Abstract
The human double minute (HDM)-2 E3 ubiquitin ligase plays a key role in p53 turnover, and has been validated pre-clinically as a target in multiple myeloma (MM) and mantle cell lymphoma (MCL). HDM-2 inhibitors are entering clinical trials, and we therefore sought to understand potential mechanisms of resistance in lymphoid models. Wild-type p53 H929 MM and Granta-519 MCL cells resistant to MI-63 or Nutlin were generated by exposing them to increasing drug concentrations. MI-63-resistant H929 and Granta-519 cells were resistant to Nutlin, while Nutlin-resistant cells displayed cross-resistance to MI-63. These cells also showed cross-resistance to bortezomib, doxorubicin, cisplatin, and melphalan, but remained sensitive to the small molecule inhibitor RITA. HDM-2 inhibitor-resistant cells harbored increased p53 levels, but neither genotoxic nor non-genotoxic approaches to activate p53 induced HDM-2 or p21. Resequencing revealed wild-type HDM-2, but mutations were found in the p53 DNA binding and dimerization domains. In resistant cells, RITA induced a G2/M arrest, up-regulation of p53 targets HDM-2, PUMA, and NOXA, and PARP cleavage. Combination regimens with RITA and MI-63 resulted in enhanced cell death compared to RITA alone. These findings support the possibility that p53 mutation could be a primary mechanism of acquired resistance to HDM-2 inhibitors in MCL and MM. Furthermore, they suggest that simultaneous restoration of p53 function and HDM-2 inhibition is a rational strategy for clinical translation.
Keywords: HDM-2, p53, Multiple Myeloma, Mantle Cell Lymphoma, MI-63, Nutlin, RITA
Introduction
The ubiquitin-proteasome pathway is a key regulator of transcription factors and cellular proteins (1). Through its role in the poly-ubiquitination of target proteins and their subsequent proteasome-mediated degradation, this system influences angiogenesis, cell-cycle progression, DNA repair, and apoptosis (2). Tumors exploit key segments of this pathway to enhance carcinogenesis and circumvent cellular surveillance strategies aimed at eliminating malignant cells. One example is over-expression of the E3 ligase Human Double Minute (HDM)-2 in 10% of human malignancies (3), depleting the tumor suppressor p53. This leads to loss of cell cycle check points and uncontrolled proliferation. A single nucleotide polymorphism within the HDM-2 promoter has also been reported to enhance HDM-2 expression and tumor formation (4).
HDM-2 binds to the N-terminus of p53 to inhibit its transcriptional activity (5, 6), and ubiquitinates it for subsequent degradation (7, 8). This can be exploited therapeutically by agents such as Nutlin and MI-63 (Fig.1A), which bind the HDM-2 p53 binding site and prevent p53 ubiquitination (9, 10). Both have shown pre-clinical activity against mantle cell lymphoma (MCL) and multiple myeloma (MM), where they stabilize p53 and enhance its pro-apoptotic effects (11–16). RITA (reactivation of p53 and induction of tumor cell apoptosis)(17)(Fig.1B), disrupts binding of p53 and HDM-2, but inhibits the interaction by binding to the HDM-2 binding site in p53 (18).
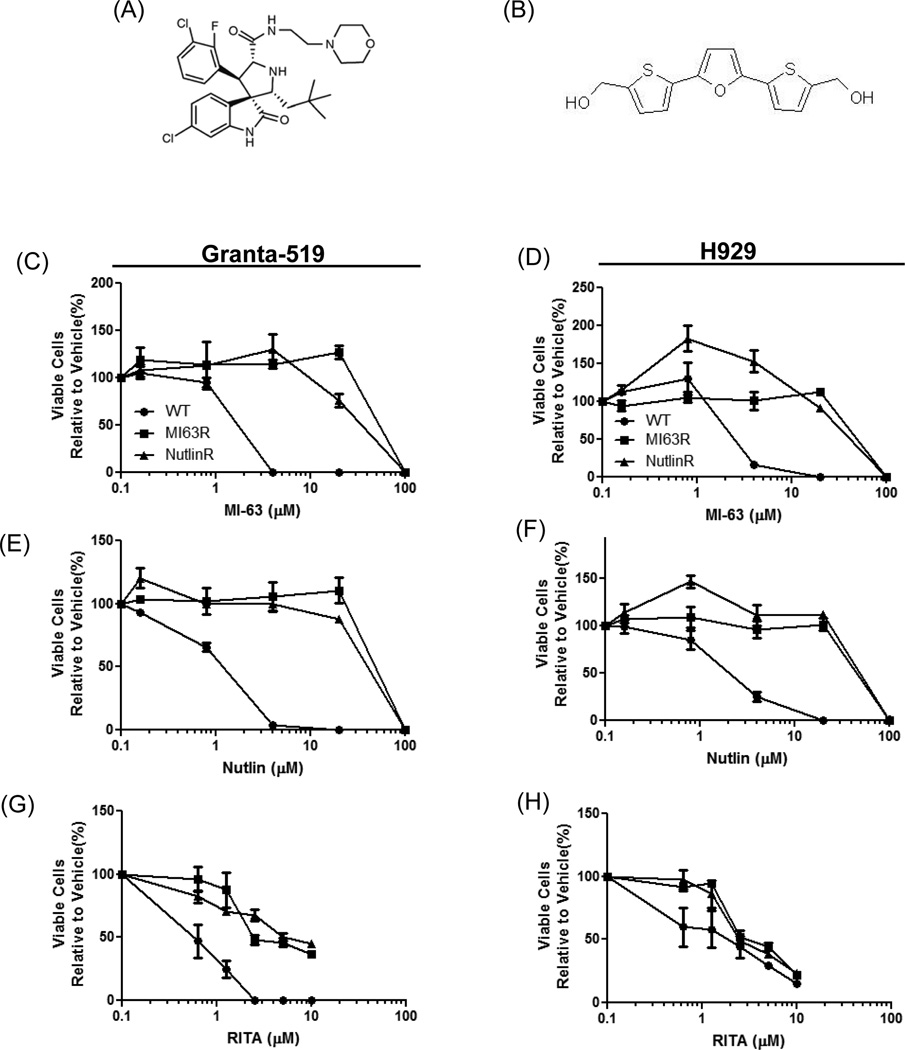
Figure 1. MI-63- and Nutlin-resistant cells show cross-resistance to HDM-2 inhibitors.
(A) Chemical structure MI-63. (B) Chemical structure RITA. Granta-519 (C, E, G) and H929 (D, F, H) wild type (WT) cells, along with their MI-63-resistant (MI63R) and Nutlin-resistant (NutlinR) counterparts, were treated for 72-hours with MI-63, Nutlin or RITA. Cell viability was determined using the WST-1 reagent, and results were expressed as the percentage viability relative to vehicle controls, which were arbitrarily set at 100%. IC50 values for MI-63, Nutlin and RITA-exposed conditions were determined, and fold changes in the IC50 were calculated relative to WT cells (Supplementary Table 1).
Targeting the proteasome/E3 ligase system has recently become a focus in anti-cancer research with novel HDM-2 inhibitors (19). The advantage of inhibiting HDM-2 could be that specific inhibition of a single E3 ligase may reduce effects, such as neuropathy, which are seen in patients treated with bortezomib. Nutlin and MI-63 have been extensively studied in lymphoid malignancies (20, 21), and at physiologic concentrations their effects are due to disruption of the p53/HDM-2 interaction (22). One disadvantage of specific small molecule inhibitors is that resistance may be more likely to develop compared to genotoxic agents, since the latter may have a broader target range (23). Studies of mechanisms of innate or primary resistance to Nutlin and MI-63 have attributed this phenotype to p53 inactivation through HDM-X overexpression (24). Acquired, or secondary Nutlin resistance has been investigated in models of osteosarcoma, neuroblastoma, rhabdomyosarcoma and melanoma (25, 26). These studies identified drug-resistant isolates, in some of which wild-type p53 was retained, while others were characterized by p53 mutations that impaired its DNA binding and transactivation. However, no data have elucidated how lymphoid malignancies acquire HDM-2 inhibitor resistance. This is especially important since these tumors show a lower rate of p53 inactivation than solid tumors (27), suggesting they may be better targets for this drug class.
We therefore sought to examine the mechanisms responsible for MI-63 and Nutlin resistance by developing models that were insensitive to these pro-apoptotic drugs. Cell lines resistant to either agent demonstrated cross-resistance to the other, and were less sensitive to bortezomib, doxorubicin, cisplatin and melphalan, but not to RITA. Analysis of these cells showed enhanced p53 expression, while genomic sequencing revealed wild-type HDM-2 but mutated p53. Exposure of resistant cells to RITA induced cell cycle arrest and p53 transcriptional targets, supporting a restoration of p53 activity. Combination studies using RITA and MI-63 showed resensitization of resistant cells to MI-63. Taken together, our data support the hypothesis that translation of HDM-2 inhibitors to the clinic for chronic treatment of patients with MCL and MM could result in selection of resistant clones with p53 point mutations. Importantly, it may be possible to circumvent resistance by using molecules that inhibit the p53/HDM-2 interaction and refold mutant p53 into a wild-type conformation.
Materials and Methods
Reagents
HDM-2 inhibitors MI-63 and MI-219 were provided by Ascenta Therapeutics (Malvern, PA). Nutlin, doxorubicin and melphalan were from Sigma-Aldrich (St. Louis, MO), RITA was from Cayman Chemical Company (Ann Arbor, MI), bortezomib from Selleck Chemicals, LLC (Houston, TX), and cisplatin from the M.D. Anderson Cancer Center Pharmacy (Houston, TX).
Cell culture and generation of MI-63- and Nutlin-resistant cells
Wild-type p53 (wtp53) Granta-519 MCL and NCI-H929 MM cell lines were from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and American Type Culture Collection (ATCC)(Manassas, VA). Cell lines were validated in the M.D. Anderson Cell Line Validation Core Facility by short tandem repeat (STR) DNA fingerprinting (May 2012) using the AmpFℓSTR Identifiler kit (Applied Biosystems; Carlsbad, CA). The STR profiles were compared to known ATCC fingerprints, the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (Nucleic Acids Research 37:D925-D932 PMCID: PMC2686526) and the M.D. Anderson fingerprint database. To generate resistance to MI-63 and Nutlin, cells were exposed to a concentration of each drug that induced a 25% proliferation inhibition for one-week, followed by a drug holiday. The drug dose was then increased every two weeks until lines were resistant to 10µM of MI-63 or Nutlin. Cells were then maintained in drug-free media and the resistant phenotype was confirmed monthly. Experiments were performed in cells that had been drug-free for at least two weeks.
Semi-quantitative and real-time PCR
RNA was isolated using the RNeasy Plus kit (Qiagen; Valencia, CA), and cDNA was synthesized using Superscript II (Invitrogen; Carlsbad, CA). Real-time PCR was performed on a StepOnePlus PCR analyzer (Applied Biosystems; Carlsbad, CA) using inventoried real time Taqman-FAM and GAPDH-VIC probes. Relative transcript expression (RQ) was determined using vehicle treated cells as a calibrator with the ΔΔCT method.
p53 and HDM-2 resequencing
Genomic DNA was extracted using the Genomic DNA extraction kit (Qiagen). Exons, introns, and regulatory regions of p53 and HDM-2 were sequenced by the M.D. Anderson DNA Analysis Core Facility. Exons were amplified using custom PCR primers, and Sanger sequencing was performed on a 3730xl DNA Analyzer using BigDye™ Terminator v3 (Applied Biosystems). Mutations analysis was performed using SeqScape® Software v2.5 (Applied Biosystems).
Immunoblotting
Protein expression was measured by immunoblot analysis as previously described (28). Antibodies against p53 were from Santa Cruz Biotechnologies (Santa Cruz, CA), anti-poly-(ADP-ribose) polymerase (PARP) was from Cell Signaling Technology (Danvers, MA), anti-p21, anti-PUMA, anti-NOXA, and anti-HDM-2 were from EMD Biosciences (Gibbstown, NJ), and anti-β-actin was from Sigma-Aldrich.
Cell cycle and cell death analysis
Cells were treated with drugs for 48-hours, fixed in 70% ethanol, and stained with propidium iodide (Sigma-Aldrich). Cell cycle data were obtained on a BD Facs Canto II flow cytometer (Becton-Dickson; Franklin Lakes, NJ) using FlowJo v.7.6.1 (Tree Star, Inc.; Ashland, OR). Cell death was measured by staining with Annexin-V Pacific Blue and TO-PRO-3 (Invitrogen).
Cell proliferation assay
The WST-1 reagent (Roche Diagnostics; Indianapolis, IN) was used to determine the effects of chemotherapeutics (29). Median inhibitory concentration (IC50) determinations were performed in 96-well plates in triplicate in 3 separate experiments performed on different days. Dose response curves were plotted using GraphPad Prism 5 (GraphPad; La Jolla, CA) showing standard error of the mean with a log scale and IC50 values calculated using the nonlinear regression competitive binding log IC50 algorithm.
Results
Generation of HDM-2 inhibitor resistant cells
To define the mechanisms through which MCL and MM could acquire HDM-2 inhibitor resistance, we exposed Granta-519 and H929 cells to increasing MI-63 and Nutlin concentrations. Median inhibitory concentrations (IC50) for wild-type Granta-519 (Granta.WT) were 2.03µM for MI-63 and 0.96µM for Nutlin (Supplemental Table 1), and for NCI-H929 (H929.WT) the IC50 was 3.30µM and 3.54µM, respectively (Supplementary Table 1). Exposure of Granta-519 cells resistant to MI-63 (Granta.MI63R) or Nutlin (Granta.NutlinR) to MI-63 or Nutlin showed that both cell lines were insensitive to MI-63 and Nutlin (Fig.1C, 1E). Granta.MI63R showed IC50’s of 60µM for MI-63 and 59.75µM for Nutlin, whereas Granta.NutlinR had an IC50 of 47.3µM for MI-63 and 44.75µM for Nutlin (Supplementary Table 1). A similar result was seen in the H929 resistant cells (Fig.1D, 1F) where H929.MI63R had an IC50 of 59.75µM to MI-63 and 56.65µM to Nutlin, whereas H929.NutlinR had an IC50 of 55.80µM for MI-63 and 47.60µM to Nutlin (Supplementary Table 1). Interestingly, enhancement of growth, particularly in H929.MI63R and H929.NutlinR, was noted in response to MI-63 or Nutlin (Fig.1D, 1F). Finally, we examined whether MI-63 and Nutlin resistance conveyed resistance to the small molecule RITA, which inhibits the p53/HDM-2 interaction by binding to p53 (18). Titration of RITA into Granta.MI63R and Granta.NutlinR cells revealed that they remained sensitive to RITA, though with a slightly increased IC50 of 1.72 and 0.84µM, respectively, compared to 0.272 µM in Granta.WT cells (Fig.1G; Supplementary Table 1). Lastly, H929.MI63R and H929.NutlinR showed some resistance to RITA, although this was not statistically significant (p>0.05), with IC50 values of 2.14µM and 1.99µM compared to 0.43µM for H929.WT cells (Fig.1H; Supplementary Table 1). These data indicate that our models were heavily cross-resistant to HDM-2 inhibitors that bind the p53 binding site on HDM-2, but not as resistant to HDM-2 inhibitors targeting the HDM-2 binding site on p53.
HDM-2 inhibitor-resistant cells demonstrate resistance against other therapeutics
We next considered whether MI-63 or Nutlin resistance could influence sensitivity to other anti-MCL (30) or MM (31) agents. Granta.MI63R and Granta.NutlinR cells displayed a slightly decreased sensitivity to doxorubicin, with an increase in the IC50 to 15.54 and 15.73nM, respectively, compared to 8.01nM in Granta.WT cells (Fig.2A; Supplementary Table 2). H929.MI63R and H929.NutlinR also showed a weak decrease in doxorubicin sensitivity (Fig.2B; Supplementary Table 2). Since the proteasome inhibitor bortezomib is effective against MCL and MM, Granta.MI63R and Granta.NutlinR cells were exposed to this agent, and showed a slight increase in resistance (Fig.2C; Supplementary Table 2). Similar results were found in H929.MI63R and H929.NutlinR cells (Fig.2D; Supplementary Table 2).
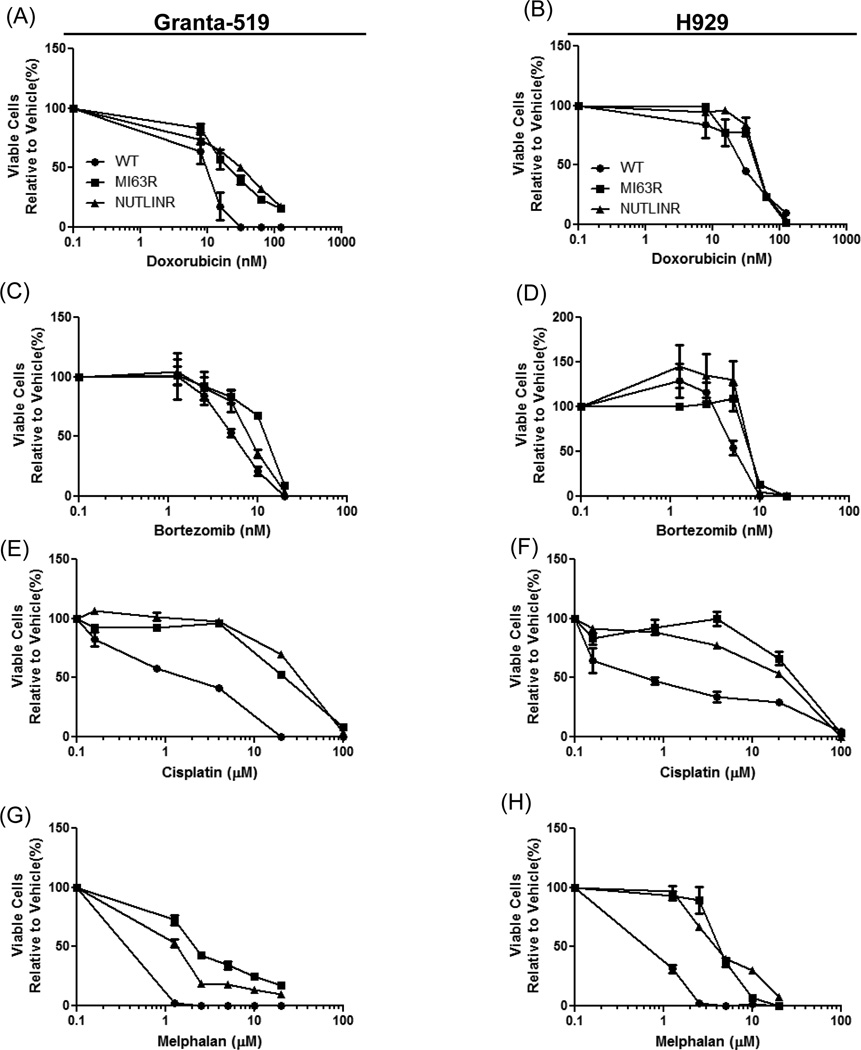
Figure 2. HDM-2 inhibitor-resistant cells show cross-resistance to some chemotherapeutics.
Granta-519 (A, C, E, G) and H929 (B, D, F, H) WT, MI63R, and NutlinR cells were treated with doxorubicin, bortezomib, cisplatin or melphalan for 72-hours. Cell viability was determined using the WST-1 reagent. IC50 values were determined for each cell line, and fold changes were calculated relative to the WT cells (Supplementary Table 2).
To further investigate whether this HDM-2 inhibitor resistance conveyed resistance to other DNA damaging agents, we examined the activity of melphalan and cisplatin. Granta.MI63R and Granta.NutlinR cells exposed to cisplatin displayed increased resistance, with IC50 values of 20 and 37.95µM, respectively, compared to 4.02µM for Granta.WT, representing a 4.9–9.4 fold increase in resistance (Fig.2E; Supplementary Table 2). H929.MI63R and H929.NutlinR cells treated with cisplatin had IC50‘s of 38.28 and 20.17µM compared to 2.55µM in the H929.WT cells (Fig.2F; Supplementary Table 2). Finally, treatment of Granta.MI63R and Granta.NutinR cells with melphalan resulted in IC50 values of 2.2 and 1.5µM, respectively, compared to 0.35µM in Granta.WT cells (Fig.2G; Supplementary Table 2). Similarly, H929.MI63R and H929.NutlinR cells were resistant to melphalan, with IC50 values of 4.77 and 2.59µM compared to 0.78µM in the H929.WT cells (Fig.2H; Supplementary Table 2), representing a 3.32- and 6.2-fold change.
Resistance to HDM-2 inhibitors is mediated by p53 point mutations
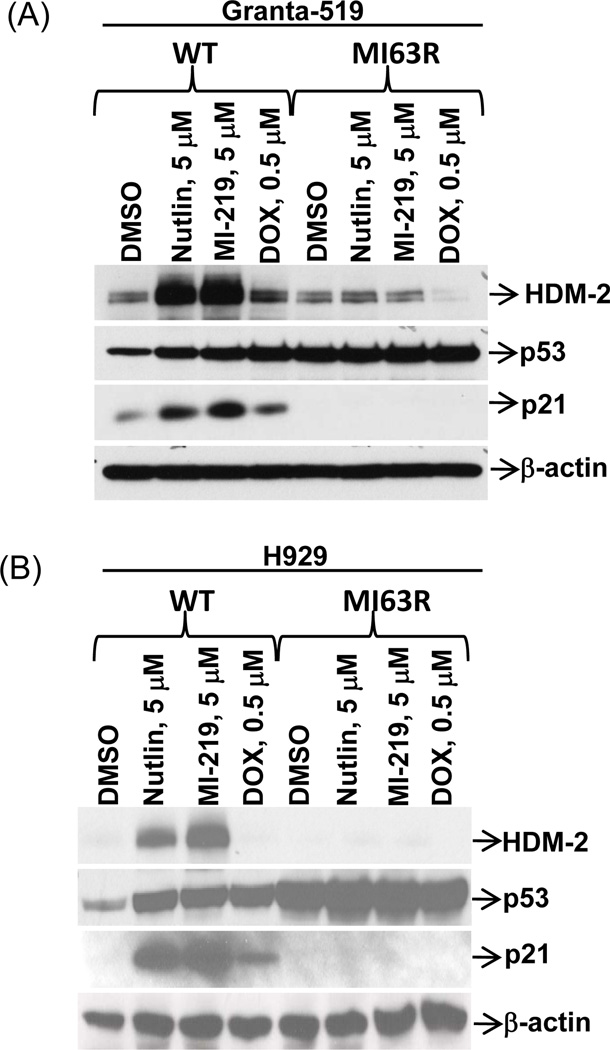
Since MI-63- and Nutlin-insensitive cells showed cross-resistance to DNA damaging agents which work in part in a p53-influenced manner, we considered the possibility that these cells had acquired p53 mutations. Granta.MI63R or H929.MI63R cells were therefore exposed to Nutlin, the in vivo MI-63 analogue MI-219, or doxorubicin, and p53, HDM-2, and p21 levels were evaluated. Both resistant cell lines showed elevated p53 levels in the vehicle controls compared to WT counterparts (Fig.3A, 3B). When Granta.WT and H929.WT cells were exposed to Nutlin or MI-219, a robust p53 increase was seen, resulting in strong HDM-2 and p21 induction. In contrast, Granta.MI63R and H929.MI63R cells showed little if any p53 increase in response to Nutlin, MI-63, or doxorubicin. Importantly, neither doxorubicin nor the HDM-2 inhibitors induced HDM-2 and p21, indicating the absence of transcriptionally active p53.
Figure 3. MI-63-resistant cells over-express p53, but cannot induce p21 in response to HDM-2 inhibition.
Granta-519 (A) and H929 (B) WT and MI63R cells were treated for 24-hours with Nutlin, MI-219, or doxorubicin (DOX), and protein lysates were subjected to Western blotting. Representative images are shown in both panels from one of three independent experiments.
We probed the mutational status of p53 by sequencing, and analysis of Granta.MI63R cells identified two mutations, Q252Q and Y205Y. Q252Q is an exon 3 inactivating nonsense mutation, while Y205Y is an exon 5 inactivating missense mutation (Supplementary Table 3). H929.MI63R and H929.NutlinR cells both carried R175H and R248Q missense mutations within exons 4 and 6, respectively. Notably, HDM-2 sequencing revealed wild-type sequences throughout, indicating drug resistance was not mediated by mutation of the MI-63 or Nutlin binding site.
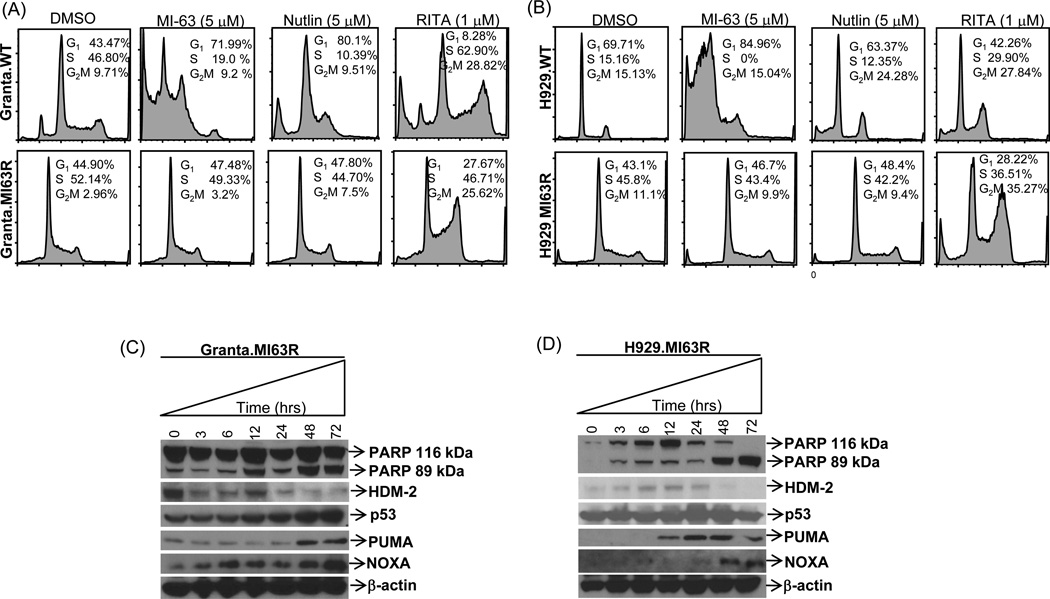
RITA induces cell cycle arrest and apoptosis in resistant cells
The unanticipated sensitivity of Granta.MI63R and H929.MI63R cells to RITA suggested that RITA may have a mechanism of action beyond inhibiting the p53/HDM-2 interaction. Treatment of Granta.WT cells with RITA resulted in G2/M cell cycle arrest compared to vehicle controls (Fig.4A, upper panel), whereas MI-63 and Nutlin induced a G1 arrest. Granta.MI63R cells showed no cell cycle changes in response to MI-63 or Nutlin but RITA induced a strong G2/M arrest (Fig.4A, lower panel). When H929.WT cells were studied, they also showed a G2/M cell cycle arrest with RITA, whereas MI-63 induced a strong G1 arrest with a sub-G1 apoptotic peak. Nutlin also induced a sub-G1 apoptotic peak and increased the G2/M fraction (Fig.4B, upper panel). Similar to the Granta model, RITA treatment of H929.MI63R cells increased the proportion of plasma cells in G2/M, whereas MI-63 and Nutlin had no effect (Fig.4B, lower panel).
Figure 4. RITA induces G2/M cell cycle arrest, and up-regulates p53 pro-apoptotic targets.
Granta-519 (A) and H929 (B) WT and MI63R resistant counterparts were treated with vehicle, MI-63, Nutlin or RITA for 48 hours, and cell cycle analysis was performed. Granta.MI63R (C) and H929.MI63R (D) cells were treated with 5µM RITA, samples were harvested at the indicated time points, and protein lysates were subjected to Western blotting.
A novel insight into RITA’s function has recently been reported by Zhao et al (32) and Messina et al (33), noting RITA could rescue the apoptosis-inducing function of mutant p53. We therefore treated MI-63-resistant cells with RITA, and evaluated its effect on p53 down-stream targets. Exposure of Granta.MI63R cells to RITA initially decreased HDM-2 expression at 3-hours, and though a small rebound was noted at 12-hours, recovery did not occur to baseline levels, and almost full abrogation of expression was visible at 48–72-hours (Fig.4C). This HDM-2 decrease coincided with an increase in p53, and up-regulation of cleaved PARP. Two other p53-influenced apoptosis markers increased, including NOXA and p53 up-regulated modulator of apoptosis (PUMA). In contrast, in H929.MI63R cells, RITA up-regulated HDM-2 at 3–24-hours but, as in Granta cells, HDM-2 expression was virtually absent by 48–72-hours (Fig.4D). Similar to Granta.MI63R cells, p53 levels also increased in H929.MI63R cells as early as 6-hours, and RITA induced apoptosis in H929.63R cells in association with increased levels of NOXA and PUMA.
RITA resensitizes HDM-2 inhibitor resistant cells to MI-63
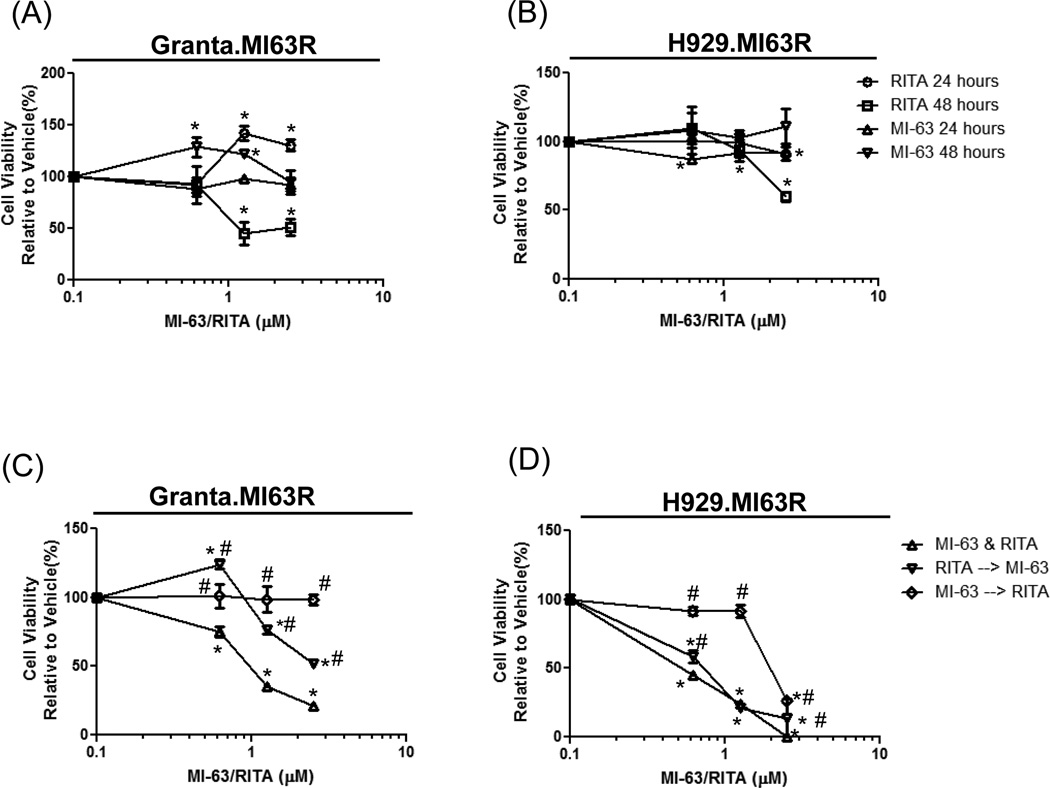
Since RITA induced the expression of PUMA and NOXA, we explored whether RITA could restore p53 function and resensitize MI-63 resistant cells to HDM-2 inhibitors. Thus, we performed combination experiments with MI-63 and RITA, and examined whether the drug exposure sequence affected the outcome. Initial studies determined the activity of RITA and MI-63 for 24- and 48-hours as single agents in the MI-63-resistant cells. RITA had no effect on cell viability at 24-hours in Granta.MI63R or H929.MI63R cells (Fig.5A, 5B). However, treatment with RITA for 48-hours reduced cell viability by 50% in Granta.MI63R and H929.MI63R at 1.25 and 2.5µM, respectively. Treatment with MI-63 had no effect at any of the concentrations used in the MI-63-resistant cells. To determine if the combination of MI-63 and RITA affected the resistant cells, RITA and MI-63 were titrated simultaneously, or cells were pre-treated with one for 24-hours, followed by addition of the other for 48-hours. When Granta.MI63R (Fig.5C, left panel) and H929.MI63R cells (Fig.5D) were treated simultaneously with RITA and MI-63, significantly greater reductions in viability were seen than with either agent alone (p<0.05 to vehicle)(Fig.5A and 5B). Sequencing studies showed that pre-treatment of Granta.MI63R (Fig.5C) or H929.MI63R (Fig.5D) cells with RITA followed by MI-63 resulted in resensitization to MI-63 (p<0.05 to vehicle and simultaneous addition). Interestingly, pre-treatment of Granta.MI63R cells with MI-63, followed later by RITA, showed no resensitization (p<0.05 to simultaneous addition), and indeed the combination was ineffective at reducing viability (Fig.5C). Studies in H929.MI63R cells showed qualitatively similar findings, particularly at lower concentrations, in that MI-63 followed by RITA induced less cell death than RITA alone (p<0.05 to simultaneous addition), supporting the possibility that this treatment sequence was antagonistic.
Figure 5. Simultaneous addition of RITA with MI-63 is synergistic in MI-63-resistant cell lines.
Granta.MI63R (A) and H929.63R (B) cells were incubated with MI-63 or RITA for 24- and 48-hours. In parallel, Granta.MI63R (C) and H929.63R (D) cells were exposed to MI-63 and RITA simultaneously, or first to MI-63 for 24-hours followed by RITA and MI-63 for 48-hours, or to RITA first for 24-hours followed later by MI-63 and RITA for 48-hours. Cell viability was determined using the WST-1 reagent, and results were expressed relative to the vehicle control. An unpaired T test was performed on conditions in panels C and D, where * designates p<0.05 relative to the vehicle control, and # designates p<0.05 relative to the simultaneous addition of RITA and MI-63.
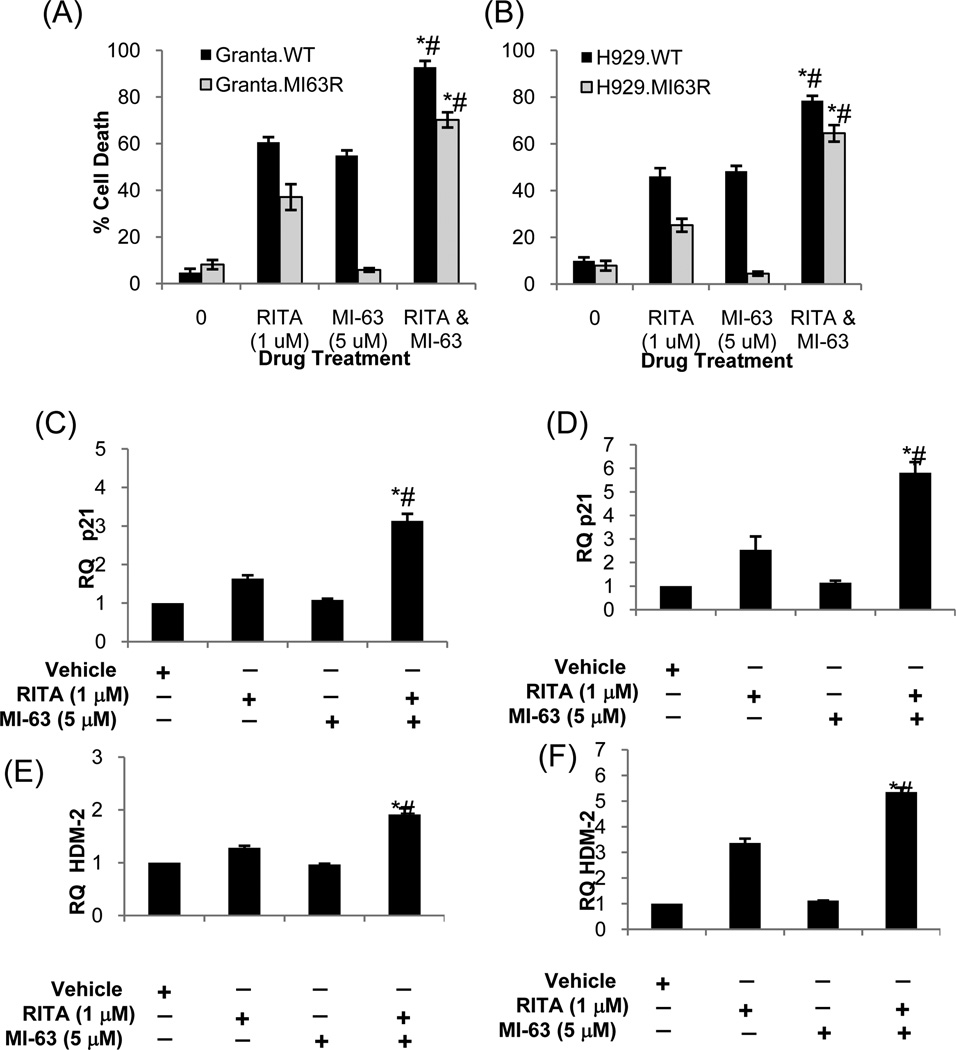
Since simultaneous addition of clinically relevant analogues of RITA and MI-63 would be a convenient treatment approach for patients, we investigated the molecular effects of this regimen. RITA induced a modest amount of apoptosis in Granta.MI63R (Fig.6A) and H929.MI63R cells (Fig.6B), albeit to a lesser degree than in their wild-type counterparts. When MI-63 was used alone, as expected, it was ineffective against either, especially in comparison to the wild-type controls. However, when RITA and MI-63 were given simultaneously, the levels of apoptosis in the HDM-2 inhibitor-resistant models resembled those in the drug-naïve, p53 wild-type counterparts (p<0.01).
Figure 6. RITA restores sensitivity to MI-63 resulting in p21 and HDM-2 induction.
Granta-519 (A) and H929 (B) WT, and MI-63R resistant counterparts were treated with RITA, MI-63, or both simultaneously for 48-hours. FACS analysis was performed with Annexin-V and TO-PRO-3, and the percentage apoptosis calculated. Granta.MI63R (C, E) and H929.MI63R (D, F) were treated with RITA, MI-63, or both simultaneously for 24-hours. RNA was extracted and cDNA was synthesized, and p21 and HDM-2 were measured by quantitative real-time PCR using the ΔΔCT method, with the vehicle treated cells used as a relative calibrator. An unpaired t-test was performed to evaluate for significance, and “*” denotes p<0.01 relative to RITA alone, while “#” denotes p<0.01 relative to MI-63 alone.
To evaluate if RITA was indeed restoring wild-type p53 function, Granta.MI63R and H929.MI63R cells were treated with RITA and MI-63, and expression of p21 and HDM-2 was analyzed by quantitative real-time PCR. RITA induced a 1.5-fold increase in p21, and a 1.25-fold increase in HDM-2 transcripts in Granta.MI63R cells (Fig.6C, 6E), while MI-63 alone failed to induce either. In contrast, the RITA/MI-63 combination increased p21 expression 3-fold, and HDM-2 nearly 2-fold. Similarly, in H929.MI63R cells, RITA induced a 2.5-fold increase in p21 and a 3-fold increase in HDM-2 (Fig.6D, 6F), whilst MI-63 failed at either. This failure was overcome by the addition of RITA and MI-63, resulting in a near 6-fold increase in p21, and a 5-fold increase in HDM-2. These data indicate that continuous exposure of MCL and MM models to the HDM-2 inhibitors MI-63 and Nutlin result in p53 point mutations as a mechanism of acquired drug resistance, and that RITA overcomes this resistance by restoring p53 function, thereby resensitizing cells to HDM-2 inhibitors.
Discussion
Inhibitors of HDM-2, including Nutlin and MI-63/MI-219, have been used as tool compounds to establish HDM-2 as a target for therapy in MCL and MM. In anticipation that HDM-2 inhibitors will be evaluated in the clinic, and since both MCL and MM are characterized by the emergence of drug resistance, it is important to develop models of such resistance to identify the responsible mechanisms. Here we report that acquired MI-63 and Nutlin resistance can be mediated by the acquisition of p53 point mutations that inactivate its activity as a transcriptional activator. DNA damaging therapeutics such as anthracyclines and radiation frequently induce p53 mutations (34, 35), but HDM-2 inhibitors are not felt to induce genotoxic stress or DNA damage (9), making this finding somewhat unexpected. Additional studies will be needed to determine if these mutations are due to a low level of genotoxic stress, outgrowth of a sub-population of mutant p53 clones present in the background of the original wild-type p53 cells, or another mechanism.
Cells with acquired resistance to one HDM-2 inhibitor were cross-resistant to another. Indeed, in cells resistant to MI-63 or Nutlin, further treatment with these inhibitors promoted cellular viability and growth (Fig.1). This may be because mutant p53 has been shown to enhance the development, maintenance, and metastatic spread of malignant cells (36–38). As HDM-2 can still ubiquitinate and degrade mutant p53 (7), HDM-2 inhibitors stabilize these p53 variants (25), and potentiate this gain of function, and potentially enhance proliferation. This suggests that the emergence of drug-resistant clones in patients could be associated with more rapid disease progression than is typically seen with other chemotherapeutics. Patients would therefore need to undergo regular measurements of their disease burden and its molecular characteristics to minimize this risk.
While HDM-2 inhibitor-resistant models showed cross-resistance to other chemotherapeutics, they remained sensitive to RITA. MI-63 and Nutlin differ in that while both bind residues Phe-19, Trp-23, and Leu-26 of the HDM-2 domain that interacts with p53, MI-63 also interacts with Leu-22 (10, 39, 40). Despite this, MI-63 was still inactive against Nutlin-resistant cells, likely demonstrating that cross-resistance to HDM-2 inhibitors is dependent upon the target binding the same p53 HDM-2 binding site. RITA, on the other hand, induced cell cycle arrest and apoptosis, and down-stream p53 targets, suggesting RITA was restoring wild-type p53 function. Confirming this possibility, simultaneous treatment with MI-63 and RITA, and pre-treatment with RITA resensitized cells to MI-63 (Fig.5, 6) with p21 and HDM-2 induction. Originally, the mechanism of action of RITA was thought to be limited to wtp53 cells (18), but recent data indicate activity against mutp53 tumors (32, 33). Indeed, RITA restored the transactivation and transrepression functions of p53 mutants, and demonstrated selectivity for mutant p53 cells (32, 33). These findings explain the ability of RITA to overcome HDM-2 inhibitor resistance, and restore MI-63 sensitivity. One interesting observation was the antagonistic effect of exposing cells to MI-63 prior to RITA, and the ability of Nutlin and MI-63 to enhance the growth of resistant cells, which may be related to the function of p53 as a tetramer. Mutant p53 molecules can disrupt the function of wild-type p53 tetrameric complexes by direct binding, resulting in p53 heterodimers that cannot form functional tetramers (41). Addition of MI-63 first may increase mutant p53 expression in the resistant cells, and when RITA is then added, the number of mutant p53 molecules may be in excess, precluding the ability of RITA to convert them to wild-type function. This may result in antagonism as the balance of mutant p53 molecules can compete off the restored wild-type p53 function. In contrast, addition of RITA prior to MI-63 would allow conversion of mutant p53 to wild-type, and thereby change the balance of dimers, ultimately resulting in wild-type function.
Inactivation of p53 through mutation is linked to decreased sensitivity to many chemotherapeutics (34, 35). Resequencing of HDM-2 and p53 in our models indicated the presence of point mutations solely within p53 DNA binding and dimerization domains. This explains the increased expression of p53, since mutant p53 has a longer half-life. Also, this helps to explain the failure of bortezomib, doxorubicin, melphalan, cisplatin, Nutlin, or MI-63/MI-219 to induce a significant amount of p21 and HDM-2. A previous study examining Nutlin-induced resistance in SJSA osteosarcoma cells found one population had acquired mutations in the p53 DNA binding and dimerization domain (25). Another population retained wtp53, but when the cells were rechallenged with Nutlin, they still underwent growth arrest (25). A more recent study which examined several cell lines with induced Nutlin resistance, including neuroblastoma, melanoma and rhabdomyosarcoma cells in which p53 mutation is infrequent, found that 80% harbored p53 mutations (42). This resistance was generated using similar methods to ours, and of particular interest was the finding that among the cell lines with induced resistance to varying genotoxic agents, only 7.14% had mutated p53 (42). This suggests that Nutlin class HDM-2 inhibitors are prone to induce p53 mutations within chronically exposed cells. All of the p53 mutations found in our cells were heterozygous, and this was also the case in the neuroblastoma Nutlin-resistant cells previously reported (26). Of particular interest was the fact that MI-63- and Nutlin-resistant H929 cells both carried the R248Q and R175H hot spot mutations, which are the most common p53 mutation in B-cell malignancies (43). The finding that both cells acquired the same mutations suggests the presence of mutated cells within the wild-type H929 cell line. These cells may be infrequent and not readily detectable, but following treatment with HDM-2 inhibitors their abundance may be increased. In contrast, the Granta-519 MI-63 and Nutlin resistant cells both had different mutations, suggesting these cells developed unique mutations not present within the wild-type parental cells, which was also observed in Nutlin resistant neuroblastoma cells (26).
The HDM-2 inhibitor R05045337 is in phase I clinical trials targeting patients with solid tumors, and with acute and chronic leukemias. Outcomes from these and other trials, and especially analyses of primary samples obtained at baseline and at the emergence of drug resistance, will further delineate mechanisms of resistance in vivo. Until then, our data suggest that p53 mutations will play a prominent role and, importantly, provide a rationale for combining HDM-2 inhibitors with p53 reactivating agents such as RITA or PRIMAMET (44). These combinations may be of particular use in MCL, where ~15% of patients have mutated p53 with a median survival of only 1.3 years (45). Similarly, p53 mutations in MM fall in the 10–20% range (46), and increase with advanced disease (47). It is exciting to note the p53 reactivating agent APR-246 recently completed phase I clinical trials (48), and has shown activity in vitro in acute myeloid leukemia (49), and malignant melanoma (50). This indicates that MM and MCL patients with baseline or acquired p53 mutations could benefit from novel chemotherapies targeting HDM-2, and that APR-246 or RITA could help to restore wild-type p53 function, leading to enhanced cellular responses to therapy.
Supplementary Material
Acknowledgments
Flow cytometry services were provided by the M. D. Anderson Flow Cytometry Core Facility. p53 and HDM-2 gene sequencing was performed by the DNA Analysis Core Facility, STR DNA fingerprinting was done by the M. D. Cancer Center Characterized Cell Line core, all of which are supported by the Cancer Center Support Grant (CA 16672).
We are grateful to Shaomeng Wang (University of Michigan) for providing MI-63 and MI-219 and Lance Leopold (Ascenta Therapeutics, Inc) for valuable suggestions.
R.J.Jones was funded by a Lymphoma Research Foundation Fellowship award (120808), and R.Z.Orlowski would like to acknowledge support from the National Cancer Institute M. D. Anderson SPORE in Multiple Myeloma (P50 CA142509).
Abbreviations
- DOX
doxorubicin
- Granta.WT
Granta-519 wild-type
- Granta.MI63R
Granta-519 MI-63 resistant cells
- Granta.NutlinR
Granta-519 Nutlin resistant cells
- HDM-2
Human Double Minute protein -2
- H929.WT
H929 wild-type
- H929.MI63R
H929 MI-63 resistant cells
- H929NutlinR
H929 Nutlin resistant cells
- IC50
Median inhibitory concentration
- MCL
mantle cell lymphoma
- MM
multiple myeloma
- mutp53
mutant p53
- PARP
poly-(ADP-ribose) polymerase
- PUMA
p53 up regulated modulator of apoptosis
- RITA
reactivation of p53 and induction of tumor cell apoptosis
- RQ
relative transcript expression
- wtp53
wild-type p53
Footnotes
Authorship
Contribution: R.J.J. designed and performed all of the research, animal experiments and wrote the manuscript; D.J.K. edited the manuscript; C.C.B assisted with data analysis; VB provided biostatistics support and help with data analysis, and R.Z.O. supervised all the research completed herein, and offered valuable suggestions and manuscript editing.
Conflict of Interest Disclosure
The authors declare no relevant financial conflicts of interest.
References
- 1.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 3.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 4.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 5.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 6.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 7.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 9.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 10.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006;49:3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 11.Jones RJ, Chen Q, Voorhees PM, Young KH, Bruey-Sedano N, Yang D, et al. Inhibition of the p53 E3 ligase HDM-2 induces apoptosis and DNA damage--independent p53 phosphorylation in mantle cell lymphoma. Clin Cancer Res. 2008;14:5416–5425. doi: 10.1158/1078-0432.CCR-08-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin L, Tabe Y, Kojima K, Zhou Y, Pittaluga S, Konopleva M, et al. MDM2 antagonist Nutlin-3 enhances bortezomib-mediated mitochondrial apoptosis in TP53-mutated mantle cell lymphoma. Cancer Lett. 2010;299:161–170. doi: 10.1016/j.canlet.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Tabe Y, Sebasigari D, Jin L, Rudelius M, Davies-Hill T, Miyake K, et al. MDM2 antagonist nutlin-3 displays antiproliferative and proapoptotic activity in mantle cell lymphoma. Clin Cancer Res. 2009;15:933–942. doi: 10.1158/1078-0432.CCR-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones RJ, Baladandayuthapani V, Neelapu S, Fayad LE, Romaguera JE, Wang M, et al. HDM-2 inhibition suppresses expression of ribonucleotide reductase subunit M2, and synergistically enhances gemcitabine-induced cytotoxicity in mantle cell lymphoma. Blood. 2011;118:4140–4149. doi: 10.1182/blood-2011-03-340323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha MN, Jiang H, Jayakar J, Reece D, Branch DR, Chang H. MDM2 antagonist nutlin plus proteasome inhibitor velcade combination displays a synergistic anti-myeloma activity. Cancer Biol Ther. 2010;9:936–944. doi: 10.4161/cbt.9.11.11882. [DOI] [PubMed] [Google Scholar]
- 16.Stuhmer T, Chatterjee M, Hildebrandt M, Herrmann P, Gollasch H, Gerecke C, et al. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood. 2005;106:3609–3617. doi: 10.1182/blood-2005-04-1489. [DOI] [PubMed] [Google Scholar]
- 17.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 18.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 19.Andreeff M, Kojima K, Padmanabhan S, Strair R, Kirschbaum M, Maslak P, et al. A Multi-Center, Open-Label, Phase I Study of Single Agent RG7112, A First In Class p53-MDM2 Antagonist, In Patients with Relapsed/Refractory Acute Myeloid and Lymphoid Leukemias (AML/ALL) and Refractory Chronic Lymphocytic Leukemia/Small Cell Lymphocytic Lymphomas (CLL/SCLL) ASH Annual Meeting Abstracts. 2010;116:657. [Google Scholar]
- 20.Secchiero P, di Iasio MG, Gonelli A, Zauli G. The MDM2 inhibitor Nutlins as an innovative therapeutic tool for the treatment of haematological malignancies. Curr Pharm Des. 2008;14:2100–2110. doi: 10.2174/138161208785294663. [DOI] [PubMed] [Google Scholar]
- 21.Saha MN, Micallef J, Qiu L, Chang H. Pharmacological activation of the p53 pathway in haematological malignancies. J Clin Pathol. 2010;63:204–209. doi: 10.1136/jcp.2009.070961. [DOI] [PubMed] [Google Scholar]
- 22.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broxterman HJ, Gotink KJ, Verheul HM. Understanding the causes of multidrug resistance in cancer: a comparison of doxorubicin and sunitinib. Drug Resist Updat. 2009;12:114–126. doi: 10.1016/j.drup.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Wahl GM, Wade M, Wong ET, Tang M, Stommel JM. Hdmx modulates the outcome of P53 activation in human tumor cells. Journal of Biological Chemistry. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 25.Aziz MH, Shen H, Maki CG. Acquisition of p53 mutations in response to the non-genotoxic p53 activator Nutlin-3. Oncogene. 2011 doi: 10.1038/onc.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaelis M, Rothweiler F, Barth S, Cinatl J, van Rikxoort M, Loschmann N, et al. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2011;2:e243. doi: 10.1038/cddis.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung KJ, Horsman DE, Gascoyne RD. The significance of TP53 in lymphoid malignancies: mutation prevalence, regulation, prognostic impact and potential as a therapeutic target. British journal of haematology. 2009;146:257–269. doi: 10.1111/j.1365-2141.2009.07739.x. [DOI] [PubMed] [Google Scholar]
- 28.Jones RJ, Dickerson S, Bhende PM, Delecluse HJ, Kenney SC. Epstein-Barr virus lytic infection induces retinoic acid-responsive genes through induction of a retinol-metabolizing enzyme, DHRS9. J Biol Chem. 2007;282:8317–8324. doi: 10.1074/jbc.M608667200. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyling M, Hiddemann W. Current treatment standards and emerging strategies in mantle cell lymphoma. Hematology / the Education Program of the American Society of Hematology American Society of Hematology. 2009:542–551. doi: 10.1182/asheducation-2009.1.542. [DOI] [PubMed] [Google Scholar]
- 31.Voorhees PM, Orlowski RZ. Emerging data on the use of anthracyclines in combination with bortezomib in multiple myeloma. Clin Lymphoma Myeloma. 2007;7(Suppl 4):S156–S162. doi: 10.3816/clm.2007.s.017. [DOI] [PubMed] [Google Scholar]
- 32.Zhao CY, Grinkevich VV, Nikulenkov F, Bao W, Selivanova G. Rescue of the apoptotic-inducing function of mutant p53 by small molecule RITA. Cell Cycle. 2010;9:1847–1855. doi: 10.4161/cc.9.9.11545. [DOI] [PubMed] [Google Scholar]
- 33.Linda Messina R, Sanfilippo M, Vella V, Pandini G, Vigneri P, Nicolosi ML, et al. Reactivation of P53 mutants by prima-1 in thyroid cancer cells. International journal of cancer. 2011 doi: 10.1002/ijc.26228. [DOI] [PubMed] [Google Scholar]
- 34.Fan S, el-Deiry WS, Bae I, Freeman J, Jondle D, Bhatia K, et al. p53 gene mutations are associated with decreased sensitivity of human lymphoma cells to DNA damaging agents. Cancer research. 1994;54:5824–5830. [PubMed] [Google Scholar]
- 35.Lai SL, Perng RP, Hwang J. p53 gene status modulates the chemosensitivity of non-small cell lung cancer cells. J Biomed Sci. 2000;7:64–70. doi: 10.1007/BF02255920. [DOI] [PubMed] [Google Scholar]
- 36.Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 37.Scian MJ, Stagliano KE, Ellis MA, Hassan S, Bowman M, Miles MF, et al. Modulation of gene expression by tumor-derived p53 mutants. Cancer research. 2004;64:7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 38.Kim E, Deppert W. Transcriptional activities of mutant p53: when mutations are more than a loss. J Cell Biochem. 2004;93:878–886. doi: 10.1002/jcb.20271. [DOI] [PubMed] [Google Scholar]
- 39.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 40.Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2;--fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 41.Hupp TR. Regulation of p53 protein function through alterations in protein-folding pathways. Cell Mol Life Sci. 1999;55:88–95. doi: 10.1007/s000180050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Design of a family-based lifestyle intervention for youth with type 2 diabetes: the TODAY study. Int J Obes (Lond) 34:217–226. doi: 10.1038/ijo.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. http://www-p53iarcfr/ [Google Scholar]
- 44.Rokaeus N, Shen J, Eckhardt I, Bykov VJ, Wiman KG, Wilhelm MT. PRIMA-1(MET)/APR-246 targets mutant forms of p53 family members p63 and p73. Oncogene. 2010;29:6442–6451. doi: 10.1038/onc.2010.382. [DOI] [PubMed] [Google Scholar]
- 45.Greiner TC, Moynihan MJ, Chan WC, Lytle DM, Pedersen A, Anderson JR, et al. p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood. 1996;87:4302–4310. [PubMed] [Google Scholar]
- 46.Portier M, Moles JP, Mazars GR, Jeanteur P, Bataille R, Klein B, et al. p53 and RAS gene mutations in multiple myeloma. Oncogene. 1992;7:2539–2543. [PubMed] [Google Scholar]
- 47.Neri A, Baldini L, Trecca D, Cro L, Polli E, Maiolo AT. p53 gene mutations in multiple myeloma are associated with advanced forms of malignancy. Blood. 1993;81:128–135. [PubMed] [Google Scholar]
- 48.Liljebris C, Mohell N, Olaisson E, Uhlin T, Wiman KG, Lehmann S. APR-246, a new class anticancer compound in phase l/lla clinical trials. ASCO Meeting Abstracts. 2010;28 TPS183. [Google Scholar]
- 49.Ali D, Jonsson-Videsater K, Deneberg S, Bengtzen S, Nahi H, Paul C, et al. APR-246 exhibits anti-leukemic activity and synergism with conventional chemotherapeutic drugs in acute myeloid leukemia cells. Eur J Haematol. 2011;86:206–215. doi: 10.1111/j.1600-0609.2010.01557.x. [DOI] [PubMed] [Google Scholar]
- 50.Bao W, Chen M, Zhao X, Kumar R, Spinnler C, Thullberg M, et al. PRIMA-1Met/APR-246 induces wild-type p53-dependent suppression of malignant melanoma tumor growth in 3D culture and in vivo. Cell Cycle. 2011;10:301–307. doi: 10.4161/cc.10.2.14538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.