstrating that liver myofibroblasts, the cells believed to be responsible for matrix formation in the liver, express CD73 is lacking. Thus we tested the hypothesis that HSC and portal fibroblasts (PF), cells that undergo differentiation into liver myofibroblasts, express CD73 in a regulated fashion. We found that CD73 is weakly expressed in quiescent HSC and PF but is markedly upregulated at the transcriptional level in myofibroblastic HSC and PF. We furthermore found that CD73 protein and its functional activity are strongly increased in fibrous septa in rats subjected to experimental fibrosis. To determine the mechanism for the upregulation of Cd73 gene, we cloned the rat Cd73 promoter and then used serial truncation and site-directed mutagenesis to identify key regulatory elements. We identified two consensus SP1 motifs and one SMAD binding site, each of which was necessary for Cd73 gene upregulation. In conclusion, activated HSC upregulate Cd73 gene expression, via specific SP1 and SMAD promoter elements, after myofibroblastic differentiation. The ecto-5′-nucleotidase/CD73 enzyme is a novel cellular marker of activated liver myofibroblasts in vivo and in vitro and thus represents a promising molecular target for antifibrotic therapies in liver diseases.
Keywords: adenosine, hepatic fibrosis, liver myofibroblasts, portal fibroblast, purinergic signaling
liver fibrosis with subsequent cirrhosis is the most common cause of liver failure. This process results from an imbalance between synthesis and degradation of extracellular matrix (ECM) (25). Liver myofibroblasts are the primary cells responsible for the extensive ECM accumulation observed in liver fibrosis. These cells derive from a variety of sources including liver nonparenchymal cells, periportal and perivascular fibroblasts, and bone marrow-derived fibrocytes (37); however, the best characterized sources of liver myofibroblasts are hepatic stellate cells (HSC) and portal fibroblasts (PF) (16). As they differentiate, liver myofibroblasts become contractile, owing to production of α-smooth muscle actin (α-SMA) and related proteins, and fibrogenic, owing to release of fibrillar collagens and other matrix proteins (16). Thus liver myofibroblasts represent excellent targets for antifibrotic therapies. Extracellular nucleosides (e.g., adenosine) and nucleotides (e.g., ATP) are potent signaling molecules that contribute to liver cellular homeostasis by regulating key physiological functions such as glucose metabolism, cholesterol transport, bile formation, and cytokine secretion (8, 18, 31, 45). These compounds exert their regulatory effects via activation of purinergic P1 (nucleoside) and P2 (nucleotide) receptors and induction of well-characterized second messengers, chiefly cAMP and cytosolic Ca2+ (4). Most liver cells express multiple P1 and P2 receptor subtype(s). Hepatocytes express A1, A2a, A2b, and A3 receptors and P2X4,7 (22) and P2Y1,2,4,6,13 receptors (9, 19), and bile duct epithelia express P2X4,7 (10, 11) and P2Y1,2,4,6,11 receptors (14). Interestingly, HSC express only A2a, A2b, and A3 receptors and switch expression of P2Y receptor subtypes after myofibroblastic differentiation (3, 15, 41). In addition to homeostatic roles, liver purinergic receptors also regulate inflammation and injury-associated processes, such as tissue repair and wound healing responses (2). Procollagen messenger RNA expression by activated fibrogenic HSC is upregulated by both UDP (through P2Y6 receptor) and adenosine (through A2a receptor) (6, 15). Additionally, the activation of A2a receptor by adenosine in myofibroblastic HSC inhibits chemotaxis and contraction while markedly increasing collagen synthesis (24, 41) and matrix metalloproteinase expression (5). Moreover, the role of extracellular adenosine as an overall immunomodulatory mediator in liver fibrosis has been demonstrated in various experimental liver fibrosis models using gene silencing or pharmacological inhibition approaches targeting specific adenosine receptor subtypes (5, 34, 35, 43). A recent report showed that mice deficient in ecto-5′-nucleotidase/CD73, the rate-limiting enzyme in hepatic adenosine production, are less prone to experimental liver fibrosis, perhaps because of changes in liver myofibroblast function (35, 36). However, the expression of CD73 by liver myofibroblast precursors (specifically HSC and PF) is unknown. Thus we tested the hypothesis that HSC and PF express ecto-5′-nucleotidase/CD73 in a regulated fashion. Here, we show that Cd73 gene expression is upregulated at the transcriptional level in HSC and PF after myofibroblastic differentiation. We also show that this transcriptional upregulation of Cd73 gene is controlled by promoter response elements for SP1 and SMAD transcription factors, providing a specific biological mechanism for this process.
EXPERIMENTAL PROCEDURES
Materials.
Tissue culture reagents were obtained from Invitrogen (Carlsbad, CA). Mouse monoclonal antibody directed against human CD73 (clone 7G2) was obtained from Invitrogen, α-SMA (clone 1A4) and β-actin (clone AC15) from Sigma-Aldrich (St. Louis, MO), and fibronectin (clone 10) from BD Transduction (San Diego, CA). Rabbit polyclonal antibody directed against β-tubulin was from Cell Signaling (Danvers, MA), and rat CD73 (r5′NT-9l) was a gift from Dr. Jean Sévigny (Laval University, Québec, Canada). All other chemicals were of the highest quality commercially available.
Animals and experimental liver fibrosis.
Male adult Sprague-Dawley rats (180–250 g; Harlan Sprague Dawley, Indianapolis, IN) were used for all experiments. Liver fibrosis was experimentally induced in adult rats by 8-wk carbon tetrachloride (CCl4) intoxication and 2-wk common bile duct ligation (BDL), as previously described (13). All procedures were approved by the Yale and University of Arkansas for Medical Sciences Institutional Animal Care and Use Committees.
Primary cell isolation and cell culture.
Primary hepatocytes, HSC, and PF were isolated from rat livers as previously described. Briefly, hepatocyte and nonparenchymal cell (NPC) fractions were obtained by in situ pronase/collagenase perfusion of livers. Rat NPC were subsequently used for isolation of HSC by density gradient centrifugation (21), and PF by serial mesh filtration of the hilar remnant (29). Resulting cell suspensions were plated onto tissue culture plastic dishes in DMEM/F-12 containing 10% FCS and antibiotics. Primary rat HSC and PF were used at day 1 (quiescent) and day 7 (myofibroblastic/activated) after plating, as previously described (21, 29). Primary activated human HSC were isolated as previously described (26) and grown in DMEM-high glucose-containing 10% FCS and antibiotics. LX-1 and LX-2 cells, human stellate cell lines resembling an activated HSC phenotype (42), were grown in DMEM-high glucose containing 2% FCS and antibiotics. All cells were maintained at 37°C, under 95% air-5% CO2.
Immunohistochemistry, enzyme histochemistry, and confocal immunofluorescence.
We fixed 6-μm sections of snap-frozen rat liver specimens, immortalized LX-2, or primary human/rat HSC (∼105 cells/coverslip) with cold acetone-10% phosphate-buffered formalin (19:1) (for immunohistochemistry and confocal immunofluorescence) or freshly prepared 4% (wt/vol) paraformaldehyde in PBS, pH 7.2 (for enzyme histochemistry). For immunohistochemistry, liver sections were stained with rabbit polyclonal anti-rat CD73 antibody r5′NT-9l (1:2,000), mouse monoclonal α-SMA antibody (1:400), or corresponding control sera, as previously described (20). For enzyme histochemistry, ectonucleotidase activities were visualized on fixed liver sections or primary human HSC by a modified Wachstein-Meisel lead phosphate method, as previously described (20). Assays were conducted with AMP (1 mM) as substrate and in the presence of tissue-nonspecific alkaline phosphatase inhibitor levamisole (5 mM; Sigma-Aldrich). All sections were counterstained with aqueous hematoxylin, and slides were mounted in Mowiol 4-88 medium (Calbiochem, La Jolla, CA). For confocal immunofluorescence, fixed immortalized LX-2 or primary human/rat HSC were incubated with mouse monoclonal anti-human CD73, α-SMA, fibronectin, and rabbit polyclonal anti-rat CD73 antibody r5′NT-9l, overnight at 4°C. Slides were then incubated with appropriate goat Alexa Fluor-conjugated anti-rabbit IgG and anti-mouse IgG antibodies (Molecular Probes, Eugene, OR) for 1 h at room temperature. Slides were subsequently incubated with TO-PRO-3 nuclear stain for 30 min at room temperature and mounted in ProLong Gold Anti-fade reagent with 4,6-diamidino-2-phenylindole (DAPI) nuclear stain medium (Molecular Probes). Slides incubated with secondary antibody alone were used as a control for specificity of fluorescence detection. We performed fluorescence microscopy using an Olympus BX51 fluorescence microscope and confocal microscopy using Zeiss LSM 510 Meta and 710 confocal imaging systems.
Immunoblot analysis.
Changes in expression of rat CD73 were determined by immunoblot using anti-rat CD73 antibody r5′NT-9l and compared with β-tubulin expression for HSC or β-actin for PF, as protein loading controls. Total proteins from HSC or PF preparations were extracted with M-PER reagent containing Halt Protease Inhibitor Single Use Cocktail (Pierce Biotechnology, Rockford, IL), concentrated by use of Amicon Ultra (10,000 MWCO; Millipore) centrifugal filter units, separated by SDS-PAGE under nonreducing conditions, and transferred onto a polyvinylidene difluoride membrane (Immobilon/Millipore, Bedford, MA). Membranes were blocked with the Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) and incubated with rabbit polyclonal anti-rat CD73 antibody r5′NT-9l, followed by goat anti-rabbit IRDye680LT, and bands were visualized by use of the Odyssey imaging system. Membranes were subsequently stripped and incubated with rabbit polyclonal anti-β-tubulin or mouse monoclonal anti-β-actin followed by goat anti-rabbit IRDye680LT or anti-mouse IRDye800 secondary antibodies (LI-COR), and bands were visualized and intensity signals quantified by use of the Odyssey imaging system.
RT-PCR.
Changes in rat Cd73 mRNA were quantitatively determined by real-time RT-PCR with an ABI PRISM 7500 sequence detection system (Applied Biosystems, Foster City, CA). cDNA was synthesized by reverse transcription by using total RNA extracted from quiescent (day 1) or myofibroblastic (day 7) HSC. Rat Cd73 and Gapdh (control) expression levels were determined by TaqMan gene expression assays Rn00571989_m1 and Rn01775763_g1 (Applied Biosystems). PCR amplification was performed under the following protocol: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 55°C for 60 s. Relative gene expression was calculated by the comparative CT method (40). To determine expression of CD73 mRNA by semiquantitative RT-PCR, cDNA was synthesized by reverse transcription using total RNA obtained commercially (human brain; Invitrogen) or extracted from immortalized (LX-1, LX-2, and HepG2) and primary human (FH11 and FH13) HSC. PCR amplification was done by using the following sets of oligonucleotide primers: for CD73, forward, 5′-TGGAACCACGTATCCATGTG-3′; reverse, 5′-ATGCTCAAAGGCCTTCTTCA-3′; for GAPDH, forward, 5′-CGACCACTTTGTCAAGCTCA-3′; reverse, 5′-AGGGGTCTACATGGCAACTG-3′.
5′RLM-RACE analysis.
The transcription start site of the rat Cd73 gene was determined RNA ligase-mediated rapid amplification of 5′ cDNA ends (5′RLM-RACE), by using FirstChoice RLM-RACE kit following the manufacturer's instructions (Ambion, Austin, TX). Total RNA was isolated from primary rat hepatocytes with TRIzol reagent (Invitrogen) and used as template for the replacement of the mRNA 5′CAP structure by a 5′RLM-RACE adaptor. The full-length RNA ligated to the adaptor was then reverse transcribed with SuperScript III First-Strand Synthesis System (Invitrogen) using oligo(dT)18 primers. Cd73 transcription start sites were analyzed by nested PCR amplification. The first amplification was done using 5′RLM-RACE and gene-specific outer primers (gsrCd73-R2) (see Table 1) at annealing temperatures of both 60 and 55°C. An aliquot of each of these first PCR products was used for an additional PCR amplification with the 5′RLM-RACE and gene-specific inner primers (gsrCd73-R1) at an annealing temperature of 60°C. PCR amplifications were performed by using the Expand High-Fidelity PCR system (Roche Biosciences, Palo Alto, CA) under the following conditions: 95°C for 1 min, 35 cycles of 95°C for 30 s, 55 or 60°C for 30 s, and 72°C for 30 s, completed by a 10-min incubation at 72°C. After analysis by agarose gel electrophoresis, a single-band PCR product from both nested amplifications was detected by staining with ethidium bromide and purified by gel extraction. The extracted samples were ligated into pCRII TOPO vector (Invitrogen) by following the manufacturer's instructions. At least 10 individual colonies from each transformation were analyzed by PCR amplification using manufacturer instructions. Eight clones originating from 55°C outer PCR and five from 60°C outer PCR were used for analysis by automated sequencing (Keck DNA Sequencing Facility, Yale University).
Table 1.
Primers used for these studies
| Primers | Sequence (5′-3′) | Location | Experiment |
|---|---|---|---|
| Gene specific outer primer gsrCd73-R2 | GTCCTTCCACACCGTTATCA | Exon 2 | 5′RACE |
| +24587/+24606 | |||
| Gene specific inner primer gsrCd73-R1 | GATGGTGCCCTGGTACTGAT | Exon 1 | 5′RACE |
| +260/+269 | |||
| prCd73 R1 | AGCGCGTTGAGCGGGTGAA | −29/−11 | 5′flanking region cloning |
| prCd73 F1 | TGCCATCACCACCAGACTAA | −2056/−2037 | 5′flanking region cloning |
| prCd73 F2 | ACCTGGTGCATTCTGGAGAT | −993/−974 | 5′flanking region cloning |
| prCd73 F3 | ACACACACACCCCAAAAGG | −735/−717 | 5′flanking region cloning |
| prCd73 F4 | CACCAAGGCAACCTCACAG | −484/−466 | 5′flanking region cloning |
| prCd73 F5 | ACGCGAACAACCTTCTCTCT | −249/−230 | 5′flanking region cloning |
| prCd73 F6 | ACTCTGGGAGTGCGTCTG | −159/−142 | 5′flanking region cloning |
| prCd73 F7 | TAGTCCACCCCTCCGCCTA | −90/−72 | 5′flanking region cloning |
| WT Cd73 probe sense | TGCGGGCGGGAGGACCTGGGG | −143/−102 | Mobility shift assay |
| CTAAAGGAGGCGGGTCTGGCC | |||
| WT Cd73 probe anti-sense | GGCCAGACCCGCCTCCTTTAG | −143/−102 | Mobility shift assay |
| CCCCAGGTCCTCCCGCCCGCA | |||
| Mutant SP1 probe sense | TGCGGTACGGAGGACCTGGGG | −143/−102 | Mobility shift assay |
| CTAAAGGAGTACGGTCTGGCC | |||
| Mutant SP1 probe anti-sense | GGCCAGACCGTACTCCTTTAG | −143/−102 | Mobility shift assay |
| CCCCAGGTCCTCCGTACCGCA | |||
| Mutant SMAD probe sense | TGCGGGCGGGAGGACCTGGGG | −143/−102 | Mobility shift assay |
| CTAAAGGAGGCGGCAGTGGCC | |||
| Mutant SMAD probe anti-sense | GGCCACTGCCGCCTCCTTTAG | −143/−102 | Mobility shift assay |
| CCCCAGGTCCTCCCGCCCGCA | |||
| Mutant SP1 + SMAD probe sense | TGCGGTACGGAGGACCTGGGG | −143/−102 | Mobility shift assay |
| CTAAAGGAGTACGCAGTGGCC | |||
| Mutant SP1 + SMAD probe anti-sense | GGCCACTGCGTACTCCTTTAG | −143/−102 | Mobility shift assay |
| CCCCAGGTCCTCCGTACCGCA |
Cloning of rat Cd73 gene putative promoter and construction of reporter vectors.
To analyze the promoter activity of the 5′ untranslated region of rat Cd73 gene, 7 discrete fragments of the promoter region with length between 2,046 and 80 base pairs (bp) were generated. An additional restriction enzyme site was added to each forward (MluI) and reverse (BglII) primer. All primer sequences and positions are listed in Table 1. For PCR amplification of the longest fragment (2,046 bp) that was expected to contain the complete promoter, 0.1 μg of commercially available rat genomic DNA (Clontech, Mountain View, CA) was used as template with the primers prCd73-F1 and prCd73-R1. Approximately 10 ng of the first plasmid was then used as template for PCR amplification of all other promoter fragments (prCd73-F2 to prCd73-F7) with the same reverse primer (prCd73-R1). All PCR amplifications were done with Expand High-Fidelity PCR System (Roche Biosciences) for maximum of fidelity of the elongation, as follows: 95°C for 2 min, 30 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 2 min 30 s, and completed by a 7-min incubation at 72°C. Amplification products were ligated into the MluI/BglII cloning site in the luciferase reporter gene pGL3-Basic vector (Promega, Madison, WI). The nucleotide sequence of each promoter fragment was confirmed by automated sequencing before use.
Site-directed mutagenesis of putative SP1- and SMAD-responsive promoter elements.
Putative promoter responsive elements for SP1 and SMAD transcription factors were identified by using the bioinformatics software tool MatInspector (Genomatix, Munich, Germany), and fragments lacking such elements were generated by site-directed mutagenesis. Site-directed mutants were generated with the QuikChangeII XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). The following sets of oligonucleotide primers were used for SP1 (1st site): forward, 5′-gagtgcgtctgcggtacggaggacctggggc-3′; reverse, 5′-gccccaggtcctccgtaccgcagacgcactc-3′; SP1 (2nd site): forward, 5′-ctggggctaaaggagtacggtctggccccgccc-3′; reverse, 5′-gggcggggccagaccgtactcctttagccccag-3′; and SMAD: forward, 5′-ctaaaggaggcggcagtggccccgccccg-3′; reverse, 5′-cggggcggggccactgccgcctcctttag-3′. PCR amplification of mutants was performed by using PfuUltra High Fidelity DNA polymerase (Stratagene) under the following parameters: 95°C for 1 min, followed by 18 cycles of 95°C for 50 s, 60°C for 50 s, and 68°C for 6 min, and then 68°C for 7 min. Each mutation was confirmed by automated sequencing before use.
Transfection of LX-2 cells.
LX-2 cells were split into 24-well plates (5 × 104/well) on the day before transfection. The basic LX-2 growth medium was replaced with serum-free Opti-MEM I (Invitrogen) just prior transfection reaction. Plasmid DNA (300 ng of each firefly luciferase reporter vector and 10 ng of Renilla luciferase vector) diluted in Opti-MEMI (50 μl) was mixed with Lipofectamine 2000 (2 μl; Invitrogen) diluted in Opti-MEMI (50 μl). The mixture was incubated for 20 min at room temperature and then added to cells in a stepwise fashion. Following a 6-h incubation at 37°C, twice-concentrated growth medium was added to cells for a 48-h incubation, before testing of transgene expression.
Luciferase activity assay.
Changes in firefly luciferase activity of transfected LX-2 cells were normalized to Renilla luciferase activity by use of the Dual-Luciferase Reporter assay system (Promega) and detected with a Synergy HT multidetection microplate reader (BioTek, Winooski, VT).
Electromobility shift assay.
Nuclear extracts from LX-2 cells were prepared by use of the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce Biotechnology) according to the manufacturer's instructions. DNA binding assays were performed with 2.5 μg of nuclear extracts by using the DIG Gel shift second generation kit (Roche Biosciences) according to the manufacturer's instructions. The DIG-labeled double-stranded oligonucleotides used as probes are described in Table 1. For competition experiments, a 125-fold molar excess of unlabeled double-stranded oligonucleotides was added to the binding reaction.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed by using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer's instructions. Briefly, nuclear lysates isolated from rat primary HSC were sonicated, and cross-linked proteins were immunoprecipitated by incubation with antibodies against Sp1 (Millipore) and various SMAD proteins as well as normal IgGs (Cell Signaling), respectively. A DNA sample from sonicated nuclear lysates that underwent proteinase K digestion only was used as internal control (input). A DNA sample immunoprecipitated by normal IgGs was used as negative control. Immunoprecipitated DNA was detected by PCR amplification using prCd73 F6 and prCd73 R1 primers (see Table 1) with Expand High-Fidelity PCR System (Roche Biosciences) at 95°C for 2 min, 35 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s, completed by a 7-min incubation at 72°C, and PCR products were analyzed by agarose gel electrophoresis.
Genetic sequence data.
A genomic DNA sequence containing previously unknown nucleic acid residues located within the cloned putative rat Cd73 promoter was identified by automated sequencing. This sequence (accession number JN792133) was deposited in the GenBank Genome Survey Sequences database (NCBI).
Statistical analysis.
Data are presented as means ± SE. Comparisons between individual groups were made with two-tailed t-tests, by use of GraphPad Prism 5 (GraphPad Software, San Diego, CA).
RESULTS
Ecto-5′-nucleotidase/CD73 protein and ecto-AMPase activity levels are altered in experimentally induced liver fibrosis.
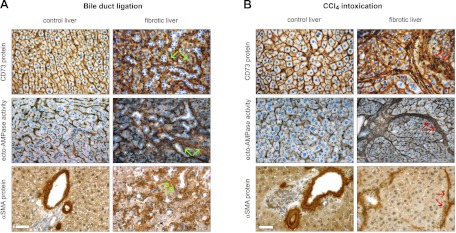
The localization of ecto-5′-nucleotidase/CD73 protein and AMP hydrolytic activity was monitored in two well-established models of liver fibrosis in rats, BDL and CCl4 intoxication. For the BDL model, in control (sham-operated) livers, ecto-5′-nucleotidase/CD73 staining was observed at the level of apical and basolateral hepatocyte membrane domains and in scattered cellular elements present in hepatic sinusoids (Fig. 1A, top left). In BDL livers, a strong staining signal for ecto-5′-nucleotidase/CD73 could be detected in fibrotic areas surrounding unstained proliferating bile ducts, and also at the level of hepatocyte membrane, although with a diffuse canalicular distribution (Fig. 1A, top right). The localization pattern of ecto-AMPase activity in BDL livers clearly paralleled that observed for ecto-5′-nucleotidase/CD73 protein (Fig. 1A, middle). For the CCl4 intoxication model, control (oil vehicle) livers exhibited ecto-5′-nucleotidase/CD73 expression profiles identical to the ones described for BDL control livers (Fig. 1B, top left). In CCl4-intoxicated livers, ecto-5′-nucleotidase/CD73 expression was mainly observed in fibrous septa surrounding regenerating hepatic nodules and also at the level of hepatocyte membrane (Fig. 1B, top right). Again, the distribution of ecto-AMPase activity paralleled the ecto-5′-nucleotidase expression pattern (Fig. 1B, middle). Interestingly, in both fibrosis models, increases in ecto-5′-nucleotidase/CD73 protein and enzymatic activity were detected in fibrotic areas that are prototypical regions in which α-SMA-positive liver myofibroblasts are detected in cirrhotic animals (Fig. 1, A and B, bottom).
Fig. 1.
CD73 protein expression and enzymatic activity redistribute to myofibroblast-rich regions in experimental liver fibrosis. A: effects of bile duct ligation (BDL) on CD73 expression and activity. Liver sections from rats subjected to BDL (2 wk) were used to visualize in situ ecto-AMPase activity, by the Wachstein/Meisel lead phosphate precipitation method, and ecto-5′-nucleotidase/CD73 and α-smooth muscle actin (α-SMA) proteins, by using a standard peroxidase-based immunohistochemistry procedure. In control animals, both ecto-AMPase activity and ecto-5′-nucleotidase/CD73 protein expression have parallel localization at the level of both canalicular and sinusoidal membrane domains of hepatocytes and the hepatic portal areas, whereas α-SMA protein is only observed at the level of the smooth muscle cell layer surrounding blood vessels. Upon hepatic fibrosis induction, ecto-AMPase activity and ecto-5′-nucleotidase/CD73 and α-SMA protein expression are seen in the vicinity of fibrotic areas surrounding proliferating bile ducts (green arrows). B: effects of CCl4 intoxication on CD73 expression and activity. The effects of CCl4-induced liver fibrosis are even more profound, as the distribution of ecto-5′-nucleotidase/CD73 protein expression and activity shifted to broad fibrotic bands surrounding hepatocyte regenerative nodules in the same locale as α-SMA-positive liver myofibroblasts (red arrows). Scale bar, 40 μM.
The myofibroblastic differentiation of activated rat HSC and PF induces upregulation of ecto-5′-nucleotidase/CD73 expression.
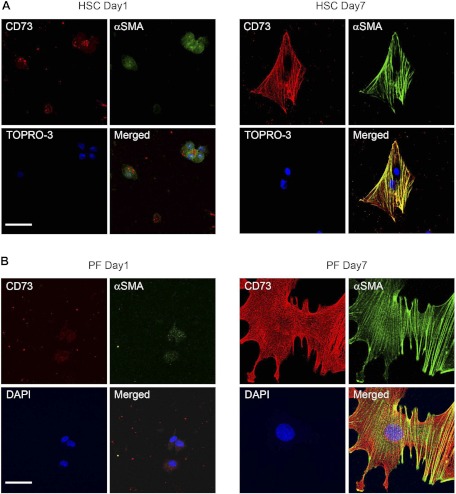
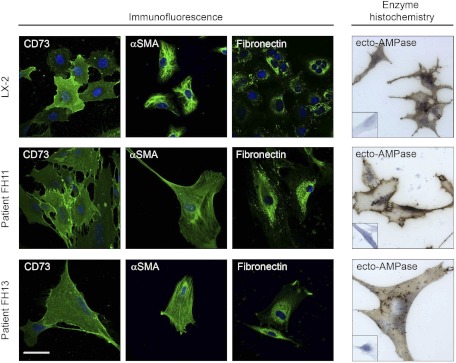
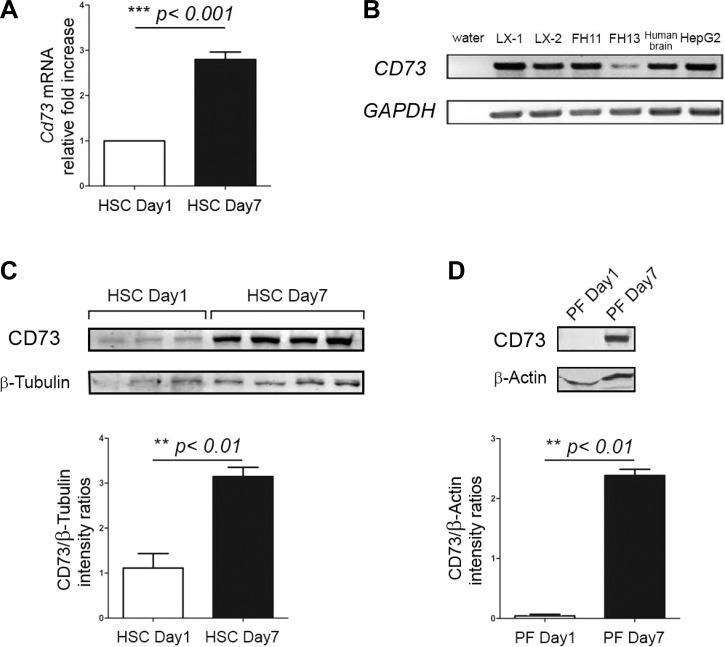
To verify the possibility that activated myofibroblasts could express ecto-5′-nucleotidase/CD73, we determined its localization in quiescent (day 1) and culture-activated (day 7) primary isolated rat HSC and PF by confocal microscopy. Expression of ecto-5′-nucleotidase/CD73 was very low in day 1 primary HSC (Fig. 2 A, HSC day 1) and absent in day 1 primary PF (Fig. 2B, PF day 1), in which quiescence was confirmed by the absence of α-SMA stress fibers. Upon culture activation, a strong increase in cell surface expression of ecto-5′-nucleotidase/CD73 was detected in both primary HSC (Fig. 2 A, HSC day 7) and PF (Fig. 2B, PF day 7). Both day 7 primary HSC and PF had clearly undergone myofibroblastic differentiation, as evidenced by their increased cell surface area, exaggerated spindle shape, and strong expression of α-SMA stress fibers. We also determined the localization of ecto-5′-nucleotidase/CD73 in immortalized LX-2 and primary isolated human FH11 and FH13 myofibroblastic human HSC, in parallel with known HSC cell markers α-SMA and fibronectin. All used human HSC expressed ecto-5′-nucleotidase/CD73 in addition to the other cell markers (Fig. 3, Immunofluorescence). When the distribution of ecto-AMPase activity was determined in those cells, brown precipitates indicative of AMP hydrolysis could be seen decorating their plasma membranes (Fig. 3, Enzyme histochemistry). We next quantified changes in Cd73 gene expression in HSC undergoing myofibroblastic differentiation at the mRNA and protein levels. Cd73 mRNA levels were significantly increased in day 7 primary HSC compared with day 1 primary HSC (Fig. 4A). Interestingly, semiquantitative RT-PCR also demonstrated that myofibroblastic LX-1, LX-2, FH11, and FH13 human HSC express CD73 mRNA (Fig. 4B). Significant increases in ecto-5′-nucleotidase/CD73 protein levels were also detected upon myofibroblastic differentiation in primary HSC (Fig. 4C) and PF (Fig. 4D). Taken together, these results strongly imply that Cd73 gene upregulation is mediated at the transcriptional level during the myofibroblastic differentiation process of activated HSC.
Fig. 2.
Upregulation and redistribution of CD73 protein in hepatic stem cells (HSC) and portal fibroblasts (PF) after culture-induced myofibroblastic differentiation. Expression of ecto-5′-nucleotidase/CD73 was examined in quiescent (day 1, top left) and activated (day 7, top right) HSC preparations, by confocal immunofluorescence. α-SMA-positive staining is seen in green and used as evidence of myofibroblastic differentiation. Ecto-5′-nucleotidase/CD73-positive staining is seen in red. TOPRO-3 labeling of nuclei is seen in blue. When merged images from day 1 and day 7 HSC are compared, high increases in expression of ecto-5′-nucleotidase/CD73 protein are observed that parallel HSC myofibroblastic differentiation. Similar changes in ecto-5′-nucleotidase/CD73 expression were noted in immunofluorescence localization experiments performed on quiescent (day 1) and myofibroblastic (day 7) PF, in which nuclei (blue) are labeled with 4,6-diamidino-2-phenylindole (DAPI) (bottom). Scale bar 20 μM.
Fig. 3.
Evidence of CD73 protein expression and enzymatic function in human HSC-derived myofibroblasts. Immortalized LX-2 myofibroblastic HSC and primary human HSC isolated from human liver explants (FH11 and FH13 HSC) were examined for the expression of ecto-5′-nucleotidase/CD73 and myofibroblast markers, α-SMA, and fibronectin by confocal immunofluorescence, and of ecto-AMPase (AMP hydrolysis) activity by enzyme histochemistry by the Wachstein/Meisel lead phosphate precipitation method as described in Fig. 2. Positive staining for ecto-5′-nucleotidase/CD73 protein as well as of the other markers can be seen in green, indicating that activated human HSC express CD73 (Immunofluorescence). DAPI-positive nuclei are seen in blue. When the ectonucleotidase activity of these cells was tested with AMP as substrate via enzyme histochemistry, brown deposits corresponding to nucleotide hydrolysis products could be detected at their surface. No signal was observed in absence of AMP substrate (all insets). Scale bar, 20 μM (immunofluorescence); scale bar, 20 μM (bright field).
Fig. 4.
Quantitative assessment of Cd73 gene upregulation in myofibroblastic HSC and PF. A: upregulation of Cd73 mRNA determined by real-time RT-PCR. Total RNA was extracted from day 1 (quiescent) and day 7 (myofibroblastic) HSC. Cd73 gene expression was determined by real-time RT-PCR and calculated relative to housekeeping Gapdh gene expression, by the 2−ΔΔCT method. [Cd73]/[Gapdh] mRNA ratios from day 1 and day 7 HSC preparation samples (n ≥ 3 for each group, 3 separate experiments) were calculated and compared. As can be seen, day 7 HSC express roughly 3× the mRNA of day 1 HSC. ***P < 0.001, HSC day 1 vs. HSC day 7. B: semiquantitative RT-PCR to determine relative expression of Cd73 mRNA in human cells. Total RNA from human immortalized LX-1, LX-2 myofibroblastic HSC, primary isolated human myofibroblastic HSC (FH11 and FH13), HepG2 hepatocellular carcinoma cells, and human brain tissue extract (positive control) was reverse transcribed, and the resulting cDNAs (lanes 2-7) were used as templates for semiquantitative PCR to detect expression of CD73 (171-bp product) and GAPDH (loading control, 228-bp product) genes. All samples are positive for expression of CD73 mRNA, except for the water control (lane 1), in which no template was added. C: upregulation of CD73 protein in myofibroblastic HSC determined by immunoblot. Total protein fractions were isolated from rat day 1 and day 7 HSC preparations and used for immunoblot to study the expression of ecto-5′-nucleotidase/CD73 and β-tubulin (loading control) proteins. A representative experiment shows a faint band (∼70 kDa) indicating low basal expression of ecto-5′-nucleotidase/CD73 that is seen in day 1 HSC, that is of greater intensity in day 7 HSC. Bottom: ratios of signal intensity [ecto-5′-nucleotidase/CD73]/[β-tubulin] protein immunoblots from day 1 and day 7 HSC (n ≥ 3 for each group) samples were calculated and compared, **P < 0.01, HSC day 1 vs. HSC day 7. D: upregulation of CD73 protein in myofibroblastic PF determined by immunoblot. Total protein fractions isolated from day 1 and day 7 PF preparations were analyzed as in C to study the expression of ecto-5′-nucleotidase/CD73 and β-actin (loading control) proteins. A representative experiment shows that expression of ecto-5′-nucleotidase/CD73 is absent in day 1 PF whereas a strong band indicative of protein expression is seen in day 7 PF. Bottom: ratios of signal intensity [ecto-5′-nucleotidase/CD73]/[β-actin] protein immunoblots from day 1 and day 7 PF (n ≥ 3 for each group) samples were calculated and compared, **P < 0.01, PF day 1 vs. PF day 7.
The cloned putative Cd73 gene promoter region is transcriptionally active.
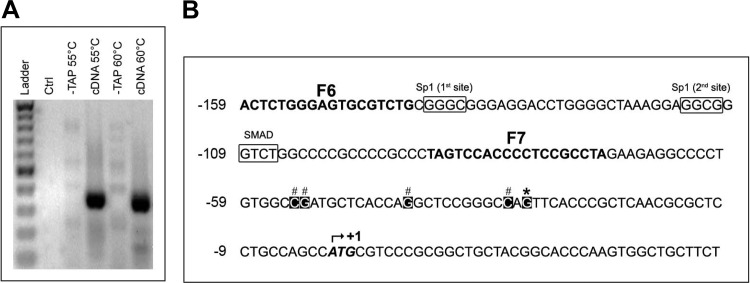
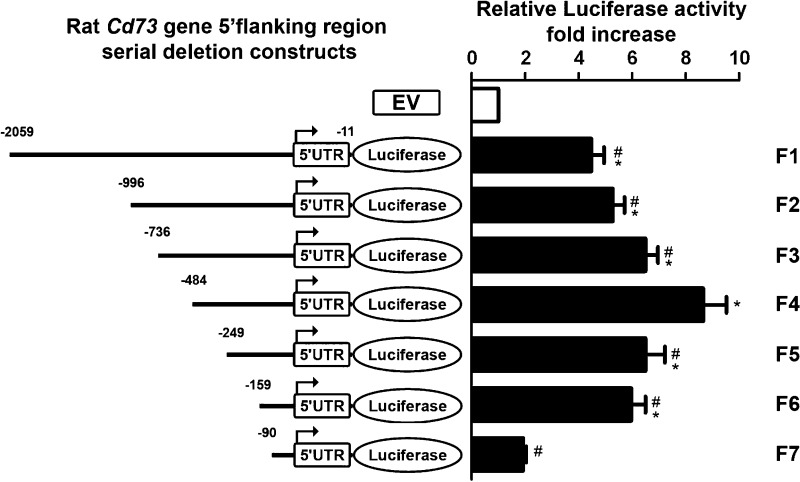
To determine the mechanism(s) responsible for the upregulated Cd73 transcription in myofibroblastic HSC, we located and cloned the putative rat Cd73 gene promoter. We first identified the transcription initiation site(s) by 5′RLM-RACE using total RNA purified from primary isolated rat hepatocytes, as described above in the experimental procedures. The first step of nested PCR amplifications was performed at 55 and 60°C annealing temperatures, using 5′RACE adaptor-ligated cDNAs as template. Then, the second step of nested PCR amplifications was performed at 60°C by using either 55 or 60°C reaction products as template, and each yielded a single band with a molecular weight of ∼300 bp (Fig. 5A). Each nested PCR product was used individually for cloning procedure followed by colony selection and insert analysis by automated sequencing. We identified a “hot spot” for the initiation of rat Cd73 gene transcription (found in 5 of 13 colonies; Fig. 5B, asterisk) located at −30 bp from ATG and alternate sites located between nucleotides −54 to −32 bp (8 of 13 colonies; Fig. 5B, pound signs). A 2,046 bp-long rat genomic DNA sequence corresponding to the putative Cd73 gene promoter was cloned into a luciferase reporter plasmid (F1) and used as template for the subsequent generation of serial truncation luciferase constructs (F2–F7). The transcriptional activity of each construct was studied by a dual-luciferase assay after transfection in immortalized LX-2 human HSC (Fig. 6). Of note, LX-2 HSC were used for these studies for several reasons: 1) LX-2 cells are well-established models of myofibroblastic HSC, 2) LX-2 cells express endogenously both CD73 mRNA and protein (Figs. 3 and 4), and 3) LX-2 cells are readily transfected with high efficiency. Of all truncation constructs generated, the F4 construct (−484 to −11 bp) displayed the highest activity that was significantly elevated compared with longer F1–F3 and shorter constructs F5–F7, suggesting the existence of both enhancer (F5–F7) and suppressor (F1–F3) elements in the cloned putative rat Cd73 promoter sequence. The luciferase activity of F7 construct was almost completely abrogated with similar levels to the ones measured for the control empty vector, designating the F6 (−159 to −11 bp) construct as the minimal promoter required for the rat Cd73 gene transcription in myofibroblastic HSC.
Fig. 5.
Cloning and characterization of the rat Cd73 promoter. A: Cd73 promoter cloning via RNA ligase-mediated rapid amplification of 5′ cDNA ends (5′RLM-RACE). Unmodified (−TAP) and modified (cDNA) cDNAs from isolated primary rat hepatocytes were used as templates for a first round of PCR amplification performed at 2 different annealing temperatures of 55 and 60°C, with 5′RLM-RACE outer and gsrCd73-R2 primers. Products of each reaction were used as templates for a nested PCR amplification performed at an annealing temperature of 60°C with 5′RLM-RACE inner and gsrCd73-R1 primers followed by electrophoretic analysis on 2% agarose gel. A negative control reaction (Ctrl), in which cDNA template was omitted, is included. B: computer-aided analysis of the putative rat Nt5e minimal promoter. The putative rat Cd73 minimal promoter nucleotide sequence was determined. The adenine of the translation start site (ATG) is indicated by an upward arrow at the nucleotide +1. Transcription start sites, deduced from the sequences of 13 individual clones originating from the outer PCR amplifications at 55 and 60°C annealing temperatures, are indicated by asterisks (*, major site) or pound signs (#, alternative sites), respectively. The prCd73-F6 to prCd73-F7 primer sequences (F6–F7) are indicated in bold type.
Fig. 6.
Expression of serial truncations of the Cd73 promoter-luciferase fusion vectors in LX2 cells. LX-2 cells were transfected with control pGL3 plasmid (empty vector, EV) or serial truncations (F2–F7) of the putative Cd73 promoter (F1 or nucleotides −2,056 to −11 bp from ATG), and luciferase activity was determined and expressed relatively to that of EV (open bar). Luciferase activity was maximal for construct F4, which was significantly higher than for longer constructs F1–F3, suggesting the presence of repressor elements in the region delimited by −735 to −484 bp from ATG of the putative promoter sequence. Moreover, F4 construct luciferase activity was noticeably decreased in shorter F5 and F6 constructs and essentially absent in the shortest construct F7, designating the F6 (−159 to −11 bp) construct as the minimal promoter required for rat Cd73 gene expression (n ≥ 3). *P < 0.001 vs. EV; #P < 0.001 vs. F4. UTR, untranslated region.
SP1 and SMAD transcription factors mediate Cd73 gene promoter activity.
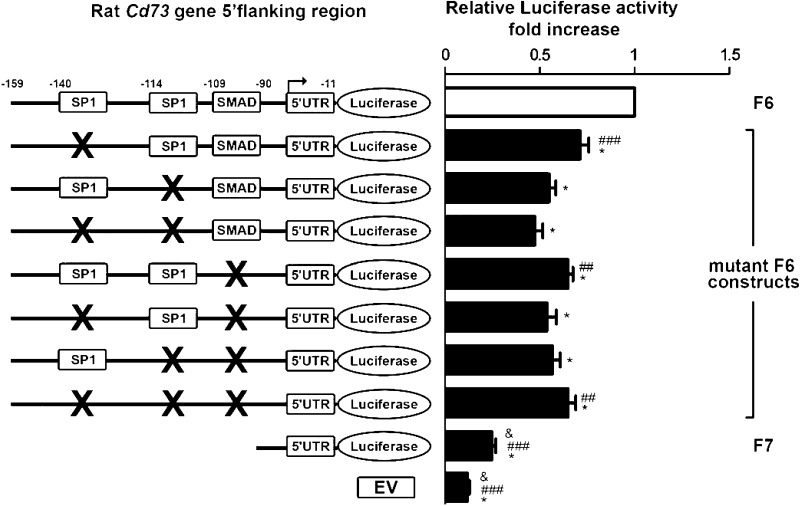
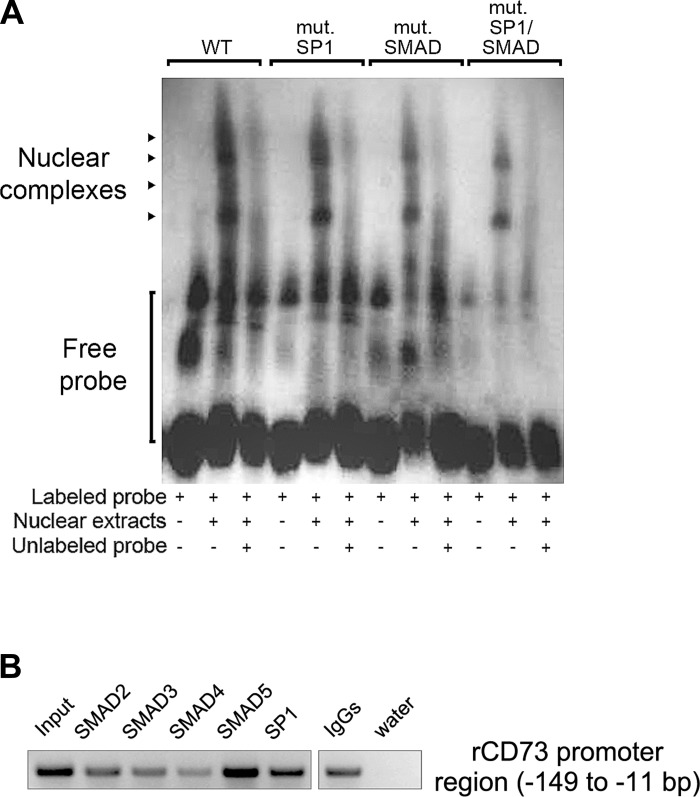
In silico analysis of the putative rat Cd73 minimal promoter F6 (−159 to −11 bp) construct revealed the presence of two SP1 (Specificity Protein 1; −140 to −137, first; −114 to −111, second) response elements and one SMAD [homologs of both Drosophila protein mothers against decapentaplegic (MAD) and the Caenorhabditis elegans protein SMA; −109 to −106] transcription factor binding site. Interestingly, SP1 and SMAD transcription factors are known downstream effectors of TGF-β signaling pathways during fibrosis (17). We evaluated the impact of individual or combined mutation(s) targeting SP1 and SMAD transcription factor binding sites on the F6 construct transcriptional activity. Wt and mutant F6 constructs were transfected in LX-2 cells, and luciferase activity was measured using a dual-luciferase assay (Fig. 7). We found that mutation of each SP1 and/or SMAD responsive element alone or in combination was sufficient to significantly decrease the F6 construct luciferase activity. These data indicate that the rat Cd73 promoter activity is mediated in a largely independent fashion by transcription factors acting at SP1 and SMAD sites in myofibroblastic HSC. To confirm our findings, we performed electrophoretic mobility shift assays of the SP1 and SMAD promoter elements using nuclear extracts from immortalized LX-2 cells (Fig. 8A). When nuclear extracts were incubated with the labeled wild-type (wt) probe, four bands corresponding to formed DNA-nuclear protein complexes were observed. Mutation of either the SMAD site or of both SP1 sites together, or mutation of the combination of all three, resulted in a significant reduction of formed DNA-nuclear protein complexes. Next, the binding specificity was confirmed by adding a 125-fold molar excess of unlabeled wt or mutated probe(s). These results demonstrate the presence of factors in LX-2 nuclear extracts that can bind SP1 and SMAD transcription factor binding sites contained in the minimal rat Cd73 promoter and regulate its transcriptional activity. Finally, ChIP assay analysis of nuclear lysates isolated from primary rat HSC showed that SP1 and (mainly) SMAD5 transcription factors can bind the putative rat Cd73 minimal promoter F6 (−159 to −11 bp) (Fig. 8B). Taken together, these data suggest that the activity of the putative minimal rat Cd73 promoter is regulated by SP1 and SMAD transcription factors in vitro and in vivo.
Fig. 7.
Use of site-directed mutagenesis to determine the functional significance of specific promoter elements in the minimal Cd73 promoter. LX-2 cells were transfected with control pGL3 plasmid (EV), wild-type (wt), or mutant F6 (nucleotides −159 to −11 from ATG) constructs, and luciferase activity was determined and expressed relatively to that of F6 (open bar). Luciferase activity is significantly decreased when mutations targeting each SP1 (−140 to −137, first; −114 to −111, second) or SMAD (−109 to −106) sites are introduced. Interestingly, mutation of SP1 (−114 to −111) site in combination with either the other SP1 (140 to −137) or single SMAD sites is sufficient to reduce, by almost 50%, F6 construct luciferase activity. Moreover, the single SMAD mutation by itself does not decrease luciferase activity to the extent of SP1 (−114 to −111) site (n ≥ 3). *P < 0.001 vs. F6; ##P < 0.01 vs. mutant SP1 (−139 and −113); ###P < 0.001 vs. mutant SP1 (−139 and −113); &P < 0.001 vs. SMAD.
Fig. 8.
EMSA experiment and chromatin immunoprecipitation (ChIP) assay to verify direct binding of SP1 and SMAD elements to the minimal Cd73 promoter. A: EMSA. Nuclear extracts obtained from LX-2 cells were subjected to EMSA using a digoxigenin-labeled probe corresponding to the wt minimal rat Cd73 promoter containing the two SP1 (nucleotides −139 to −136 and −113 to −110) and the SMAD (−108 to −105) motifs. In addition, wt probes containing mutation(s) targeting the two SP1 and single SMAD sites alone or in combination were used. Competitive analysis was performed in the presence of a 125-fold molar excess of unlabeled competitor, as indicated. Formation of DNA-protein complexes are indicated by arrowheads. A representative image of 3 independent experiments is shown. B: ChIP assay. Nuclear lysates isolated from primary rat HSC were subjected to ChIP using antibodies against SP1 and various SMAD transcription factors, and PCR analysis with prCd73 F6 and prCd73 R1 primer set. Strong 170-bp PCR products are observed in wells containing DNA material immunoprecipitated with antibodies directed against SP1 and (mainly) SMAD5 transcription factors. The “input” and “normal IgGs” represent internal and negative controls, respectively. A representative image of 2 independent experiments is shown.
DISCUSSION
In the present work, we tested the hypothesis that the ecto-5′nucleotidase/CD73, the rate-limiting enzyme for production of adenosine, is upregulated by HSC and PF upon myofibroblastic differentiation. We found that this indeed occurs and that this upregulation is mediated at the transcriptional level by specific transcription factors that are biologically relevant to liver myofibroblast function. CD73 is an attractive target for liver fibrosis research. Prior studies demonstrated that experimental liver fibrosis in mice increases hepatic Cd73 mRNA levels (35, 36). Cd73-deficient mice are resistant to experimental liver fibrosis, suggesting that CD73 plays a critical role in the modulation of liver fibrogenesis (35, 36). These findings are of great potential importance; however, they are limited by the lack of examination of the cell-specific expression of CD73. Specifically, it could be surmised that the reduction in fibrosis was due to differences in function of inflammatory or antigen-producing cells, changes in hepatocellular death, or some other mechanism(s). The present study suggests that the role of CD73 in liver fibrosis is mediated, at least in part, at the level of liver myofibroblasts. Although this is not the focus of the present article, it is noteworthy to mention that liver parenchymal cells (or hepatocytes) also express CD73 protein (as observed in Fig. 1) and could contribute, to a certain extent, to CD73-related biological pathways.
There are good data supporting the potential pathophysiological relevance of CD73 upregulation in liver myofibroblasts. The functional importance of ecto-5′-nucleotidase/CD73 is due to its enzymatic activity; CD73 is the rate-limiting step in generation of extracellular adenosine (27). Adenosine is not secreted into the extracellular space in a regulated fashion; however, ATP does undergo such secretion (7). Thus CD73 controls extracellular levels of adenosine, a potent modulator of fibrosis and inflammation in the liver (39). In liver injury, alterations in cytokine release, fatty acid metabolism, acute toxic liver injury, ischemia-reperfusion damage, and toxin-induced liver fibrosis are modulated by adenosine. Adenosine effects cell changes via signaling through its four G protein-coupled receptors: A1, A2a, A2b, and A3 (5, 30, 33–35, 43). Of particular relevance to the present study, adenosine regulates a number of fibrogenic functions in myofibroblastic HSC. HSC express A2a, A2b, and A3 receptors, and adenosine regulates chemotaxis, contraction, matrix metalloproteinase production, and collagen production in HSC (24, 32, 35, 41). Thus expression of CD73 likely represents the mechanism by which adenosinergic signals are generated to regulate downstream fibrogenic effects in HSC. In contrast, a recent study showed that Cd73 gene knockdown in immortalized mouse HSC enhanced collagen production, migratory capacity, and mRNA expression of adenosine-producing tissue nonspecific alkaline phosphatase (TNAP) ectoenzyme (1), which may argue against this hypothesis. Hence the in vivo role of CD73 expressed by liver myofibroblasts remains unknown. In the present study, we found that ecto-5′-nucleotidase/CD73 protein expression is increased in both myofibroblastic HSC (from low basal levels) and activated portal fibroblasts (from negligible levels) upon culture activation, in agreement with its observed distribution within the scar-laden areas in BDL and CCl4-intoxicated cirrhotic rat livers. Interestingly, a recent study describing a large-scale proteome analysis of primary activated rat HSC reported Cd73 among the genes whose expression is highly increased upon cell activation (28). We also found that primary human myofibroblastic HSC and LX-2 cells express CD73. This suggests that CD73 upregulation could represent a common mechanistic pathway triggered in HSC and PF undergoing myofibroblastic differentiation, across rat and human species. This is in stark contrast to the observed changes in the ecto-ATPase NTPDase2, which is upregulated in myofibroblastic differentiation of HSC but downregulated in myofibroblastic differentiation of PF (12, 13, 15, 44). Thus direct study of promoter elements of both of these genes is of biological relevance in liver fibrosis research.
Here we identified and cloned the putative rat Cd73 promoter and demonstrated that it contains functional response elements for both SP1 and SMAD transcription factors. Interestingly, single mutation of these sites reduced promoter activity, suggesting that each element is functional. Moreover, although combined mutations of each SP1 site with the SMAD substantially reduced promoter activity, triple mutation did not reduce it further, suggesting that each SP1 and SMAD response element is sufficient to upregulate Cd73 gene expression. We further confirmed by EMSA the contribution of SP1 and SMAD signaling to the rat Cd73 promoter activity and identified by ChIP assay SP1 and SMAD5 (among others), as transcription factors that are recruited to and can interact with the putative rat Cd73 promoter region. Both SP1 and SMAD transcription factors act as downstream effector molecules or as coactivators mediating TGF-β-induced gene activation during fibrosis (17). TGF-β is one of the best-characterized modulators in the pathogenesis of liver fibrosis (23). Its biological action controls both inflammation and fibrogenesis within the liver. TGF-β modulates transcriptional activity of critical liver myofibroblast target genes, including procollagen α1, matrix metalloproteinase-11, cyclin D1, and α-SMA (17, 46). Although we have not tested this here, the rat Cd73 gene expression may also by regulated by TGF-β signaling. Interestingly, the mouse Cd73 gene expression is induced in splenic T cells, bone marrow-derived dendritic cells, and peritoneal macrophages, upon in vitro stimulation with TGF-β (38).
In conclusion, we have shown that Cd73 gene expression increases in both HSC and PF during myofibroblastic differentiation. Since CD73 is the rate-limiting enzyme in generation of adenosinergic signaling, and adenosinergic signals are strongly profibrogenic, we propose that Cd73 gene upregulation provides a novel and biologically relevant mechanism by which liver myofibroblasts may transduce profibrogenic signals. Future studies will be performed to directly test this hypothesis in vitro and in vivo.
GRANTS
This work was supported by the National Institutes of Health NIH/NIDDK R01 DK076735 and R01 DK070849 grants to J. A. Dranoff, NIH DK6047402, DK071745, and R56 DK092128 grants to M. B. Bansal, and the American Liver Foundation Roger L. Jenkins, M.D. Postdoctoral Research Fellowship Award to M. Fausther.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
The authors acknowledge technical assistance from Feng Hong (Mount Sinai School of Medicine) with isolation of primary human HSC and from Mary Kearney (Yale School of Medicine) with culture of primary human HSC.
REFERENCES
- 1. Andrade CM, Lopez PL, Noronha BT, Wink MR, Borojevic R, Margis R, Lenz G, Battastini AM, Guma FC. Ecto-5′-nucleotidase/CD73 knockdown increases cell migration and mRNA level of collagen I in a hepatic stellate cell line. Cell Tissue Res 344: 279–286, 2011 [DOI] [PubMed] [Google Scholar]
- 2. Beldi G, Enjyoji K, Wu Y, Miller L, Banz Y, Sun X, Robson SC. The role of purinergic signaling in the liver and in transplantation: effects of extracellular nucleotides on hepatic graft vascular injury, rejection and metabolism. Front Biosci 13: 2588–2603, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benitez-Rajal J, Lorite MJ, Burt AD, Day CP, Thompson MG. Phospholipase D and extracellular signal-regulated kinase in hepatic stellate cells: effects of platelet-derived growth factor and extracellular nucleotides. Am J Physiol Gastrointest Liver Physiol 291: G977–G986, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci 64: 1471–1483, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan ES, Montesinos MC, Fernandez P, Desai A, Delano DL, Yee H, Reiss AB, Pillinger MH, Chen JF, Schwarzschild MA, Friedman SL, Cronstein BN. Adenosine A(2A) receptors play a role in the pathogenesis of hepatic cirrhosis. Br J Pharmacol 148: 1144–1155, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Che J, Chan ES, Cronstein BN. Adenosine A2A receptor occupancy stimulates collagen expression by hepatic stellate cells via pathways involving protein kinase A, Src, and extracellular signal-regulated kinases 1/2 signaling cascade or p38 mitogen-activated protein kinase signaling pathway. Mol Pharmacol 72: 1626–1636, 2007 [DOI] [PubMed] [Google Scholar]
- 7. Corriden R, Insel PA. Basal release of ATP: an autocrine-paracrine mechanism for cell regulation. Sci Signal 3: re1, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixon CJ, White PJ, Hall JF, Kingston S, Boarder MR. Regulation of human hepatocytes by P2Y receptors: control of glycogen phosphorylase, Ca2+, and mitogen-activated protein kinases. J Pharmacol Exp Ther 313: 1305–1313, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Dixon CJ, Woods NM, Webb TE, Green AK. Evidence that rat hepatocytes co-express functional P2Y1 and P2Y2 receptors. Br J Pharmacol 129: 764–770, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doctor RB, Matzakos T, McWilliams R, Johnson S, Feranchak AP, Fitz JG. Purinergic regulation of cholangiocyte secretion: identification of a novel role for P2X receptors. Am J Physiol Gastrointest Liver Physiol 288: G779–G786, 2005 [DOI] [PubMed] [Google Scholar]
- 11. Dolovcak S, Waldrop SL, Fitz JG, Kilic G. 5-Nitro-2-(3-phenylpropylamino)benzoic acid (NPPB) stimulates cellular ATP release through exocytosis of ATP-enriched vesicles. J Biol Chem 284: 33894–33903, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dranoff JA, Kruglov EA, Robson SC, Braun N, Zimmermann H, Sévigny J. The ecto-nucleoside triphosphate diphosphohydrolase NTPDase2/CD39L1 is expressed in a novel functional compartment within the liver. Hepatology 36: 1135–1144, 2002 [DOI] [PubMed] [Google Scholar]
- 13. Dranoff JA, Kruglov EA, Toure J, Braun N, Zimmermann H, Jain D, Knowles AF, Sévigny J. Ectonucleotidase NTPDase2 is selectively down-regulated in biliary cirrhosis. J Investig Med 52: 475–482, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Dranoff JA, Masyuk AI, Kruglov EA, LaRusso NF, Nathanson MH. Polarized expression and function of P2Y ATP receptors in rat bile duct epithelia. Am J Physiol Gastrointest Liver Physiol 281: G1059–G1067, 2001 [DOI] [PubMed] [Google Scholar]
- 15. Dranoff JA, Ogawa M, Kruglov EA, Gaca MD, Sévigny J, Robson SC, Wells RG. Expression of P2Y nucleotide receptors and ectonucleotidases in quiescent and activated rat hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol 287: G417–G424, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dranoff JA, Wells RG. Portal fibroblasts: underappreciated mediators of biliary fibrosis. Hepatology 51: 1438–1444, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellenrieder V. TGFbeta regulated gene expression by Smads and Sp1/KLF-like transcription factors in cancer. Anticancer Res 28: 1531–1539, 2008 [PubMed] [Google Scholar]
- 18. Fabre AC, Malaval C, Ben Addi A, Verdier C, Pons V, Serhan N, Lichtenstein L, Combes G, Huby T, Briand F, Collet X, Nijstad N, Tietge UJ, Robaye B, Perret B, Boeynaems JM, Martinez LO. P2Y13 receptor is critical for reverse cholesterol transport. Hepatology 52: 1477–1483, 2010 [DOI] [PubMed] [Google Scholar]
- 19. Fabre AC, Vantourout P, Champagne E, Terce F, Rolland C, Perret B, Collet X, Barbaras R, Martinez LO. Cell surface adenylate kinase activity regulates the F(1)-ATPase/P2Y (13)-mediated HDL endocytosis pathway on human hepatocytes. Cell Mol Life Sci 63: 2829–2837, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fausther M, Lecka J, Soliman E, Kauffenstein G, Pelletier J, Sheung N, Dranoff JA, Sevigny J. Co-expression of ecto-5′-nucleotidase/CD73 with specific NTPDases differentially regulates adenosine formation in the rat liver. Am J Physiol Gastrointest Liver Physiol 302: G447–G459, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedman SL, Rockey DC, McGuire RF, Maher JJ, Boyles JK, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology 15: 234–243, 1992 [DOI] [PubMed] [Google Scholar]
- 22. Gonzales E, Prigent S, Abou-Lovergne A, Boucherie S, Tordjmann T, Jacquemin E, Combettes L. Rat hepatocytes express functional P2X receptors. FEBS Lett 581: 3260–3266, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Gressner AM, Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J Cell Mol Med 10: 76–99, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hashmi AZ, Hakim W, Kruglov EA, Watanabe A, Watkins W, Dranoff JA, Mehal WZ. Adenosine inhibits cytosolic calcium signals and chemotaxis in hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol 292: G395–G401, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol 6: 425–456, 2011 [DOI] [PubMed] [Google Scholar]
- 26. Hong F, Tuyama A, Lee TF, Loke J, Agarwal R, Cheng X, Garg A, Fiel MI, Schwartz M, Walewski J, Branch A, Schecter AD, Bansal MB. Hepatic stellate cells express functional CXCR4: role in stromal cell-derived factor-1alpha-mediated stellate cell activation. Hepatology 49: 2055–2067, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol Ther 107: 1–30, 2005 [DOI] [PubMed] [Google Scholar]
- 28. Ji J, Yu F, Ji Q, Li Z, Wang K, Zhang J, Lu J, Chen L, EQ, Zeng Y, Ji Y. Comparative proteomic analysis of rat hepatic stellate cell activation: a comprehensive view and suppressed immune response. Hepatology 56: 332–349, 2012 [DOI] [PubMed] [Google Scholar]
- 29. Kruglov EA, Jain D, Dranoff JA. Isolation of primary rat liver fibroblasts. J Investig Med 50: 179–184, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Lee HT, Kim M, Joo JD, Gallos G, Chen JF, Emala CW. A3 adenosine receptor activation decreases mortality and renal and hepatic injury in murine septic peritonitis. Am J Physiol Regul Integr Comp Physiol 291: R959–R969, 2006 [DOI] [PubMed] [Google Scholar]
- 31. Minagawa N, Nagata J, Shibao K, Masyuk AI, Gomes DA, Rodrigues MA, Lesage G, Akiba Y, Kaunitz JD, Ehrlich BE, Larusso NF, Nathanson MH. Cyclic AMP regulates bicarbonate secretion in cholangiocytes through release of ATP into bile. Gastroenterology 133: 1592–1602, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohamadnejad M, Sohail MA, Watanabe A, Krause DS, Swenson ES, Mehal WZ. Adenosine inhibits chemotaxis and induces hepatocyte-specific genes in bone marrow mesenchymal stem cells. Hepatology 51: 963–973, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Odashima M, Otaka M, Jin M, Komatsu K, Wada I, Matsuhashi T, Horikawa Y, Hatakeyama N, Oyake J, Ohba R, Linden J, Watanabe S. Selective A2A adenosine agonist ATL-146e attenuates acute lethal liver injury in mice. J Gastroenterol 40: 526–529, 2005 [DOI] [PubMed] [Google Scholar]
- 34. Ohta A, Sitkovsky M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature 414: 916–920, 2001 [DOI] [PubMed] [Google Scholar]
- 35. Peng Z, Borea PA, Varani K, Wilder T, Yee H, Chiriboga L, Blackburn MR, Azzena G, Resta G, Cronstein BN. Adenosine signaling contributes to ethanol-induced fatty liver in mice. J Clin Invest 119: 582–594, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng Z, Fernandez P, Wilder T, Yee H, Chiriboga L, Chan ES, Cronstein BN. Ecto-5′-nucleotidase (CD73)-mediated extracellular adenosine production plays a critical role in hepatic fibrosis. FASEB J 22: 2263–2272, 2008 [DOI] [PubMed] [Google Scholar]
- 37. Popov Y, Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology 50: 1294–1306, 2009 [DOI] [PubMed] [Google Scholar]
- 38. Regateiro FS, Howie D, Nolan KF, Agorogiannis EI, Greaves DR, Cobbold SP, Waldmann H. Generation of anti-inflammatory adenosine by leukocytes is regulated by TGF-beta. Eur J Immunol 41: 2955–2965, 2011 [DOI] [PubMed] [Google Scholar]
- 39. Robson SC, Schuppan D. Adenosine: tipping the balance towards hepatic steatosis and fibrosis. J Hepatol 52: 941–943, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008 [DOI] [PubMed] [Google Scholar]
- 41. Sohail MA, Hashmi AZ, Hakim W, Watanabe A, Zipprich A, Groszmann RJ, Dranoff JA, Torok NJ, Mehal WZ. Adenosine induces loss of actin stress fibers and inhibits contraction in hepatic stellate cells via Rho inhibition. Hepatology 49: 185–194, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut 54: 142–151, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang P, Han Z, Chen P, Zhu L, Wang S, Hua Z, Zhang J. A contradictory role of A1 adenosine receptor in carbon tetrachloride- and bile duct ligation-induced liver fibrosis in mice. J Pharmacol Exp Ther 332: 747–754, 2010 [DOI] [PubMed] [Google Scholar]
- 44. Yu J, Lavoie EG, Sheung N, Tremblay JJ, Sévigny J, Dranoff JA. IL-6 downregulates transcription of NTPDase2 via specific promoter elements. Am J Physiol Gastrointest Liver Physiol 294: G748–G756, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu J, Sheung N, Soliman EM, Spirli C, Dranoff JA. Transcriptional regulation of IL-6 in bile duct epithelia by extracellular ATP. Am J Physiol Gastrointest Liver Physiol 296: G563–G571, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang W, Ou J, Inagaki Y, Greenwel P, Ramirez F. Synergistic cooperation between Sp1 and Smad3/Smad4 mediates transforming growth factor beta1 stimulation of alpha 2(I)-collagen (COL1A2) transcription. J Biol Chem 275: 39237–39245, 2000 [DOI] [PubMed] [Google Scholar]