tRNA Levels and Codon Bias Affect the Fidelity and Efficiency of Translation
The two major steps of gene translation are transcription of the gene to mRNA molecules, and their translation to proteins by the ribosomes. The elongation part of translation includes the iterative decoding of the gene codons by the ribosome with the aid of tRNA molecules. Each codon is recognized by a set of tRNA molecules that are charged with the amino acid encoded by it (Alberts et al., 2002), and the translation time of a codon is usually positively correlated with the abundance of the tRNA molecules recognizing it. Thus, increasing expression levels of the tRNA molecules recognizing a codon, or replacing a codon with a different one recognized by tRNA molecules with higher expression levels, should usually have positive effect on its translation rate (Gustafsson et al., 2004; Tuller et al., 2010).
Missense errors in translation occur at a rate of 1 per 103–104 (Ogle and Ramakrishnan, 2005; Kramer and Farabaugh, 2007), i.e., assuming average protein length of 400 codons, around 18% of the proteins contain at least one missense substitution. In addition, roughly 10–50% of random substitutions disrupt protein function, usually due to loss of folding mutations (Pakula and Sauer, 1989; Markiewicz et al., 1994; Guo et al., 2004; Bloom et al., 2006). It is known that the speed by which codons are translated can also affect the rate of missense translation errors, and thus the folding of the translated protein, resulting in misfolded toxic proteins (Akashi, 1994; Bloom et al., 2006; Zhou et al., 2009). Thus, the probability of missense translation errors of codons that are recognized by tRNA genes with lower concentrations is usually higher than in codons with higher concentrations, since in these cases with higher probability a wrong tRNA replaces the right one (Akashi, 1994; Zhou et al., 2009).
Global Up-Regulation of tRNA Levels in Cancer
It is known that in many cancerous cells there is an increased growth rate that is regulated by signals related to proliferation, metabolism, and protein synthesis (White, 2005; Gillies et al., 2008; Jones and Thompson, 2009; Mei et al., 2010b; Cairns et al., 2011). This phenomenon may be partially caused by global up-regulation of tRNA molecules. The trigger for these global signals can be the down-regulation of retinoblastoma proteins, p53 and ARF, which cause up-regulation of RNA polymerases I and III, and oncoproteins such as Myc that stimulate the transcription of rRNA and tRNA genes (Cabarcas and Schramm, 2011).
Indeed, the effect of tRNA on tumorigenesis has been previously reported (Berns, 2008; Pavon-Eternod et al., 2009). For example, Pavon-Eternod et al. (2009) used tRNA chips for measuring the expression levels of tRNA molecules to show that in breast cancer there is global over-expression of tRNA species. Specifically, the expression levels of nuclear-encoded tRNAs increase by up to threefold, and mitochondrial-encoded tRNAs increase by up to fivefold in breast cancer. It was also shown that in general these changes maintain the ranking of the expression levels of tRNA genes, as there is significant correlation between the tRNA levels in cancerous and healthy cells (Mahlab et al., 2012). Similar results were obtained for other components of the translation machinery (such as aminoacyl-tRNA synthetases; Vellaichamy et al., 2009).
In this subsection we emphasized the global changes in the expression levels of tRNA genes. However, it was also reported that specific pathways and genes relevant to cancer undergo increased differential regulation of translation in cancer due to point mutations in specific genes’ coding sequences, or genomic changes that effect tRNA levels. For example, it was shown that tRNA isoacceptor over-expression may increase the translational efficiency of genes relevant to cancer development, progression (Pavon-Eternod et al., 2009), and apoptosis (Mei et al., 2010a,b). In addition, it was demonstrated that many of the gain-of function and dominant-negative mutations in the tumor suppressor gene TP53 increase its translation efficiency also when considering the cancerous changes in the tRNA pool (Waldman et al., 2009).
tRNA Genes, Translation Fidelity, and Neurodegeneration
As mentioned above, mistranslation-induced protein misfolding is a dominant constraint on coding sequence evolution (Drummond and Wilke, 2008).
Misfolded proteins is one of the causes of neurodegeneration (Dobson, 2003; Selkoe, 2003; Ross and Poirier, 2004; Lee et al., 2006). For example, it was shown that low levels of mischarged tRNAs can lead to an intracellular accumulation of misfolded proteins in neurons (Lee et al., 2006); similarly, mutations in other components of the translation machinery, such as Aminoacyl-tRNA synthetases, may lead to similar problems (Antonellis and Green, 2008). Indeed, it was shown that mutations in broadly expressed genes involved in translation and protein folding produce brain-specific phenotypes (Zhao et al., 2005; Lee et al., 2006), suggesting that neural tissues are more sensitive to protein misfolding; thus, this sensitivity corresponds to lower mutation rates (or evolutionary rates) in animal neuronal genes (Zhang and Li, 2004; Lee et al., 2006; Wang et al., 2007; Drummond and Wilke, 2008; Tuller et al., 2008). These results suggest that the disruption of translational fidelity in terminally differentiated neurons leads to the accumulation of misfolded proteins and cell death, and provides a novel mechanism underlying neurodegeneration.
The Diagnostic Potential of tRNA Genes and Coding Sequence Mutations and Corresponding Challenges
The results reviewed in this paper suggest that the expression levels of tRNA genes can be used as biomarkers for diseases such as cancer. One challenge related to this point is to develop robust and efficient approaches for measuring tRNA levels. This is not trivial due to two major reasons: first, tRNA molecules undergo many RNA modifications making the mapping of their deep sequencing reads more challenging (Gustilo et al., 2008; Mahlab et al., 2012). Second, the strong folding of the tRNA molecules decreases their hybridization to DNA chips. Currently, one of the most reliable approaches for measuring tRNA levels is by DNA chips designed specifically for this purpose by Prof. Tao Pan (Dittmar et al., 2006; Pavon-Eternod et al., 2009). However other methods for measuring tRNA levels, e.g., by using liquid chromatography mass spectrometry and signature digestion products (Castleberry and Limbach, 2010), are available.
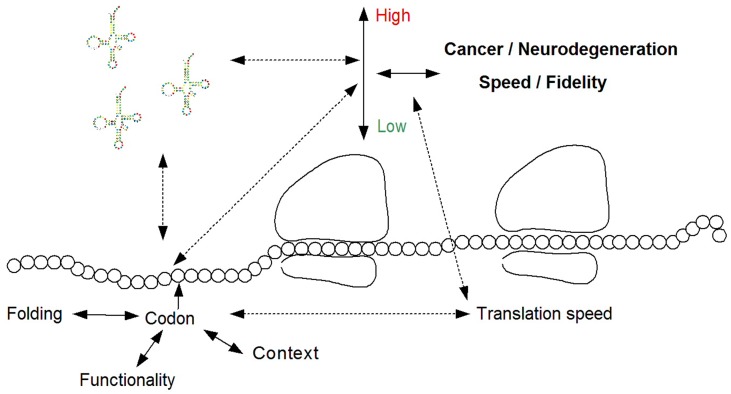
In addition, diagnosis related to cancer and neurodegeneration can be performed based on non-synonymous, but also synonymous mutations and SNPs, by predicting their effect on the translation rate of a gene. Such a diagnostic tool should consider the adaptation of the mutated codon, or the codon with a SNP, to the human tRNA pool; this can be done by measuring the tRNA levels in the relevant tissue (as mentioned above) or by using a proxy such as the tRNA copy number (Mahlab et al., 2012). In addition, the diagnostic tool should consider amongst others the region of the mutation/SNP within the coding sequence, the codons, and nucleotides surrounding it, and also the effect of the relevant codon on the folding of the mRNA and the charge of the protein it encodes (Tuller et al., 2011). To this end, biophysical predictors of translation efficiency can be employed (Tuller et al., 2010). Finally, the diagnosis should be based on the functionality of the gene; for example, the effect of elevated/decreased adaptation of a codon to the tRNA pool is expected to be opposite for tumor suppressors and oncogenes (Figure 1).
Figure 1.
The relation between translation efficiency and the pathogenesis of cancer and neurodegeneration, and the possible diagnosis of these diseases based on tRNA molecules, and/or the effect of the tRNA pool on mutations and SNPs. Such a diagnostic tool should consider the tRNA pool and the adaptation of the mutated codon to it, the biophysics of translation including the context of the codon and the effect of the mutation on the mRNA folding, and the functionality of the mutated protein.
References
- Akashi H. (1994). Synonymous codon usage in Drosophila melanogaster: natural selection, and translational accuracy. Genetics 136, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2002). Molecular Biology of the Cell. New York: Garland Science [Google Scholar]
- Antonellis A., Green E. D. (2008). The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet. 9, 87–107 10.1146/annurev.genom.9.081307.164204 [DOI] [PubMed] [Google Scholar]
- Berns A. (2008). A tRNA with oncogenic capacity. Cell 133, 29–30 10.1016/j.cell.2008.03.017 [DOI] [PubMed] [Google Scholar]
- Bloom J. D., Labthavikul S. T., Otey C. R., Arnold F. H. (2006). Protein stability promotes evolvability. Proc. Natl. Acad. Sci. U.S.A. 103, 5869–5874 10.1073/pnas.0510098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabarcas S., Schramm L. (2011). RNA polymerase III transcription in cancer: the BRF2 connection. Mol. Cancer 10, 47. 10.1186/1476-4598-10-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns R. A., Harris I. S., Mak T. W. (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]
- Castleberry C. M., Limbach P. A. (2010). Relative quantitation of transfer RNAs using liquid chromatography mass spectrometry and signature digestion products. Nucleic Acids Res. 38, e162. 10.1093/nar/gkq578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K. A., Goodenbour J. M., Pan T. (2006). Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221. 10.1371/journal.pgen.0020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C. M. (2003). Protein folding and misfolding. Nature 426, 884–890 10.1038/nature02261 [DOI] [PubMed] [Google Scholar]
- Drummond D. A., Wilke C. O. (2008). Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 10.1016/j.cell.2008.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies R. J., Robey I., Gatenby R. A. (2008). Causes, and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 49(Suppl. 2), 24S–42S 10.2967/jnumed.107.047258 [DOI] [PubMed] [Google Scholar]
- Guo H. H., Choe J., Loeb L. A. (2004). Protein tolerance to random amino acid change. Proc. Natl. Acad. Sci. U.S.A. 101, 9205–9210 10.1073/pnas.0405878101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S., Minshull J. (2004). Codon bias, and heterologous protein expression. Trends Biotechnol. 22, 346–353 10.1016/j.tibtech.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Gustilo E. M., Vendeix F. A., Agris P. F. (2008). tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 11, 134–140 10.1016/j.mib.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. G., Thompson C. B. (2009). Tumor suppressors, and cell metabolism: a recipe for cancer growth. Genes Dev. 23, 537–548 10.1101/gad.1756509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E. B., Farabaugh P. J. (2007). The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96 10.1261/rna.294907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Beebe K., Nangle L. A., Jang J., Longo-Guess C. M., Cook S. A., et al. (2006). Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 10.1038/nature05096 [DOI] [PubMed] [Google Scholar]
- Mahlab S., Tuller T., Linial M. (2012). Conservation of the relative tRNA composition in healthy, and cancerous tissues. RNA 18, 640–652 10.1261/rna.030775.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz P., Kleina L. G., Cruz C., Ehret S., Miller J. H. (1994). Genetic studies of the lac repressor. XIV. Analysis of. 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J. Mol. Biol. 240, 421–433 10.1006/jmbi.1994.1458 [DOI] [PubMed] [Google Scholar]
- Mei Y., Stonestrom A., Hou Y. M., Yang X. (2010a). Apoptotic regulation and tRNA. Protein Cell 1, 795–801 10.1007/s13238-010-0107-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G. et al. (2010b). tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 37, 668–678 10.1016/j.molcel.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J. M., Ramakrishnan V. (2005). Structural insights into translational fidelity. Annu. Rev. Biochem. 74, 129–177 10.1146/annurev.biochem.74.061903.155440 [DOI] [PubMed] [Google Scholar]
- Pakula A. A., Sauer R. T. (1989). Genetic analysis of protein stability, and function. Annu. Rev. Genet. 23, 289–310 10.1146/annurev.ge.23.120189.001445 [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M. R., Pan T. (2009). tRNA over-expression in breast cancer, and functional consequences. Nucleic Acids Res. 37, 7268–7280 10.1093/nar/gkp787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. A., Poirier M. A. (2004). Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–S17 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- Selkoe D. J. (2003). Folding proteins in fatal ways. Nature 426, 900–904 10.1038/nature02264 [DOI] [PubMed] [Google Scholar]
- Tuller T., Kupiec M., Ruppin E. (2008). Evolutionary rate and gene expression across different brain regions. Genome Biol. 9, R142. 10.1186/gb-2008-9-9-r142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T., Veksler-Lublinsky I., Gazit N., Kupiec M., Ruppin E., Ziv-Ukelson M. (2011). Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol. 12, R110. 10.1186/gb-2011-12-11-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T., Waldman Y. Y., Kupiec M., Ruppin E. (2010). Translation efficiency is determined by both codon bias, and folding energy. Proc. Natl. Acad. Sci. U.S.A. 107, 3645–3650 10.1073/pnas.0909910107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellaichamy A., Sreekumar A., Strahler J. R., Rajendiran T., Yu J., Varambally S., et al. (2009). Proteomic interrogation of, androgen action in prostate cancer cells reveals roles of aminoacyl tRNA synthetases. PLoS One 4, e7075. 10.1371/journal.pone.0007075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman Y. Y., Tuller T., Sharan R., Ruppin E. (2009). TP53 cancerous mutations exhibit selection for translation efficiency. Cancer Res. 69, 8807–8813 10.1158/0008-5472.CAN-09-1653 [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Chien H. C., Osada N., Hashimoto K., Sugano S., Gojobori T., et al. (2007). Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 5, e13. 10.1371/journal.pbio.0050013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J. (2005). RNA polymerases I, and III, growth control, and cancer. Nat. Rev. Mol. Cell. Biol. 6, 69–78 10.1038/nrm1551 [DOI] [PubMed] [Google Scholar]
- Zhang L., Li W. H. (2004). Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol. Biol. Evol. 21, 236–239 10.1093/molbev/msh010 [DOI] [PubMed] [Google Scholar]
- Zhao L., Longo-Guess C., Harris B. S., Lee J. W., Ackerman S. L. (2005). Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat. Genet. 37, 974–979 10.1038/ng1620 [DOI] [PubMed] [Google Scholar]
- Zhou T., Weems M., Wilke C. O. (2009). Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 26, 1571–1580 10.1093/molbev/msp070 [DOI] [PMC free article] [PubMed] [Google Scholar]