Abstract
Human immunodeficiency virus (HIV) primarily infects glial cells in the central nervous system (CNS). Recent evidence suggests that HIV-infected individuals who abuse drugs such as methamphetamine (METH) have higher viral loads and experience more severe neurological complications than HIV-infected individuals who do not abuse drugs. The aim of this study was to determine the effect of METH on HIV expression from the HIV long terminal repeats (LTR) promoter and on an HIV integrated provirus in microglial cells, the primary host cells for HIV in the CNS. Primary human microglial cells immortalized with SV40 T-antigen (CHME-5 cells) were co-transfected with an HIV LTR reporter and the HIV Tat gene, a key regulator of viral replication and gene expression, and exposed to METH. Our results demonstrate that METH treatment induced LTR activation, an effect potentiated in the presence of Tat. We also found that METH increased the nuclear translocation of the nuclear factor kappa B (NF-κB), a key cellular transcriptional regulator of the LTR promoter, and the activity of an NF-κB-specific reporter plasmid in CHME-5 cells. The presence of a dominant-negative regulator of NF-κB blocked METH-related activation of the HIV LTR. Furthermore, treatment of HIV-latently infected CHME-5 (CHME-5/HIV) cells with METH induced HIV expression in a dose-dependent manner, and nuclear translocation of the p65 subunit of NF-κB. These results suggest that METH can stimulate HIV gene expression in microglia cells through activation of the NF-κB signaling pathway. This mechanism may outline the initial biochemical events leading to the observed increased neurodegeneration in HIV-positive individuals who use METH.
Keywords: HIV, microglia, methamphetamine, NF-kappa B (NF-κB), nuclear translocation
Introduction
The relationship of methamphetamine (METH) abuse and human immunodeficiency virus (HIV) infection extends beyond behavioral and social interactions. METH, a potent central nervous system (CNS) stimulant is associated with increased incidence of HIV infection (Buchacz et al, 2005; Freeman et al, 2011). HIV infected individuals that abuse METH exhibit increased cognitive deficits compared to HIV-negative individuals that abuse METH or HIV-positive individuals that do not abuse METH (Rippeth et al, 2004). A case study showed accelerated HIV- associated dementia in an HIV-positive patient abusing METH (Nath et al, 2001).
The molecular mechanisms by which METH exerts its effect in the CNS are not well defined. The potent neurotoxic effect of METH seems to primarily target dopaminergic neurons and part of its toxicity has been associated with glial activation (Guilarte et al, 2003; LaVoie et al, 2004; Thomas et al, 2004). Humans who abuse METH have increased binding of a radiotracer for activated microglia compared to control subjects (Sekine et al, 2008). Collectively, the data suggest that METH influences microglial activity.
In relationship to HIV, a study using proton magnetic spectroscopy found that METH+/HIV+ individuals had decreased neuronal function and increased glial activation compared to METH−/HIV+ individuals (Chang et al, 2005). Consistent with the in vivo imaging, post-mortem analysis of METH+/HIV+ brains revealed lower expression of dendritic and synaptic markers, decreased number of interneurons, and increased expression of markers on activated microglia and astrocytes compared to METH−/HIV+ brains (Langford et al, 2003). In rodents, Tat protein administered intrastriatally 24 h prior to intraperitoneal METH injections synergistically reduces dopamine levels and decreases dopamine transporter binding in the striatum (Cass et al, 2003; Maragos et al, 2002; Theodore et al, 2006a). This synergistic toxicity is partially mediated through tumor necrosis factor α (TNFα) upregulation (Theodore et al, 2006b). Collectively, these data support that the combination of METH consumption and HIV infection act synergistically to promote with neuronal dysfunction and degeneration.
The expression of HIV proteins is controlled by DNA elements located on the long terminal repeats (LTR) sequence of the HIV genome. The HIV genome is a single-stranded RNA. Upon infection of the host cell, the HIV genome is reverse-transcribed into cDNA which enters the nucleus where it integrates into the host genomic DNA. The transcription of this HIV proviral DNA is regulated by viral and cellular factors that interact with LTR sequences of the HIV genome. Cellular transcription factors, specifically activator protein 1 (AP-1), nuclear factor of activated T cells (NF-AT1), nuclear factor – interleukin 6 (NF-IL6), SP-1, upstream stimulating factor (USF), E26 transforming sequence-binding factors (Ets-1), T cell factor 1 alpha (TCF-1α), and nuclear factor kappa B (NF-κB), have binding sites located within the LTR and have been implicated in regulating HIV LTR expression (Copeland, 2005; Pereira et al, 2000; Tang et al, 1999). METH has been shown to increase the activity and binding of several HIV LTR-relevant cellular transcription factors including AP-1 (Sheng et al, 1996; Wang et al, 1992), NF-AT (Jayanthi et al, 2005), and NF-κB (Asanuma and Cadet, 1998).
Evidence from in vitro and in vivo studies also indicates that METH increases LTR activity. For example, individuals who abuse METH have a higher plasma HIV load (Ellis et al, 2003). Also, METH was shown to increase replication of the feline immunodeficiency virus in astrocytes (Gavrilin et al, 2002). A recent study demonstrated that METH could increase replication of HIV in peripheral monocytes (Toussi et al, 2009). However, the ability of METH to influence HIV LTR activity in microglia, cells that typically harbor HIV proviral DNA in brain (Lee et al, 1993a; Lee et al, 1993b)), has not been reported. In the current study, we examined the effects of METH on HIV transcription in human microglial cells. Our findings suggest that the HIV LTR in human microglia cells in vitro can be activated by methamphetamine. METH-induced expression from the HIV LTR requires NF-κB signaling.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
The dual reporter plasmid, pLTRC-Luc-EGFP (Ravi and Mitra, 2007) was generously provided by Dr. Debashis Mitra (National Center for Cell Science, India). pC-Tat.BL43.CS (pC-Tat) and pE-Tat.BL43.CS (pE-Tat) were provided by the NIH AIDS Reagent Program. pGL4.30 [luc2P/NFAT-RE/Hygro], pGL4.32[luc2P/NF-κB-RE/Hygro], Luciferase 1000 Assay system, and CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) were purchased from Promega (Madison, WI). The IκB dominant negative (IκBDN) construct, previously described (Sanchez et al, 2003), was generously provided by Dr. Sanjay Maggirwar (University of Rochester). Nuclear Extraction Kits and TransAM NF-κB p65 were purchased from Active Motif (Carlsbad, CA. (+) METH-HCl (referred to as “METH” hereafter), fetal bovine serum (FBS), tissue necrosis factor α (TNFα), and lipopolysaccharide (LPS) were obtained from Sigma-Aldrich (St Louis, MO). DC Assay was purchased from BioRad (Hercules, CA).
Cell Culture and Treatment
The CHME-5 cell line was created by transfecting human fetal microglia with the large T antigen of the simian virus 40 (Janabi et al, 1995). Cells were maintained in Dulbecco’s minimal essential medium, high glucose (DMEM HG) supplemented with 5% FBS and 1% penicillin/streptomycin based on previous studies (Cox et al, 2009).
For transfection, cells were plated in clear or opaque 96 well plates (Corning Costar, Sigma Aldrich) at 8×104 cells/well and transfected 14–16 h post-plating using Lipofectamine 2000 (Invitrogen, CA). For luciferase experiments, pLTRC-Luc-EGFP and Tat expressing plasmids were kept constant and, 4 h after transfection, METH was added at indicated concentrations. Cells were assayed 24 h post-transfection unless otherwise noted.
For obtaining HIV-latently infected CHME-5, cells were plated on a 6-well plate at a density of (1×106 cells/well) for 48 h prior to infection with vesicular stomatitis virus G-(VSVG) pseudotyped HIVs bearing a fragment of HIV-1pNL4-3, containing Tat, Rev, Env, and Vpu, cloned into the pHR' backbone (Dull et al, 1998; Pearson et al, 2008), plus Nef adjacent to the reporter gene d2E green fluorescence protein (GFP) inserted next to Env. The viral particles were produced by the triple transfection of 293T cells using lipofectamine, as described previously (Kim et al, 2006), and the vector titer was determined by the infection of 1×106 CHME-5 cells with a serial dilution of the harvested medium supernatant. Briefly, the plate containing cells and virus was spinoculated in a swing-bucket centrifuge at 3000 × g for 1.5 h at room temperature, and incubated at 37°C in 5% CO2 for 48 h prior to trypsin treatment and fluorescence-activated cell sorting (FACS). GFP+ cells were further cultured and allowed to enter into a latent state (GFP expression below 5%) for four weeks. Latency of CHME-5/HIV cells was characterized by re-activating HIV expression (GFP; see below) and evaluating nuclear translocation of NF-κB p65 and two of its phosphorylated forms (S536 and S468) by Western blot (see below) in the presence of TNFα (50 ng/mL) for 0.5, 2, 4, 8, and 16 h. Latent CHME-5/HIV cells were then untreated or treated with increasing concentration of METH (0, 50 and 300 µM). GFP fluorescence was imaged using identical acquisition setting among groups within a given experiment. Images were acquired using a Nikon TE2000 inverted scope equipped with a DS-QiMc camera and controlled by NIS Elements software (Nikon). Again, treatment with 50 ng/mL TNFα (Pearson et al, 2008) was used as a positive control for reactivation. For testing NF-κB dependence of METH-mediated activation of HIV, CHME-5/HIV cells were pre-treated for 1 h with either 100 µM of IKKγ NEMO binding domain inhibitory peptide or equivalent amount of the control peptide (Imgenex, CA) dissolved in dimethyl sulfide (DMSO) prior to incubation with 600 µM of METH for 16 h.
LTR and NF-κB Reporter Assays
To measure the production of firefly (Photinus pyralis) luciferase from the transfected pLTRC-Luc-EGFP plasmid, the Luciferase 1000 Assay System was used for all experiments. Briefly, CHME-5 cells were transfected in an opaque 96-well plate, as described above, using either pC-Tat or pE-Tat and pLTRC-Luc-EGFP at a 1:8 mass ratio. Four hours post-transfection, cells were treated with vehicle, 100, 300, 500, 700 or 1000 µM of METH. At 24 h post-treatment, culture medium was removed and 50 µL/well of 1x Reporter Lysis Buffer was added to cells for 2 minutes at room temperature. Luciferase activity was read at constant settings using Synergy 2 plate reader (Biotek, VT). To visualize GFP from pLTRC-Luc-EGFP, CHME-5 cells were plated as described above in clear 96 well plates. At 24 h post-treatment, cells were fixed with 4% paraformaldehyde for 1 h at room temperature and stained with the nuclear dye 4',6-diamidino-2-phenylindole (DAPI) and mouse anti-GFP (Roche Applied Science, IN). With camera settings constant across treatment groups, GFP and DAPI fluorescence as well as brightfield images were captured using a Nikon TE2000 inverted scope equipped with a DS-QiMc camera and controlled by NIS Elements software (Nikon), as described above.
Similarly, CHME-5 cells plated in opaque 96 well plates and transfected with either pGL4.30[luc2P/NFAT-RE/Hygro] or pGL4.32[luc2P/NF-κB-RE/Hygro] reporter vectors were treated with METH for 24 h and the luciferase activity was measured using the Luciferase 1000 assay system as described above.
MTS Assay
CHME-5 cells were plated and transfected with pLTRC-Luc-EGFP and pC-Tat, and treated with METH, as described above. Cell viability was assessed using CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (MTS assay). Viability was measured 24 h post-treatment by removing half of the media (95 µL to account for evaporation) and replaced with 20 µL of 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)/phenazine ethosulfate (PMS) solution. Plates were incubated at 37°C for 45 minutes in the presence of the MTS/PMS stable solution, and read at an absorbance of 490 nm using the Synergy 2 plate reader.
NF-κB Translocation Assays
NF-κB translocation from cytosol to nucleus was assessed by measuring NF-κB p65 presence in nuclear extracts following METH treatment. In the first set of experiments, CHME-5 cells were plated on 60 mm-diameter dishes at 7×106 cells/plate for 14 to 16 h, and then treated with 500 µM METH for 30 minutes, 1, 4 or 24 h. Nuclear extracts were prepared according to the Nuclear Extraction Kit (Active Motif). Extracts were stored at -80°C until protein quantification by a detergent compatible protein assay (Biorad). Samples were diluted 1:15, after determining an optimal dilution factor, and subjected to an NF-κB p65 ELISA (Active Motif). Plates were read at 450 nm with a reference wave of 655 nm using Synergy 2 plate reader. To block NF-κB translocation, CHME-5 cells were plated at 8×104 cells/well in an opaque 96-well plate, and transfected with pLTRC-Luc-EGFP with or without pIκBDN (Sanchez et al, 2003). At 4 h post-transfection, cells were treated with 500 µM of METH for 24 h and luciferase activity measured, as described above.
In the second set of experiments, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDSPAGE)/Western blot analysis (carried out as described elsewhere) of NF-κB p65 was used to assess activation and nuclear translocation of NF-κB p65 in CHME-5/HIV cells upon treatment with 600 µM METH. For this, 7×106 cells were plated and incubated for 16 h prior to treatment for 30 minutes, 16 and 24 h. After two washings with PBS, cells were collected in 500 µL of Buffer A [10 mM Hepes/KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA)] in the presence of phenylmethylsulfonyl fluoride (PMSF; 1mM), dithiothreitol (DTT; 1mM), 0.5% nonyl phenoxypolyethoxylethanol (NP-40), and protease/phosphatase inhibitors tablets. Nuclei were collected after centrifugation at 1,500 × g for 10 minutes at 4°C, and three cycles of washing in Buffer A. Finally, nuclei were re-suspended in 100 µL of Buffer B (20 mM Hepes/KOH, pH 7.9, 25% glycerol, 420 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2) containing PMSF (1 mM), DTT (1 mM), and inhibitors tablets, and centrifuged at 20,000 × g for 15 minutes at 4°C. Supernatant was then collected, its protein concentration measured by Bradford assay, and subjected to SDS-PAGE/Western blot using the Santa Cruz Biotechnology (CA) antibodies against NF-κB p65, TFIIH p62, and SPT-5, as loading control. These primary antibodies were bound by the IRDye 800CW donkey anti-rabbit secondary antibody, and the membranes were scanned and analyzed using the Odyssey® Infrared Imaging System (LI-COR Biosciences, NE).
Statistical Analysis
The student’s t-test and either one- or two-way ANOVA were used for statistical comparison. Bonferroni tests or Newman-Keuls tests were used for post-hoc analysis of one- and two-way ANOVA, respectively. Data are presented as mean ± S.E.M.; p-values of <0.05 were considered significant.
RESULTS
METH induces HIV LTR expression in CHME-5 cells
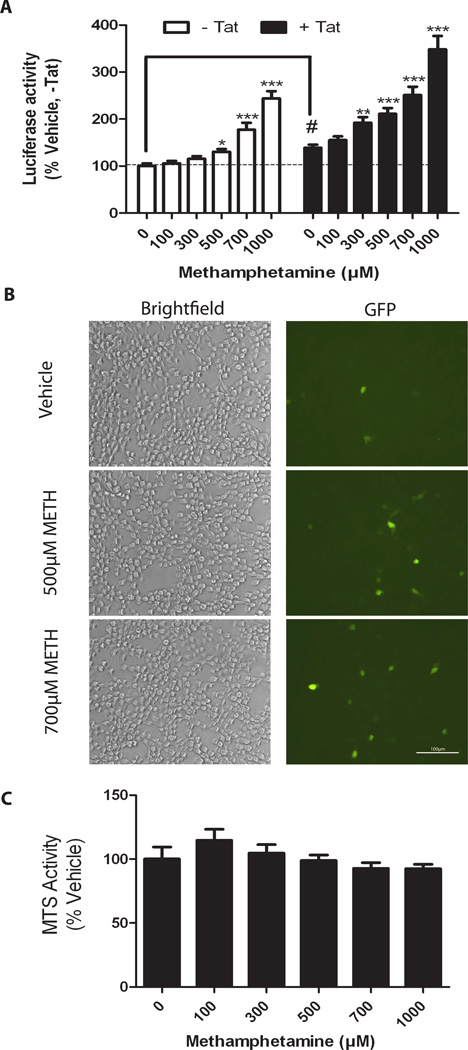
CHME-5 cells were co-transfected with the reporter plasmid pLTRC-Luc-EGFP together with either pC-Tat (+Tat) or a control plasmid, pE-Tat (−Tat). As expected, Tat alone significantly elevated LTR activity (Figure 1A, “#” p<0.001, t-test, -Tat vs Tat). In the absence of Tat (−Tat), METH caused a significant, dose-dependent increase in luciferase expression (Figure 1A, p<0.0001, One-way ANOVA, white bars). In the presence of Tat, a similar response was observed, with the dose response curve shifted towards the left (Figure 1A, p<0.0001, Oneway ANOVA, black bars), suggesting a synergistic/additive interaction between METH and Tat in enhancing LTR promoter activity. Consistent with the luciferase data, METH at a dose of 700 µM METH increased the expression of GFP in the presence of Tat (Figure 1B, right panel, GFP).
Figure 1. METH induces luciferase and GFP expression from HIV-LTR-Luc-IRES-GFP in CHME-5 cells.
A, CHME-5 cells were co-transfected with pLTR-Luc-IRES-GFP reporter together with a control (−Tat) or Tat-expressing plasmid (+Tat). Cells were treated with METH for 4 h post-transfection and assayed for luciferase activity at 24 h post-transfection. METH significantly increased activity independently of Tat (*p<0.05, **p<0.01, ***p<0.001, One-way ANOVA SNK post-hoc versus 0 METH); (# p<0.001, Student t-test versus No Tat, 0 METH). B, CHME-5 cells were transfected with pLTR-Luc-IRES-GFP together with pC-Tat and treated with 0, 500, and 700 µM of METH for 24 h and stained with mouse anti GFP (indicated to left of the Brightfield panels). Brightfield (left panel) and GFP fluorescence (right panel) were imaged for each treatment group. METH increased GFP fluorescence at 500 and 700 µM METH compared to vehicle-treated cells. C, METH did not significantly alter viability (MTS assay) of CHME-5 cells treated with indicated doses of METH.
The effect of METH on cell survival was quantitatively measured using an MTS assay at 24 h post-treatment. These results show no significant change in viability among treatment groups co-transfected with pLTRC-Luc-EGFP and pC-Tat (Figure 1C). Similarly, no apparent change in cell morphology was observed (Figure 1B, left panel, Brightfield).
METH activates NF-κB in CHME-5 cells
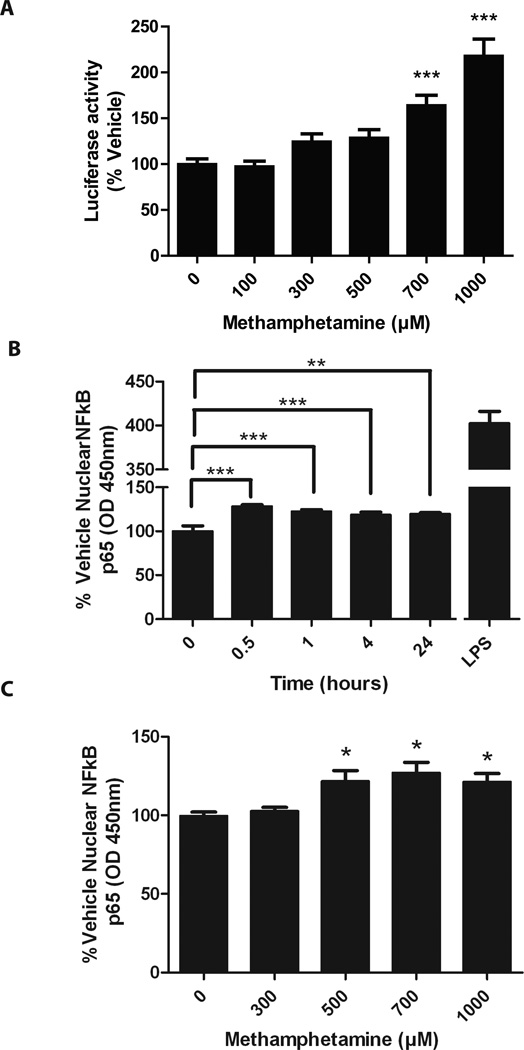
NF-κB, a cellular transcription factor that can undergo translocation to the nucleus upon various cellular or extracellular stimuli (Ghosh and Karin, 2002), is a major and potent regulator of HIV LTR transcription (Mallardo et al, 1996). We next examined the effect of METH on NF-κB activation in CHME-5 cells using a luciferase reporter plasmid that is dependent on NF-κB. Treatment with METH increased luciferase activity from the NF-κB reporter in a dose-dependent manner (Figure 2A, p<0.0001, One-way ANOVA), suggesting that METH alone increases NF-κB activity in CHME-5 cells. NF-κB activation typically occurs within minutes to an hour of exposure to external stimuli. Due to the known ability of NF-κB to translocate from the cytoplasm to the nucleus upon activation, we measured nuclear levels of NF-κB following METH treatment (500 µM) of CHME-5 cells by ELISA of the p65 subunit after nuclear lysis. Our results demonstrated that METH significantly increased nuclear NF-κB levels as early as 30 minutes and remained elevated up to 24 h post-treatment (Figure 2B). METH dose-dependently increased nuclear NF-κB protein levels at 1 h (Figure 2C, p=0.0105, One-way ANOVA).
Figure 2. METH increases activity of cellular transcription factors in CHME-5 cells.
A, CHME-5 cells were transfected with a reporter plasmid containing minimal promoter with NF-κB response elements, and treated with METH for 24 h. METH treatment significantly increased luciferase activity (One-way ANOVA Dunnet Test post-hoc versus 0 METH, *p<0.05, ***p<0.001). B, CHME-5 cells were treated with 500 µM METH for various times. Nuclear extracts were prepared and assayed for NF-κB p65 by ELISA. NF-κB p65 was significantly elevated in nuclear extract as early as 30 minutes and remained elevated at 24 h. LPS treatment for 1 h was used as a positive control, (p<0.001, One-way ANOVA Dunnet post-hoc versus 0 METH, **p<.01,***p<0.001). C, CHME-5 cells were treated with various amounts of METH for 1 h. Nuclear extracts were prepared and assayed for p65 by ELISA. NF-κB was significantly elevated in nuclear extracts treated with 500 µM, 700 µM, and 1000 µM METH. (p=0.01, One-way ANOVA Dunnet post-hoc versus 0 METH, **p<0.01).
NF-κB activity is necessary for METH induction of HIV LTR
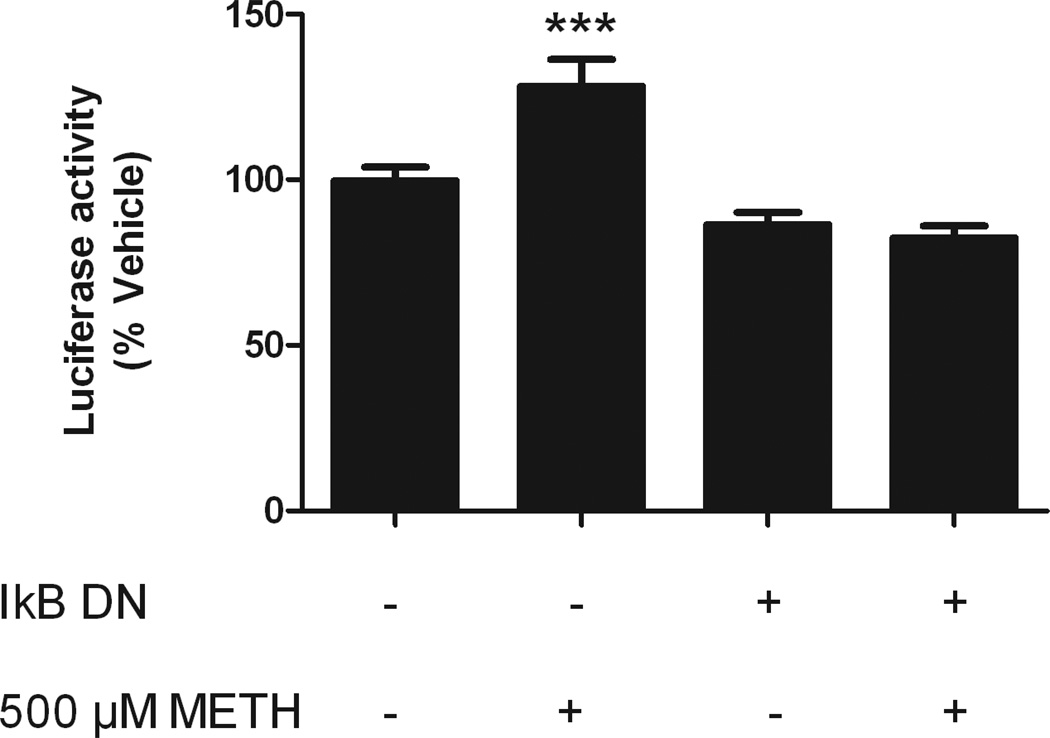
We next investigated whether the METH-related increase of NF-κB activity was necessary for the observed induction of HIV LTR by METH. The nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha (IκBα) mutant S32/36A (IκB-DN) lacks the phosphorylation site and cannot dissociate from NF-κB, therefore preventing NF-κB translocation and activity (Sanchez et al, 2003). CHME-5 cells were co-transfected with the pLTRC-Luc-EGFP and IκB-DN plasmids, and then treated with 500 µM METH. As evidenced by the difference in luciferase activity, the METH-related increase in HIV LTR activity was blocked by the presence of the IκB-DN (Figure 3), consistent with the cellular role of wild type IκBα, and further suggesting that METH-mediated induction of HIV LTR in CHME-5 cells occurs through NF-κB activation. There was a slight decrease (~15%) in basal activity caused by transfection of IκB-DN compared to control cells that was independent of the presence of methamphetamine (Figure 3, p<0.05, t-test).
Figure 3. NF-κB signaling is necessary for METH-mediated induction of HIV-LTR-Luc-IRES-GFP.
CHME-5 cells were co-transfected with pLTR-Luc-IRES-GFP and pI-κBDN, and treated with 500 µM of METH. Luciferase activity was measured at 24 h post-treatment. IκBDN significantly decreased luciferase activity. (p<0.001 One-way ANOVA, *** p<0.001 SNK post-hoc versus other groups).
METH reactivates HIV transcription in CHME-5 cells latently infected with HIV vector in an NF-κB-dependent manner
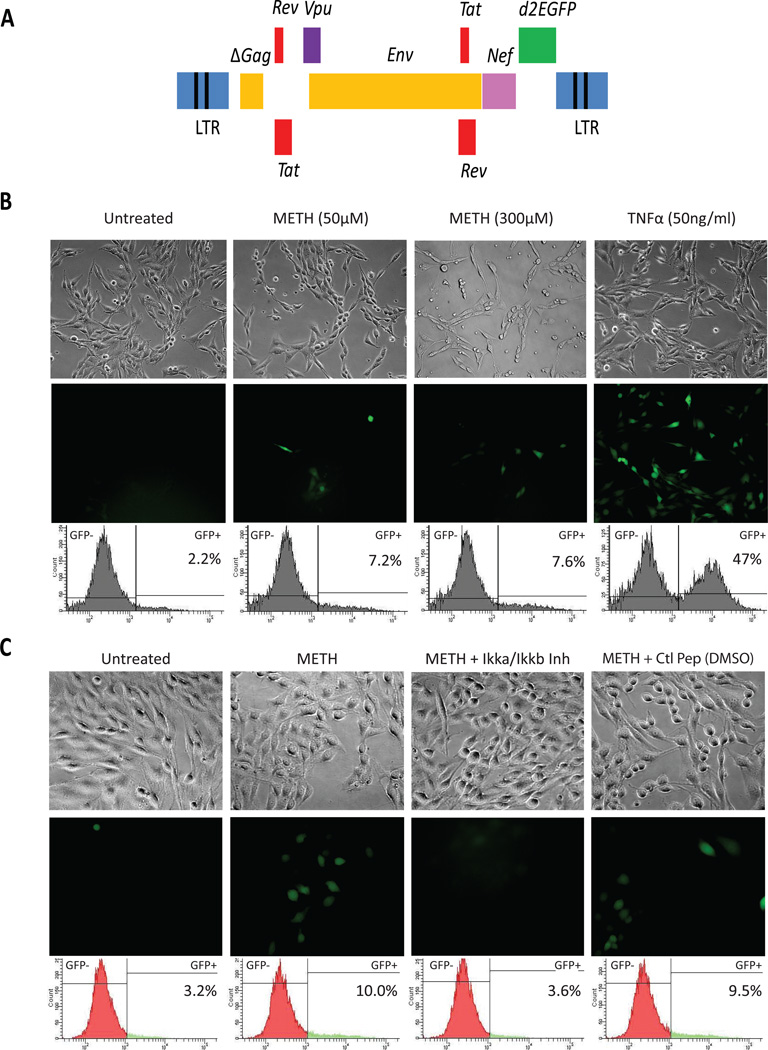
A lentivirus vector expressing d2EGFP was used to develop a mixed population of HIV-infected CHME-5 cells (Figure 4A). Reactivation of latent HIV genome was demonstrated using a 50 ng/mL TNFα treatment for 0.5, 2, 4, 8, and 16 hours. Images of GFP fluorescence were acquired at the specified time points and indicated a substantial increase in the fraction of cells expressing GFP (Supplemental Figure 1A). In addition, Western blot analysis demonstrated that treatment with TNFα increased NF-κB p65 and its phosphorylated forms S536 and S468 nuclear translocation as early as 30 minutes, and which fluctuates over time, similar to what has been observed in re-activated latently infected T-cells (Pearson et al., 2008; Supplemental Figure 1B). Once integrated into the genome, this proviral vector, like wild-type HIV, is cis-regulated by the HIV proteins Tat and Rev, and therefore is able to maintain a positive feedback circuit that regulates HIV transcription for as long as Tat levels surpass a necessary threshold (Pearson et al, 2008). The fluorescence reporter d2EGFP is cloned next to the Nef gene to facilitate monitoring of HIV expression (Figure 4A). These CHME-5/HIV cells were exposed to increasing doses of METH (0, 50, and 300 µM) for 16 h. Our result (Figure 4B) demonstrates that METH was capable of inducing, without apparent toxicity (Brightfield), HIV expression at doses as low 50 µM as depicted by the images showing increasing number of cells expressing HIV (GFP positive cells). TNFα (50 ng/ml) treatment, used as positive control, showed a substantial increase in the fraction of cells expressing HIV. To confirm the result shown in Figure 3, indicating that METH-mediated activation of HIV LTR in transfected CHME-5 cells depends on NF-κB activation, CHME-5/HIV cells were pre-treated with the IKKγ NEMO binding domain inhibitory peptide (Ikka/Ikkb), which inhibits NF-κB activity by interfering with the IKK complex formation (Imgenex), prior to incubation with METH. The result of this experiment (Figure 4C) further confirmed that NF-κB activation is necessary for the METH-induced activation of HIV since the inhibitory peptide prevented METH from activating HIV in CHME-5/HIV cells, an effect not observed with the control peptide (ctl peptide).
Figure 4. METH reactivates CHME-5 cells latently infected with HIV.
A, Genomic organization of the HIV lentiviral vector. A fragment of HIV-1pNL4-3, containing Tat, Rev, Env, Vpu, and Nef with the reported gene d2EGFP, is cloned into the pHR’ backbone. The resulted plasmid was used to produce the VSVG HIV as described previously (Kim et al, 2006). B Reactivation of HIV by METH. CHME-5 cells were latently infected with HIV (CHME-5/HIV) and incubated either in the absence (untreated) or in the presence of increasing doses of METH, as indicated above each set of brightfield and GFP-fluorescence pictures. Cells were reactivated with TNFα (50 ng/mL), as positive, control. FACS plots show increase in the % of GFP+ cells in response to METH or TNFα. C, Reactivation of HIV by METH depends on the activation of NF-κB. CHME-5/HIV cells were pre-incubated with 100 µM of either an inhibitory of the IKK complex (Ikka/Ikkb) or a control peptide (ctl peptide) prior to incubation with METH (600 µM).
METH induces NF-κB nuclear translocation in HIV-latently infected CHME-5 cells
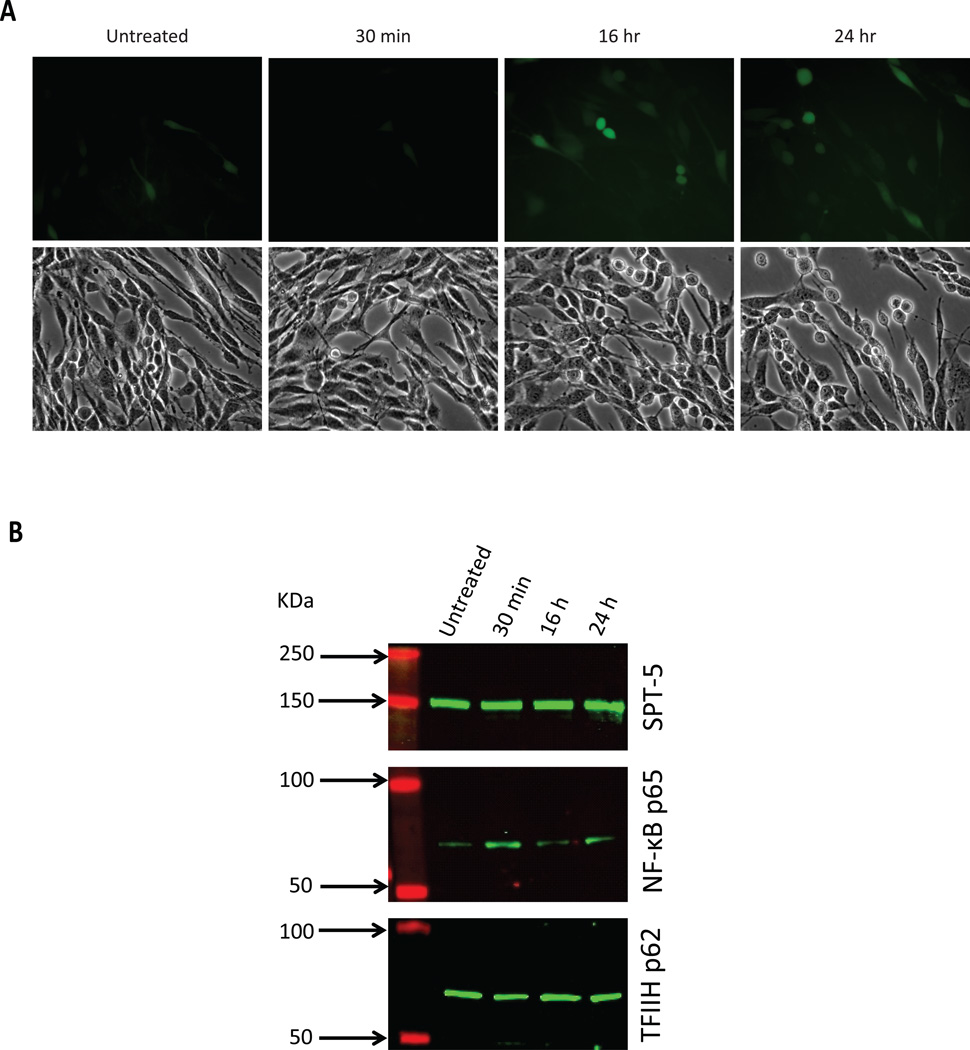
To confirm our observation that NF-κB translocates to the nucleus in CHME-5 cells transfected with pLTRC-Luc-EGFP (Figure 2), we also evaluated NF-κB nuclear translocation in CHME-5/HIV cells by Western blot analysis of nuclear fractions isolated from either untreated or METH-treated CHME-5/HIV cells. The CHME-5/HIV cell cultures used to extract the nuclear proteins are shown by microphotographs on Figure 5A (untreated vs. 30 minutes, 16 h, and 24 h). Treatment of CHME-5/HIV cells with 600 µM METH increased NF-κB p65 nuclear presence after a 30-minutes exposure (Figure 5B) as evidenced by the increased band intensity of NF-κB p65 at 30 minutes. Nuclear NF-κB p65 was found essentially at basal levels after 16 h and 24 h post-stimulation since the band intensities are comparable to that of untreated cells. For loading control, nuclear extracts were blot against SPT-5, a constitutive nuclear protein; no significant variation in the expression of SPT-5 was observed across the time points examined. In addition, for comparison and control, we have also evaluated expression of TFIIH p62, a transcription factor constitutively present in the nucleus and recruited, like NF-κB p65, to the viral promoter during emergence from latency (Kim et al, 2006).
Figure 5. Nuclear translocation of NF-κB in HIV-latently infected CHME-5 cells treated with METH.
A, Reactivation of CHME-5/HIV by METH. Cells were incubated either in the absence (untreated) or in the presence of METH (600 µM) for 30 minutes, 16 h and 24 h as indicated next to each set of Brightfield and GFP-fluorescence pictures. B, NF-κB p65 Western blot of nuclear extracts isolated from untreated and METH-treated cells. MW markers and time of treatment are indicated above the blots; the weight of markers is indicated in KDa (red bands), and the presence of proteins is indicated by the green bands. SPT-5 and TFIIH (indicated to the right of the corresponding blot) were used as control.
DISCUSSION
In the brain, HIV establishes latent or active infection primarily in astrocytes and microglia cells where viral proteins are produced and shed (Kaul et al, 2001). Viral proteins such as Tat and gp120, and the immune response to viral proteins are thought to be primary contributors to HIV-related neurodegeneration (Kaul and Lipton, 2006). Importantly, it is now widely accepted that METH abuse by HIV-infected individuals leads to increased neurological deficits and neuronal dysfunction (Nath et al, 2001; Rippeth et al, 2004). In the current study, we examined the effects of METH on activation of the HIV LTR, which controls viral replication in the human host cells, including microglia. We have reported here a dose-dependent activation of the HIV LTR by METH that was independent of the presence of the HIV protein Tat in CHME-5 microglial cells transfected with an LTR-containing vector. METH also activated NF-κB, a cellular transcription factor and well established regulator of the HIV LTR. Disruption of NF-κB signaling blocked the induction of HIV LTR and consequently HIV gene expression. In addition, we have demonstrated here, using CHME-5 cells infected with HIV, that METH caused increased transcription of the HIV genome integrated as part of the human host genome, more closely recreating in vivo conditions. Under these conditions, METH was also capable of inducing NF-κB nuclear translocation, allowing transcription. Collectively, our data suggest that METH directly induces HIV expression in human microglia cells through activation of NF-κB.
We found that METH, applied at concentrations lower that those reported to happen in the brain of users, increased the activation of NF-κB signaling in human microglia cells. We detected significant effects in the 300–700 µM range in transfected cells and even below 50 µM in infected microglial cells. Estimated level in the spleen of single users is between 100 to 400 µM and binge users between 240 to 1144 µM (Tallóczy et al, 2008). Based on distribution and pharmacokinetic studies of METH in the human body, METH concentration in the spleen reaches its peak in 3.5 minutes and in the brain in 9 minutes; peak concentration in the spleen is twice that in brain (Volkow et al, 2010). However, they found the brain showed one of the slowest clearance rates. Taken together, it is feasible that METH doses in the human brain may occur in the 30–500 µM range, where we observed an effect of METH on HIV LTR in microglial cells.
The HIV LTR controls the expression of the viral genes, and the ability of METH to increase the activity of the HIV LTR in microglial cells would increase the production of viral proteins and possibly viral particles in the brain. The viral proteins gp120 and Tat have been shown to promote neurodegeneration (King et al, 2006; Mocchetti et al, 2007), and the combined exposure of METH and Tat and/or gp120 acts synergistically to promote neurotoxicity (Cadet and Krasnova, 2007). It is therefore plausible that one of the mechanisms that METH abuse in HIV-infected individuals increases neurodegeneration is through the increase viral gene expression. Our data supports the possibility that METH can act directly on human microglial cells and increase the activity of the HIV LTR, increasing viral protein production without necessarily increasing viral load.
Our findings are consistent with a previous study showing that METH can increase HIV replication in peripheral monocytes through an NF-κB-related mechanism (Toussi et al, 2009). The importance of NF-κB in regulating HIV gene expression is well documented, and other studies suggest METH can increase NF-κB toxicity in the brain (Asanuma and Cadet, 1998). Our data show that NF-κB is essential for the METH-related increase in HIV LTR activity, however, we also found a more modest but significant increase in transcriptional activity of NF-AT (results not shown), a cellular transcription factor that can influence the HIV LTR. This finding suggests that other cellular transcription factors may be altered by METH and influence viral transcription from the HIV LTR. We are presently investigating the role of METH in inducing epigenetic changes at the level HIV promoter leading to HIV reactivation as well as its specificity for particular cellular signaling pathways. Also, additional studies examining METH-related changes to the microglia transfactome are currently underway.
“Microglial activation” is a general term for the phenotypic changes of microglial cells in response to perturbations to the brain such as those that occur following injury or pathological conditions e.g. stroke, seizures, and viral infection. Activated microglia exhibit increased proliferation, hypertrophy, increased migration, increased phagocytosis, and increased production of pro-inflammatory molecules such as cytokines and chemokines (Tambuyzer et al, 2009). METH has previously been shown to activate microglia in rodent brain (Guilarte et al, 2003; LaVoie et al, 2004; Thomas et al, 2004), and humans who abuse METH have increased binding of a radiotracer for activated microglia compared to control subjects (Sekine et al, 2008). The mechanisms of microglial activation by METH are not known, but it is thought to involve glutamate receptor signaling (Thomas and Kuhn, 2005). We found that METH increases activation of rat primary microglial cells (Supplemental Figure 2), which suggests that microglia, in the absence of neurons capable of both releasing and responding to glutamate, can be activated by METH. In human microglial cells, we found that METH increased nuclear NF-κB and activation of a NF-κB-dependent reporter. In addition to being a primary cellular transcription factor for the HIV LTR, NF-κB contributes to the production of pro-inflammatory cytokines in activated microglia cells (Mattson, 2005). Therefore, it is plausible that HIV-infected microglial cells exposed to METH may contribute to local neurodegeneration by both the production of viral proteins and promoting local neuroinflammation through NF-κB activity and/or, more importantly, by the synergistic interaction between METH and the viral proteins. Additional studies are needed to examine role of NF-κB in METH and HIV-related neurodegeneration, and whether METH is capable to interact, directly or indirectly, with viral proteins in ways that may explain the increased HIV-related neurologic disorders observed in HIV patients consuming psychostimulants such as METH.
In conclusions, we show that METH can directly increase HIV LTR activity in human microglial cells in vitro through an NF-κB-dependent mechanism. These findings support a role of METH in promoting HIV gene expression in the progression of HIV-associated neurodegeneration.
Supplementary Material
Acknowledgements
The following reagents were obtained through the AIDS Research and Reference Program, Division of AIDS, NIAID, NIH: pCTatBL43.CS and pETatBL43.CS from Dr. Udaylumar Ranga. This work was supported by the Intramural Research Program at the National Institute on Drug Abuse and DP1 DA028869 to JK.
References
- Asanuma M, Cadet J. Methamphetamine-induced increase in striatal NF-kappaB DNA-binding activity is attenuated in superoxide dismutase transgenic mice. Brain Res Mol Brain Res. 1998;60:305–309. doi: 10.1016/s0169-328x(98)00188-0. [DOI] [PubMed] [Google Scholar]
- Buchacz K, McFarland W, Kellogg TA, Loeb L, Holmberg SD, Dilley J, Klausner JD. Amphetamine use is associated with increased HIV incidence among men who have sex with men in San Francisco. AIDS. 2005;19:1423–1424. doi: 10.1097/01.aids.0000180794.27896.fb. [DOI] [PubMed] [Google Scholar]
- Cadet JL, Krasnova IN. Interactions of HIV and methamphetamine: cellular and molecular mechanisms of toxicity potentiation. Neurotox Res. 2007;12:181–204. doi: 10.1007/BF03033915. [DOI] [PubMed] [Google Scholar]
- Cass W, Harned M, Peters L, Nath A, Maragos W. HIV-1 protein Tat potentiation of methamphetamine-induced decreases in evoked overflow of dopamine in the striatum of the rat. Brain Res. 2003;984:133–142. doi: 10.1016/s0006-8993(03)03122-6. [DOI] [PubMed] [Google Scholar]
- Chang L, Ernst T, Speck O, Grob CS. Additive effects of HIV and chronic methamphetamine use on brain metabolite abnormalities. Am J Psychiatry. 2005;162:361–369. doi: 10.1176/appi.ajp.162.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland K. Modulation of HIV-1 transcription by cytokines and chemokines. Mini Rev Med Chem. 2005;5:1093–1101. doi: 10.2174/138955705774933383. [DOI] [PubMed] [Google Scholar]
- Cox C, Cao S, Lu Y. Enhanced detection and study of murine norovirus-1 using a more efficient microglial cell line. Virol J. 2009;6:196. doi: 10.1186/1743-422X-6-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R, Childers M, Cherner M, Lazzaretto D, Letendre S, Grant I Group HNRC. Increased human immunodeficiency virus loads in active methamphetamine users are explained by reduced effectiveness of antiretroviral therapy. J Infect Dis. 2003;188:1820–1826. doi: 10.1086/379894. [DOI] [PubMed] [Google Scholar]
- Freeman P, Walker BC, Harris DR, Garofalo R, Willard N, Ellen JM Team ftATNfHAIb. Methamphetamine Use and Risk for HIV Among Young Men Who Have Sex With Men in 8 US Cities. Arch Pediatr Adolesc Med. 2011;165:736–740. doi: 10.1001/archpediatrics.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilin M, Mathes L, Podell M. Methamphetamine enhances cell-associated feline immunodeficiency virus replication in astrocytes. J Neurovirol. 2002;8:240–249. doi: 10.1080/13550280290049660. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Guilarte TR, Nihei MK, McGlothan JL, Howard AS. Methamphetamine-induced deficits of brain monoaminergic neuronal markers: distal axotomy or neuronal plasticity. Neuroscience. 2003;122:499–513. doi: 10.1016/s0306-4522(03)00476-7. [DOI] [PubMed] [Google Scholar]
- Janabi N, Peudenier S, Héron B, Ng KH, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- Jayanthi S, Deng X, Ladenheim B, McCoy M, Cluster A, Cai N, Cadet J. Calcineurin/NFAT-induced up-regulation of the Fas ligand/Fas death pathway is involved in methamphetamine-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2005;102:868–873. doi: 10.1073/pnas.0404990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410:988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- Kaul M, Lipton SA. Mechanisms of neuronal injury and death in HIV-1 associated dementia. Curr HIV Res. 2006;4:307–318. doi: 10.2174/157016206777709384. [DOI] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 2006;25:3596–3604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JE, Eugenin EA, Buckner CM, Berman JW. HIV tat and neurotoxicity. Microbes Infect. 2006;8:1347–1357. doi: 10.1016/j.micinf.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Langford D, Adame A, Grigorian A, Grant I, McCutchan JA, Ellis RJ, Marcotte TD, Masliah E Group HNRC. Patterns of selective neuronal damage in methamphetamine-user AIDS patients. J Acquir Immune Defic Syndr. 2003;34:467–474. doi: 10.1097/00126334-200312150-00004. [DOI] [PubMed] [Google Scholar]
- LaVoie M, Card J, Hastings T. Microglial activation precedes dopamine terminal pathology in methamphetamine-induced neurotoxicity. Exp Neurol. 2004;187:47–57. doi: 10.1016/j.expneurol.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Lee SC, Hatch WC, Liu W, Brosnan CF, Dickson DW. Productive infection of human fetal microglia in vitro by HIV-1. Ann N Y Acad Sci. 1993a;693:314–316. doi: 10.1111/j.1749-6632.1993.tb26295.x. [DOI] [PubMed] [Google Scholar]
- Lee SC, Hatch WC, Liu W, Kress Y, Lyman WD, Dickson DW. Productive infection of human fetal microglia by HIV-1. Am J Pathol. 1993b;143:1032–1039. [PMC free article] [PubMed] [Google Scholar]
- Mallardo M, Dragonetti E, Baldassarre F, Ambrosino C, Scala G, Quinto I. An NF-kappaB site in the 5'-untranslated leader region of the human immunodeficiency virus type 1 enhances the viral expression in response to NF-kappaB-activating stimuli. J Biol Chem. 1996;271:20820–20827. doi: 10.1074/jbc.271.34.20820. [DOI] [PubMed] [Google Scholar]
- Maragos WF, Young KL, Turchan JT, Guseva M, Pauly JR, Nath A, Cass WA. Human immunodeficiency virus-1 Tat protein and methamphetamine interact synergistically to impair striatal dopaminergic function. J Neurochem. 2002;83:955–963. doi: 10.1046/j.1471-4159.2002.01212.x. [DOI] [PubMed] [Google Scholar]
- Mattson MP. NF-kappaB in the survival and plasticity of neurons. Neurochem Res. 2005;30:883–893. doi: 10.1007/s11064-005-6961-x. [DOI] [PubMed] [Google Scholar]
- Mocchetti I, Nosheny RL, Tanda G, Ren K, Meyer EM. Brain-derived neurotrophic factor prevents human immunodeficiency virus type 1 protein gp120 neurotoxicity in the rat nigrostriatal system. Ann N Y Acad Sci. 2007;1122:144–154. doi: 10.1196/annals.1403.010. [DOI] [PubMed] [Google Scholar]
- Nath A, Maragos W, Avison M, Schmitt F, Berger J. Acceleration of HIV dementia with methamphetamine and cocaine. J Neurovirol. 2001;7:66–71. doi: 10.1080/135502801300069737. [DOI] [PubMed] [Google Scholar]
- Pereira L, Bentley K, Peeters A, Churchill M, Deacon N. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi D, Mitra D. HIV-1 long terminal repeat promoter regulated dual reporter: potential use in screening of transcription modulators. Anal Biochem. 2007;360:315–317. doi: 10.1016/j.ab.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Rippeth JD, Heaton RK, Carey CL, Marcotte TD, Moore DJ, Gonzalez R, Wolfson T, Grant I, Group H. Methamphetamine dependence increases risk of neuropsychological impairment in HIV infected persons. J Int Neuropsychol Soc. 2004;10:1–14. doi: 10.1017/S1355617704101021. [DOI] [PubMed] [Google Scholar]
- Sanchez JF, Sniderhan LF, Williamson AL, Fan S, Chakraborty-Sett S, Maggirwar SB. Glycogen synthase kinase 3beta-mediated apoptosis of primary cortical astrocytes involves inhibition of nuclear factor kappaB signaling. Mol Cell Biol. 2003;23:4649–4662. doi: 10.1128/MCB.23.13.4649-4662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Ouchi Y, Sugihara G, Takei N, Yoshikawa E, Nakamura K, Iwata Y, Tsuchiya K, Suda S, Suzuki K, Kawai M, Takebayashi K, Yamamoto S, Matsuzaki H, Ueki T, Mori N, Gold M, Cadet J. Methamphetamine causes microglial activation in the brains of human abusers. J Neurosci. 2008;28:5756–5761. doi: 10.1523/JNEUROSCI.1179-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng P, Ladenheim B, Moran T, Wang X, Cadet J. Methamphetamine-induced neurotoxicity is associated with increased striatal AP-1 DNA-binding activity in mice. Brain Res Mol Brain Res. 1996;42:171–174. doi: 10.1016/s0169-328x(96)00192-1. [DOI] [PubMed] [Google Scholar]
- Tallóczy Z, Martinez J, Joset D, Ray Y, Gácser A, Toussi S, Mizushima N, Nosanchuk J, Nosanchuk J, Goldstein H, Loike J, Sulzer D, Santambrogio L. Methamphetamine inhibits antigen processing, presentation, and phagocytosis. PLoS Pathog. 2008;4:e28. doi: 10.1371/journal.ppat.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambuyzer BR, Ponsaerts P, Nouwen EJ. Microglia: gatekeepers of central nervous system immunology. J Leukoc Biol. 2009;85:352–370. doi: 10.1189/jlb.0608385. [DOI] [PubMed] [Google Scholar]
- Tang H, Kuhen K, Wong-Staal F. Lentivirus replication and regulation. Annu Rev Genet. 1999;33:133–170. doi: 10.1146/annurev.genet.33.1.133. [DOI] [PubMed] [Google Scholar]
- Theodore S, Cass W, Maragos W. Methamphetamine and human immunodeficiency virus protein Tat synergize to destroy dopaminergic terminals in the rat striatum. Neuroscience. 2006a;137:925–935. doi: 10.1016/j.neuroscience.2005.10.056. [DOI] [PubMed] [Google Scholar]
- Theodore S, Cass WA, Nath A, Steiner J, Young K, Maragos WF. Inhibition of tumor necrosis factor-alpha signaling prevents human immunodeficiency virus-1 protein Tat and methamphetamine interaction. Neurobiol Dis. 2006b;23:663–668. doi: 10.1016/j.nbd.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Thomas D, Kuhn D. MK-801 and dextromethorphan block microglial activation and protect against methamphetamine-induced neurotoxicity. Brain Res. 2005;1050:190–198. doi: 10.1016/j.brainres.2005.05.049. [DOI] [PubMed] [Google Scholar]
- Thomas D, Walker P, Benjamins J, Geddes T, Kuhn D. Methamphetamine neurotoxicity in dopamine nerve endings of the striatum is associated with microglial activation. J Pharmacol Exp Ther. 2004;311:1–7. doi: 10.1124/jpet.104.070961. [DOI] [PubMed] [Google Scholar]
- Toussi S, Joseph A, Zheng J, Dutta M, Santambrogio L, Goldstein H. Short communication: Methamphetamine treatment increases in vitro and in vivo HIV replication. AIDS Res Hum Retroviruses. 2009;25:1117–1121. doi: 10.1089/aid.2008.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ, Shumay E, Telang F, Thanos PK, Alexoff D. Distribution and pharmacokinetics of methamphetamine in the human body: clinical implications. PLoS One. 2010;5:e15269. doi: 10.1371/journal.pone.0015269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Watanabe Y, Osugi T, Ikemoto M, Hirata M, Miki N. In situ DNA-protein binding: a novel method for detecting DNA-binding activity of transcription factor in brain. Neurosci Lett. 1992;146:25–28. doi: 10.1016/0304-3940(92)90163-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.