Abstract
Clear cell-type renal cell carcinomas (clear RCC) are characterized almost universally by loss of heterozygosity on chromosome 3p, which usually involves any combination of three regions: 3p25-p26 (harboring the VHL gene), 3p12-p14.2 (containing the FHIT gene), and 3p21-p22, implying inactivation of the resident tumor-suppressor genes (TSGs). For the 3p21-p22 region, the affected TSGs remain, at present, unknown. Recently, the RAS association family 1 gene (isoform RASSF1A), located at 3p21.3, has been identified as a candidate lung and breast TSG. In this report, we demonstrate aberrant silencing by hypermethylation of RASSF1A in both VHL-caused clear RCC tumors and clear RCC without VHL inactivation. We found hypermethylation of RASSF1A's GC-rich putative promoter region in most of analyzed samples, including 39 of 43 primary tumors (91%). The promoter was methylated partially or completely in all 18 RCC cell lines analyzed. Methylation of the GC-rich putative RASSF1A promoter region and loss of transcription of the corresponding mRNA were related causally. RASSF1A expression was reactivated after treatment with 5-aza-2′-deoxycytidine. Forced expression of RASSF1A transcripts in KRC/Y, a renal carcinoma cell line containing a normal and expressed VHL gene, suppressed growth on plastic dishes and anchorage-independent colony formation in soft agar. Mutant RASSF1A had reduced growth suppression activity significantly. These data suggest that RASSF1A is the candidate renal TSG gene for the 3p21.3 region.
Loss of heterozygosity (LOH) of chromosome 3p is the most common event in clear cell-type renal cell carcinomas (clear RCC). It involves one or more of the three major commonly deleted regions (1–3) that may be subdivided further by using pioneering stringent criteria for LOH analysis (4–6).
The most frequent form of hereditary RCC is the familial VHL cancer syndrome (7). The VHL gene was mapped to 3p25 and was isolated by positional cloning (8). It was inactivated by intragenic and genomic deletions and various kinds of mutations in 100% of analyzed VHL families and in a large portion of sporadic clear RCC (9). Inactivation of VHL by promoter hypermethylation (10) in sporadic clear RCC was noted also and was observed quite frequently (20%). However, in other sporadic clear RCC (reaching in some studies 30–50%) and some non-VHL clear RCC families (11, 12), VHL was not affected (see review in ref. 13).
Another commonly deleted region was found at 3p12-p14. Although the overall frequency of the specific LOH is not high, chromosome transfer experiments indicated that the 3p12-p14 region could suppress the tumorigenic properties of some clear RCC cell lines (14), implying the presence of a gene or genes involved in the origin and/or development of clear RCC. This role may be attributed to the FHIT gene at 3p14.3 (15), and/or other tumor-suppressor genes (TSGs) residing in 3p12.
Multiple studies have identified LOH on 3p21-p22 as the most frequent 3p loss in renal tumor development (4, 5, 16, 17). Recent data indicate that this region consists of two distinct subregions: centromeric 3p21.3 and telomeric 3p21.3-p22, with both regions being deleted frequently (5). The same broad region (3p21-p22) was implicated in tumorigenesis of several other histologically different malignancies, including homozygous deletions at 3p21.3 in lung and breast tumors (4, 18).
Recently, a new RAS effector homolog gene, RASSF1, was located within the critical lung and breast cancer deletion region at centromeric 3p21.3 (18, 19). Two distinct GC-rich promoters produced RASSF1A and RASSF1C transcripts. RASSF1A mRNA was missing in most analyzed small-cell lung cancer cell lines because of methylation of the RASSF1A putative promoter region (19–21). Interestingly, the RASSF1A protein interacts with the DNA repair protein, XPA (19). As seen with the mouse Ras effector protein Nore1 and its rat ortholog Maxp1, the human RASSF1A isoform (but not RASSF1C) has high homology to the cysteine-rich diacylglycerol/phorbolester-binding domain, also known as the protein kinase C conserved region 1. Reexpression of the RASSF1A transcript in lung carcinoma cells reduced colony formation, suppressed anchorage-independent growth, and inhibited tumor formation in nude mice (19, 20).
In this report, we addressed the possible role of this putative lung and breast TSG in renal tumorigenesis. Our data suggest that the RASSF1A gene is a candidate renal TSG for the centromeric 3p21.3 region.
Materials and Methods
Cell Lines.
The 786-0 and ACHN RCC cell lines were purchased from ATCC. A498, CAKI 1, CAKI 2, HN4, HN51, KH39, KRC/Y, TK10, and TK164 were described in ref. 5. The L24 line and L&U3-L&U6 somatic-cell hybrid clones were established as reported (22).
Analysis of RASSF1A Promoter Methylation.
Bisulfite DNA treatment has been described (22). Amplification of bisulfite-converted RASSF1A promoter sequences was carried with an upstream primer B-Ras-E-1D, GTA GGT TAA GTG TGT TGT TTT AGT A, and a downstream primer B-Ras-E-4R, ACC AAA AAC CAA CTA CCR TAT AAA ATT. Nested PCR was conducted by using primers B-Ras-E-2D, AGT ATA GTA AAG TTG GTT TTT AGA AAT A (upstream), and B-Ras-E-6R, AAT ACC AAC TCC CRC AAC TCA ATA AAC TCA (downstream). For amplification of RASSF1C bisulfite-treated promoter sequences, we used primers BISLU7–1D, AGT GAT GAG GTT ATT TTT GGG GG; BISLU7–1R, CTC TTA ACT ACA ATA ACC ACT ACT C (first PCR); BISLU7–2D, TTG TTT TAA TGA GAT AAG AGT TAG ATT; and BISLU7–2R, TCA TAC TAC TCC AAA TCA TTT CAA AAA (nested PCR). Each PCR amplification consisted of 35 cycles; 94°C for 30 sec, 58°C for 30 sec, and 72°C for 40 sec. For combined bisulfite-restriction analysis (COBRA), PCR products were cut with BstUI or MspI and separated on a 4–20% TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) polyacrylamide gel (Invitrogen). The principles of the COBRA procedure were described by Xiong and Laird (23).
For sequencing of PCR products obtained from bisulfite-treated genomic DNA, the fragments were subcloned into the pCR2.1 plasmid (TA cloning kit, Invitrogen) and several clones for each cell line were sequenced by using a 373 stretch DNA sequencer (Applied Biosystems).
Methylation-specific PCR was performed as described (20).
RASSF1A Expression Study.
The 380-bp HindIII/SmaI fragment of the RASSF1A cDNA insert that included exons 1α and 2αβ (19) was subcloned into pBluescript II SK(+) vector. A 20-bp deletion was created within the cDNA insert by digestion of the plasmid with XcaI/MscI and religation. The resulting plasmid was cut with SmaI, and artificial run-off RNA was generated by using the T7 MEGAscript kit (Ambion, Austin, TX). VHL group 7 cDNA was digested with BstEII and XbaI. A BstEII/XbaI fragment, containing exon 2 and part of exon 3 cDNA, was subcloned into the pBluescript II SK(+) vector. A 40-bp deletion was created within the cDNA insert by digestion of the plasmid with BspEI/Tth111I and religation. The resulting plasmid was cut with SacII, and artificial run-off RNA was generated by using the T7 MEGAscript kit (Ambion). To study expression of the RASSF1A and VHL genes, predetermined amounts of these RNAs were added to 1.5 μg of total RNA or 0.5 μg of poly(A)+ RNA.
Reverse transcription was conducted by using the RETROscript kit (Ambion). cDNA was used for PCR amplification (95°C for 30 sec and 72°C for 40 sec, using from 20 to 35 cycles, depending on the amount of PCR product). The primers used were F2.1A-1D, AGC GCC CAA AGC CAG CGA AGC ACG G and F2.1A-1R, CCC GCA ACA GTC CAG GCA GAC GAG C. When higher sensitivity was required, a nested PCR was performed by using the primers F2.1A-2D, TGA GCT CAT TGA GCT GCG GGA GCT GGC A and F2.1A-2R, GGC AGC GGT AGT GGC AGG TGA ACT TGC A. PCR products were separated in a 4–20% TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) polyacrylamide gel (Invitrogen).
Mutation Screening.
PCR amplification of exons 1α and 2αβ was performed by using genomic primers 1Fwup, CTC TAG CAC AGT AAA GCT GGC; 2ARv, AGC CAC CTC TGC TCA TCT GTG G, and nested primer 1Fw, CTC TCC TCA GCT CCT TCC (for exon 1); plus intronic primers 4Fw, CTG GGG CTT CCC TTC AAT CTC; 4Rv, AGC TCC AGC GAC CGC TTC; and 3ARv, CAA GCG CAC AAG AGT GGC CTC for exon 2. The cycling conditions were 4 min at 94°C, 30 sec at 94°C, 30 sec at 60°C, and 1 min at 72°C, for 35 cycles, followed by 7 min at 72°C. The PCR-amplified DNA was cloned and sequenced.
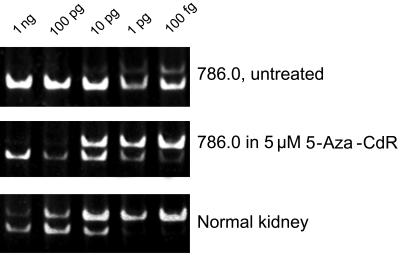
Reexpression of RASSF1A After 5-Aza-2′-deoxycytidine (5-Aza-CdR) Treatment.
RCC cells (786-0) were treated with 5 μM of 5-Aza-CdR for 4 days, and the total RNA from these cells was analyzed along with RNA from untreated 786-0 cells and normal kidney RNA, purchased from Invitrogen. For each of these three samples, 5 reverse transcription (RT) reactions were set up, each containing different amounts of artificial RASSF1A RNA (1 ng, 100 pg, 10 pg, 1 pg, or 100 fg). Total cellular RNA (1.5 μg) was used per RT reaction. PCR was performed as described, and the products were run on 4–20% polyacrylamide gels (Invitrogen).
Cell-Growth Assays.
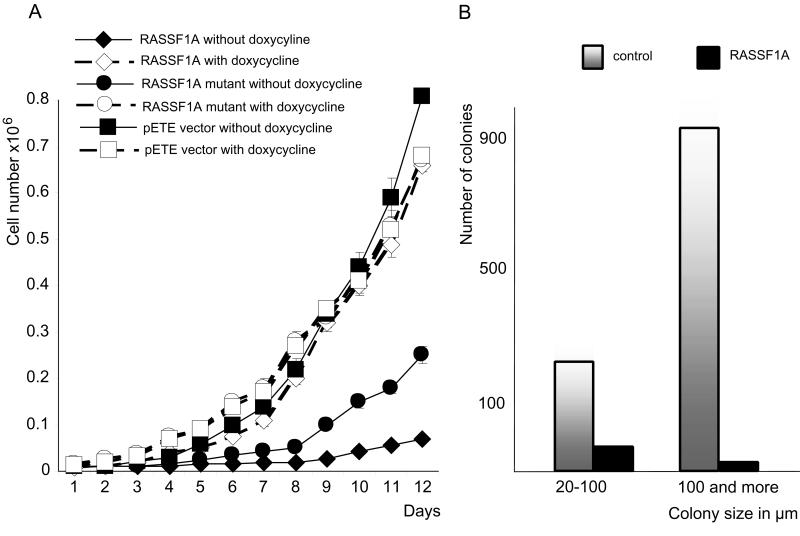
RASSF1A cDNA was cloned into the pcDNA3.1 (Invitrogen) expression vector and transfected into KRC/Y RCC cells by using lipofectamine. Geneticin-resistant colonies were expanded and 5 × 104 cells were mixed with 1.5 ml of agar (0.3% in DMEM) and overlaid on 5 ml of 0.5% plating agar in DMEM (in 60 × 15-mm Petri dishes). After 10 days, the plates were stained with neutral red, and colonies with a diameter of greater than 100 μm and less than 100 μm were counted separately. To study growth on plastic Petri dishes, wild-type and mutant RASSF1A, containing Cys65Arg and Val211Ala amino acid changes were, cloned into a pETE episomal tetracycline-regulated vector (J.L. and E.Z., unpublished results). These constructs and the original pETE vector were transfected into a KRC/Y cell line constitutively producing tTA (A.P. and E.Z., unpublished results). In the absence of tetracycline or doxycycline (dox), RASSF1A genes were expressed, whereas in the presence of 500 ng/ml of dox, no expression occurred. G4180-resistant clones were selected and wild-type RASSF1A clone 1, mutant RASSF1A clone 2, and pETE clone 1 were used in further experiments. Growth of the cells was monitored daily in the presence and absence of dox. Cells from 3 parallel dishes were counted.
Results
RASSF1A Is Hypermethylated in Primary Clear RCC Tumors and in RCC Cell Lines.
A moderately GC-rich putative promoter of the RASSF1A transcript was located immediately downstream from the 3′ end of the BLU gene (Fig. 4A, which is published as supplemental data on the PNAS web site, www.pnas.org). Promoter methylation was analyzed by COBRA or methylation-specific PCR by using DNA samples from primary clear RCC tumors and matching normal kidney DNA. Normal kidney DNA was isolated from visually unaffected renal parenchyma, as shown by staining with hematoxylin and eosin. Control normal tissue was free of tumor cells, whereas contamination of tumor by normal cells was less than 30%. RASSF1A–methylation-specific PCR analysis was performed as described elsewhere (20). For COBRA analysis of the putative RASSF1A promoter, we used four BstUI CGCG-recognition sites (Fig. 4B, which is published as supplemental data on the PNAS web site), which remain preserved only after bisulfite treatment if both CG dinucleotides (CpG) within the DNA recognition sequence are methylated. The inability of BstUI to cut the PCR products would indicate that the original genomic fragment is unmethylated. The same genomic fragment also contains MspI sites (CCGG), which would be eliminated after bisulfite conversion in both methylated and unmethylated DNA. Therefore, the MspI digest was used to ensure that this conversion was complete.
We found complete or partial methylation in 39 of 43 tumor samples (91%). In the remaining four tumors, no mutations or single-nucleotide polymorphisms were found within exons 1α and 2αβ after sequencing. For 29 of these 43 samples (67%), the normal kidney DNA was available from the same patient. According to methylation-specific PCR, in 27 of these 29 tumor-normal pairs (93%), tumor DNA was hypermethylated always, compared with the matching normal DNA, which in some cases also was methylated but to a lesser extent. Six of these tumor-normal bisulfite-treated DNA pairs were amplified by PCR, subcloned, and sequenced (Fig. 5, which is published as supplemental data on the PNAS web site). In each pair, tumor DNA was hypermethylated, compared with normal matching kidney DNA. Contamination of tumor with normal kidney DNA in these preparations cannot be excluded, thus the actual difference in these pairs is likely to be even greater. As we could observe partial methylation in normal control DNA (DNA from normal tissue in close proximity to tumor tissue), these results indicate that silencing the RASSF1A promoter might be an early event in the carcinogenesis of some clear RCC. A partial or complete methylation was observed for all 18 studied RCC lines (Fig. 1).
Figure 1.
Correlation of promoter methylation and RASSF1A expression in clear RCC tumor cell lines. (A) COBRA. For COBRA analysis, PCR fragments were amplified by using bisulfite-treated genomic DNA and nested primers shown in Fig. 4B, which is published as supplemental data on the PNAS web site. PCR fragments were digested with BstUI or MspI. BstUI cuts revealed the presence of methylated CpGs in the original DNA. The absence of MspI cuts confirmed proper conversion of unmethylated cytidines. (B) Sequencing of bisulfite-treated amplified DNA. For detailed methylation analysis of the COBRA PCR products, the fragments were subcloned, and several plasmid clones were sequenced for each cell line. Yellow and green squares represent unmethylated and methylated CpGs, respectively. Thirty-two CpGs located between nested COBRA primers (Fig. 4B, which is published as supplemental data on the PNAS web site) are numbered, starting from NT 17782. CpGs located within BstUI-cutting sites are highlighted in gray. The positions of the 5′ end of RASSF1A cDNA and the RASSF1A initiation codon are marked. (C) RT-PCR quantitation of the RASSF1A transcript. The upper band in each gel shows control RT-PCR using 1.5 μg of normal kidney mRNA per lane. The lower gel shows RT-PCR amplification using mRNA from the eight RCC cell lines (same conditions). The lower band in each gel corresponds to the different amount (1 ng, 100pg, 10 pg, or 1 pg) of artificial truncated RASSF1A RNA added to the RT reaction. Upper bands are produced from the endogenous RASSF1A mRNA. The percentage of endogenous RASSF1A mRNA was calculated for each cell line and shown on the left.
RASSF1A Hypo- and Hypermethylated Alleles Are Maintained Simultaneously in Somatic-Cell Hybrids.
We studied RASSF1A transcription and methylation in RCC somatic-cell hybrid clones that were created earlier for VHL studies (22). Both parental lines (UOK121 and UMRC6) contained multiple copies of chromosome 3 (5–6 copies; data not shown). We found that in UOK121 cells, the RASSF1A transcript was expressed, and at least some of the copies of chromosome 3 carried a hypomethylated RASSF1A promoter. In UMRC6 cells, the gene was silenced and methylated. The presence of chromosome 3 from both parental cell lines in somatic-cell hybrids was confirmed by using an informative single-nucleotide polymorphism located within 3′ untranslated region (UTR) of the VHL gene (24). Analysis of the cell hybrids indicated that RASSF1A maintained normal expression in one of four clones comparable to that of the parental UOK121 line. Sequencing of bisulfite-treated DNA from this hybrid revealed the presence of both heavily methylated and hypomethylated copies of the RASSF1A promoter (Fig. 6, which is published as supplemental data on the PNAS web site). This fact may point to various cis elements potentially important for maintenance of a stable methylation pattern in this region. Both hypo- and hypermethylated alleles also are maintained in the duplicated chromosomes of the partially methylated cell lines HN51, A498, CAKI 2, and TK164 (data not shown).
RASSF1C Promoter Is Not Methylated in RCC Cell Lines and Primary Renal Tumors.
The CpG islands that coincided with exon 1α of RASSF1A and exon 2γ of the RASSF1C transcription units (Fig. 4A, which is published as supplemental data on the PNAS web site) have similar sizes and GC contents and are 4 kb apart. However, we found, that in contrast to RASSF1A, this island never was methylated in the same set of primary renal tumors. Consequently, the RASSF1C transcript is always present in these cells (data not shown). Interestingly, the RASSF1C has been reported to be silenced in some ovarian tumor samples (25).
Transcriptional Silencing of RASSF1A Correlates with DNA Methylation.
RASSF1A expression was analyzed in 18 RCC cell lines by Northern blot analysis and quantitative RT-PCR. We found a hypermethylated promoter and low (or absent) RASSF1A expression in nine lines (ACHN, HN4, HN51, CAKI 2, UMRC6, 786-0, RFX393, UOK108, and UOK127).
Seven cell lines had partial methylation of the promoter and moderate to normal expression levels of the RASSF1A transcript (KH39, CAKI 1, TK164, KRC/Y, UOK102, UOK121 and UOK143). The remaining two cell lines, TK10 and A498, expressed high levels of RASSF1A. For three RASSF1A-expressing cell lines (TK10, KRC/Y, and CAKI 1), sequencing of the first two RASSF1A exons was performed. In TK10 and KRC/Y, a nonconservative change in the SH3 domain was found (Lys-21→Gln-21). In CAKI 1, another change in the SH3 domain was detected (Glu-6→Asp-6).
In several cell lines (UMRC6, 786-0, KRC/Y, RFX393, UOK108, UOK121, UOK127, and UOK143), the interrelation between RASSF1A expression and promoter methylation was analyzed in detail. To determine the methylation status of each CpG dinucleotide, COBRA PCR fragments were subcloned and several clones from each cell line were sequenced (Fig. 1 A and B). To estimate the level of RASSF1A expression in normal kidney and tumor samples by quantitative RT-PCR, a predetermined amount of artificial RASSF1A RNA carrying a 20-bp deletion was added to the RNA samples before RT. After RT-PCR, the artificial RNA with a 20-bp deletion yielded a shorter PCR fragment. The amount of native and artificial RASSF1A RNA in the sample was estimated by comparing the intensities of the normal and control PCR bands (Fig. 1C).
According to the RT-PCR quantitation, RASSF1A mRNA is expressed in normal kidney at the level of 0.01% of the total mRNA level. The expression level in the cell lines carrying methylated RASSF1A promoter, including UMRC6, 786-0, RFX393, UOK108, and UOK127, was below 0.001%. In three “semimethylated” cell lines, expression varied between 0.001–0.02% (Fig. 1C). In general, the extent of methylation of the GC-rich genomic region at the 5′ end correlated with RASSF1A transcriptional repression.
Reexpression of RASSF1A in RCC Cells After 5-Aza-CdR Treatment.
In the 786-0 RCC cell line, RASSF1A transcription is silenced and the putative promoter region is methylated heavily (Fig. 1). RASSF1A transcription was reactivated after incubation of 786-0 cells for 3 days in the medium containing 5 μM of the DNA methylation inhibitor 5-Aza-CdR (Fig. 2). RT-PCR quantitation revealed a 50- to 100-fold increase in RASSF1A mRNA expression. Therefore, transcriptional repression is at least in part mediated by DNA methylation.
Figure 2.
Reexpression of RASSF1A gene in 786-0 cells. The cells were grown in 5 μM 5-Aza-CdR for 4 days. Total RNA (1.5 μg ) was used per RT reaction. The amount of artificial truncated RASSF1A RNA added per each RT reaction is indicated on the top.
Reexpression of RASSF1A in tTA-Engineered KRC/Y Cells Suppresses Growth on Plastic Surfaces.
Cells were transfected with the episomal pETE–RASSF1A constructs, selected and grown on plastic Petri dishes in the presence and absence of dox. They were monitored daily by counting cells from triplicate dishes (see Materials and Methods). As shown (Fig. 3A), the wild-type allele drastically suppressed cell growth in the absence of dox (83% suppression on day 6 and 91% on day 12) compared with the empty control vector, whereas the mutant gene was much less effective (64% suppression on day 6 and 69% on day 12). In the presence of dox, no difference in growth rate was detected among these clones.
Figure 3.
Suppression of KRC/Y cell growth by RASSF1A reexpression. (A) Growth suppression of RASSF1A-transfected cells on plates. (B) Colony count of RASSF1A-transfected and control cells in soft agar.
Reexpression of RASSF1A in KRC/Y Cells Suppresses Anchorage-Independent Growth.
The expressing construct pcDNA3.1-RASSF1A was transfected into KRC/Y cells. G418-resistant RASSF1A-expressing clones were expanded. The numbers of small (size 20–100 μm) and large (size 100 μm and more) colonies were decreased for the RASSF1A-expressing cells after 10 days of growth in soft agar. In general, the number of colonies formed by RASSF1A-transfected cells was only 5% of those from the control cells (Fig. 3B). After 15 days, the original KRC/Y cells comprised a continuous layer of cells. In contrast, RASSF1A-transfected clones expanded at a much lower rate, forming spheroid bodies (Fig. 7, which is published as supplemental data on the PNAS web site, www.pnas.org).
Discussion
Allelic losses on 3p12-p14, 3p21-p22, and 3p25-p26 are the most frequent events in the genesis of nonpapillary RCC. The FHIT gene, located within the first region at 3p14.2, was down-regulated in more than 50% of clear RCC tumors examined (15). The VHL tumor-suppressor gene identified at 3p25 was inactivated in 100% of VHL families and in a large percentage of sporadic clear RCC (8–10). To date, no clear RCC candidate TSGs have been found in the 3p21-p22 region. This region is now subdivided into the centromeric 3p21.3 and the telomeric 3p21.3-p22 subparts (5).
Here we present, to our knowledge, the first evidence to support the potential role of the candidate lung and breast cancer TSG, RASSF1A, in the development of clear RCC tumors. Surprisingly, this 3p21.3 gene (from centromeric subregion 3p21.3) was involved in both types of clear RCC, i.e., with and without VHL inactivation.
The RASSF1A gene was hypermethylated in 91% of primary clear RCC tumors (39 of 43 tumors). In 27 of 29 available tumor-normal pairs, the RASSF1A promoter was hypermethylated in tumor DNA. In normal matching DNA from tumor-surrounding tissues, the RASSF1A promoter was methylated also albeit to a much lesser extent, suggesting that silencing of it might be a causative early event in the carcinogenesis of some clear RCC preceding or after LOH.
In RCC cell lines, hypermethylation of the RASSF1A promoter region correlated with transcriptional down-regulation of the RASSF1A mRNA. Interestingly, the nearby RASSF1C promoter was found unmethylated in all renal tumor samples and cell lines. Both RASSF1A and RASSF1C predicted proteins show high homology (55% identity) to the mouse RAS effector protein Nore1 and to the rat orthologous protein Maxp1 (19). Nore1 interaction with Ras is GTP-dependent and follows receptor activation. However, RASSF1A and RASSF1C proteins have different NH2 termini. The RASSF1C N-terminal sequence has no similarity to any known proteins, whereas the N-terminal part of RASSF1A contains SH3 (residues 1–51) and diacylglycerol/phorbolester-binding (DAG) domain (residues 52–101) domains in addition to the common RAS-association domain (19, 20). All of these domains may involve the RASSF1A protein in signal-transduction pathways and define its important tumor-suppressor properties. Finding amino acid changes (Lys-21→Gln-21 and Glu-6→Asp-6) in three RASSF1A-expressing clear RCC cell lines (this work) and multiple changes in the SH3-, DAG-, and RAS-association domains in lung cancer cells (19–21) strengthens this possibility.
RASSF1A suppressed the growth of KRC/Y cells in Petri dishes and anchorage-independent growth in soft agar. The specificity of these activities was supported by the absence of suppression in the presence of dox when the transgenes were not expressed. Moreover, the mutant RASSF1A with mutations in diacylglycerol/phorbolester-binding- and RAS-associated domains possessed significantly reduced growth-suppressing activity.
Suppression of RASSF1A transcription did not correlate with the status of the VHL gene. In RFX393, UOK108, UOK121, UOK127, and UOK143 cell lines, the VHL gene was silenced by hypermethylation (10, 22), but the RASSF1A was hypermethylated only in RFX393, UOK108, and UOK127. RASSF1A wild-type mRNA was expressed in the remaining “VHL-methylated” lines UOK121 and UOK143. UMRC6 and 786-0 carried mutations of the VHL gene (8, 9) and methylated (silenced) RASSF1A alleles. Finally, KRC/Y cells had a normally expressed VHL gene and expressed RASSF1A that encodes a nonconservative amino acid change. Simultaneous inactivation of both genes in one cell line would imply that VHL and RASSF1A proteins participate in different tumorigenic pathways. Indeed, multiple 3p LOH (at 3p25-p26 and 3p21-p22) are common in nonpapillary RCC (4, 5, 17) and can occur at different stages of tumor development in the same tumor. Inactivation of RASSF1A in clear RCCs that possessed a normally expressed VHL implies an important role of the gene in the origin of this type of clear RCC. On the other hand, its inactivation in VHL-caused tumors suggests a role in tumor development. Thus, the RASSF1A gene could play a dual role in carcinogenesis, being causally involved in the origin of some clear RCCs and driving progression of some VHL-caused clear RCCs.
Coexistence of hypo- and hypermethylated gene copies in the same cell or partial methylation of the same copy can cause only partial down-regulation of the RASSF1A transcript in a large proportion of RCC cell lines and primary tumors. However, our growth-suppression experiments indicate that even a partial RASSF1A repression, sometimes in conjunction with amino acid changes, may provide important selective advantages for the growth of affected cells.
It seems that clear RCC is characterized by genetic heterogeneity with different pathologically indistinguishable forms caused by separate TSGs. They were described originally as conforming to the two-hit paradigm (26, 27). However, recent data suggest that multiple hits are required in clear RCC development (28). These hits may include different tumor-suppressor genes located on chromosomes 3p25-p26, 3p21-p22, (subregions, centromeric 3p21.3 and 3p21.3-p22) and 3p12-p14.
Numerous studies have shown that 3p21-p22 LOH is the most frequent loss in sporadic nonpapillary RCC, although 3p25-p26 and 3p12-p14 losses usually occur earlier in some tumor development. When multiple LOH are present, tumors with LOH on 3p21-p22 also carry LOH on 3p14 or 3p25 (5, 16, 17), suggesting that LOH on 3p21-p22 is an important event in RCC progression. However, another study of multiple tumors from a single patient established that early 3p allelic losses (adenoma but not carcinoma-related) occurred predominantly in the 3p25 or the 3p12-p14 regions, but not in 3p21–3p22 (29). On the other side, the non-VHL hereditary clear RCC locus was excluded by linkage analysis from chromosome 3p, implying that losses of 3p genes are not involved in the origin of this particular hereditary form of clear RCC (12).
The present data suggest that RASSF1A is the candidate renal 3p21.3 TSG. More studies are necessary to elucidate the role of RASSF1A in other multiple types of human cancer.
Supplementary Material
Acknowledgments
We thank Dr. A. Danilkovitch-Miagkova for technical advice. UOK102, UOK108, UOK121, UOK127, and UOK143 clear RCC cell lines were kindly provided by M. W. Linehan (National Cancer Institute, Bethesda, MD). The UMRC6 line was provided by H. B. Grossman (Southwest Oncology Group, San Antonio, TX). RASSF1A cDNA was provided by R. Dammann and G. Pfeifer (Beckman Institute, Pasadena, CA).
This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-56000. We acknowledge the National Cancer Institute for allocation of computing time and staff support at the Frederick Biomedical Supercomputer Center. K.D. was supported in part by the Dutch Cancer Society and the Karel Frederik Stichting. The Karolinska Institute Group was funded by grants from the Swedish Cancer Society, Karolinska Institute, Ingabritt och Arne Lundbergs Forskningsstiftelse, and by the Royal Swedish Academy of Science (to E.Z. and to G.K.). J.D.M. was supported by National Cancer Institute Grants CA71618 and SPORE P50 CA70907, and by the G. Harold and Leila Y. Mathers Charitable Foundation. E.J.S. was supported by National Cancer Institute Grant CA19104.
Abbreviations
- RASSF1
RAS association family 1 gene
- LOH
loss of heterozygosity
- RCC
renal cell carcinoma
- COBRA
combined bisulfite-restriction analysis
- 5-Aza-CdR
5-aza-2′-deoxycytidine
- dox
doxycycline
- RT
reverse transcription
- TSG
tumor-suppressor gene
- CpG
CG dinucleotide within DNA sequence
References
- 1.Lubinski J, Hadaczek P, Podolski J, Toloczko A, Sikorski A, McCue P, Druck T, Huebner K. Cancer Res. 1994;54:3710–3713. [PubMed] [Google Scholar]
- 2.Yamakawa K, Morita R, Takahashi E, Hori T, Ishikawa J, Nakamura Y. Cancer Res. 1991;51:4707–4711. [PubMed] [Google Scholar]
- 3.Van der Hout A H, van der Vlies P, Wijmenga C, Li F P, Oosterhuis J W, Buys C H. Genomics. 1991;11:537–542. doi: 10.1016/0888-7543(91)90060-r. [DOI] [PubMed] [Google Scholar]
- 4.Braga E, Pugacheva E, Basov I, Ermilova V, Kazubskaya T, Mazurenko N, Kisseljov F, Liu J, Garkavtseva R, Zabarovsky E R, et al. FEBS Lett. 1999;454:215–219. doi: 10.1016/s0014-5793(99)00807-8. [DOI] [PubMed] [Google Scholar]
- 5.Alimov A, Kost-Alimova M, Liu J, Li C, Bergerheim U, Imreh S, Klein G, Zabarovsky E R. Oncogene. 2000;19:1392–1399. doi: 10.1038/sj.onc.1203449. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Zabarovska V, Braga E, Alimov A, Klein G, Zabarovsky E R. FEBS Lett. 1999;462:121–128. doi: 10.1016/s0014-5793(99)01523-9. [DOI] [PubMed] [Google Scholar]
- 7.Melmon K L, Rosen S W. Am J Med. 1964;36:595–617. doi: 10.1016/0002-9343(64)90107-x. [DOI] [PubMed] [Google Scholar]
- 8.Latif F, Tory K, Gnarra J, Yao M, Duh F-M, Orcutt M-L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1994;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 9.Gnarra J R, Tory K, Weng Y, Schmidt L, Wei M H, Li H, Latif F, Liu S, Chen F, Duh F-M, et al. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 10.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M, et al. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teh B T, Giraud S, Sari N F, Hii S I, Bergerat J P, Larsson C, Limacher J M, Nicol D. Lancet. 1997;349:848–849. doi: 10.1016/S0140-6736(05)61751-5. [DOI] [PubMed] [Google Scholar]
- 12.Woodward E R, Clifford S C, Astuti D, Affara N A, Maher E R. J Med Genet. 2000;37:348–353. doi: 10.1136/jmg.37.5.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van den Berg A, Buys C H. Genes Chromosomes Cancer. 1997;19:59–76. doi: 10.1002/(sici)1098-2264(199706)19:2<59::aid-gcc1>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez Y, El-Naggar A, Pathak S, Killary A M. Proc Natl Acad Sci USA. 1994;91:3383–3387. doi: 10.1073/pnas.91.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadaczek P, Siprashvili Z, Markiewski M, Domagala W, Druck T, McCue P A, Pekarsky Y, Ohta M, Huebner K, Lubinski J. Cancer Res. 1998;58:2946–2951. [PubMed] [Google Scholar]
- 16.Van den Berg A, Hulsbeek M M F, de Jong D, Kok K, Veldhuis P M, Roche J, Buys C H. Genes Chromosomes Cancer. 1996;15:64–72. doi: 10.1002/(SICI)1098-2264(199601)15:1<64::AID-GCC9>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Maestro M L, del Barco V, Sanz-Casla M T, Moreno J, Adrover E, Izquierdo L, Zanna I, Fernandez C, Redondo E, Blanco J, Resel L. Oncology. 2000;59:126–130. doi: 10.1159/000012149. [DOI] [PubMed] [Google Scholar]
- 18.Lerman M I, Minna J D. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 19.Dammann R, Li C, Yoon J-H, Chin P L, Bates S, Pfeifer G P. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 20.Burbee D G, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Virmani A, Bader S, Sekido Y, et al. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agathanggelou A, Honorio S, Macartney D P, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw J A, et al. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmin I, Geil L, Ge H, Bengtsson U, Duh F-M, Stanbridge E J, Lerman M I. Oncogene. 1999;18:5672–5679. doi: 10.1038/sj.onc.1202959. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Z, Laird P W. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne S J, Richards F M, Maher E R. Hum Mol Genet. 1994;3:390. doi: 10.1093/hmg/3.2.390. [DOI] [PubMed] [Google Scholar]
- 25.Vos M D, Ellis C A, Bell A, Birrer M J, Clark G J. J Biol Chem. 2000;275:35669–35672. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 26.Erlandsson R, Boldog F, Sumegi J, Klein G. Cancer Genet Cytogenet. 1988;36:197–202. doi: 10.1016/0165-4608(88)90145-8. [DOI] [PubMed] [Google Scholar]
- 27.Maher E R, Yates J R, Ferguson-Smith M A. J Med Genet. 1990;27:311–314. doi: 10.1136/jmg.27.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein W D, Stein A D. J Clin Epidemiol. 1995;48:767–777. doi: 10.1016/0895-4356(94)00192-s. [DOI] [PubMed] [Google Scholar]
- 29.Van den Berg A, Dijkhuizen T, Draijers T G, Hulsbeek M M F, Maher E R, van den Berg E, Strokel S, Buys C H. Genes Chromosomes Cancer. 1997;19:228–232. doi: 10.1002/(sici)1098-2264(199708)19:4<228::aid-gcc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.