Abstract
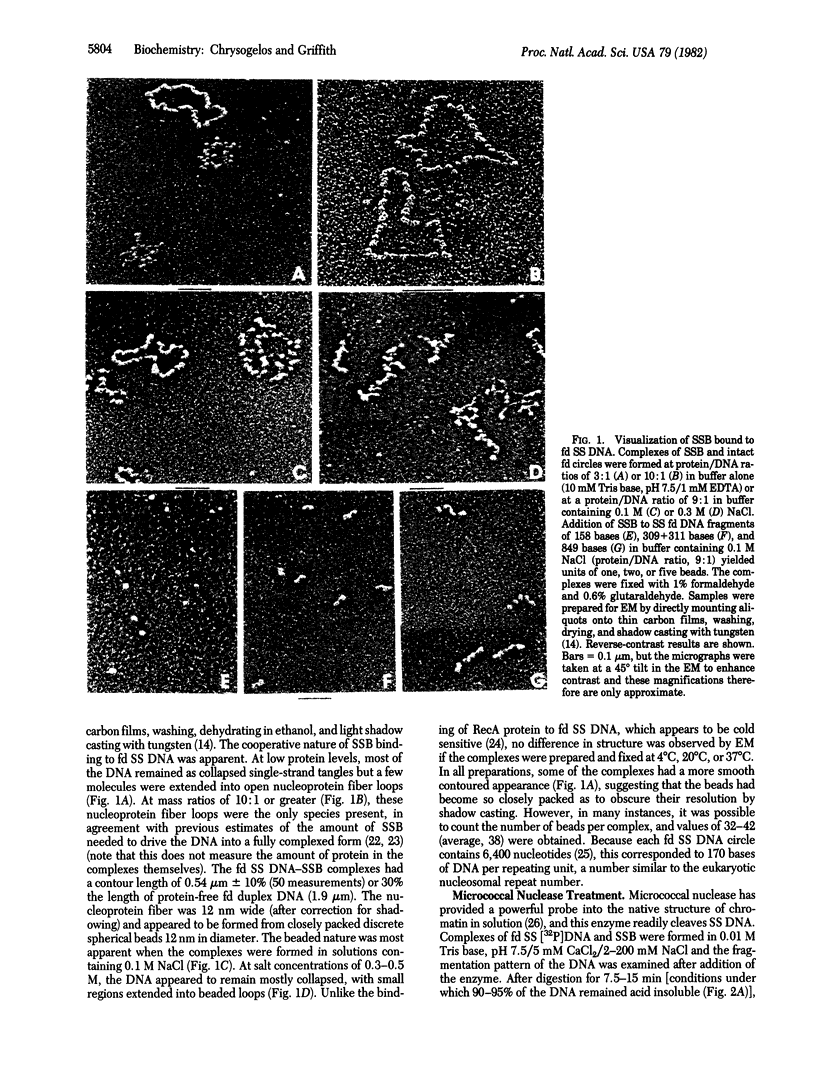
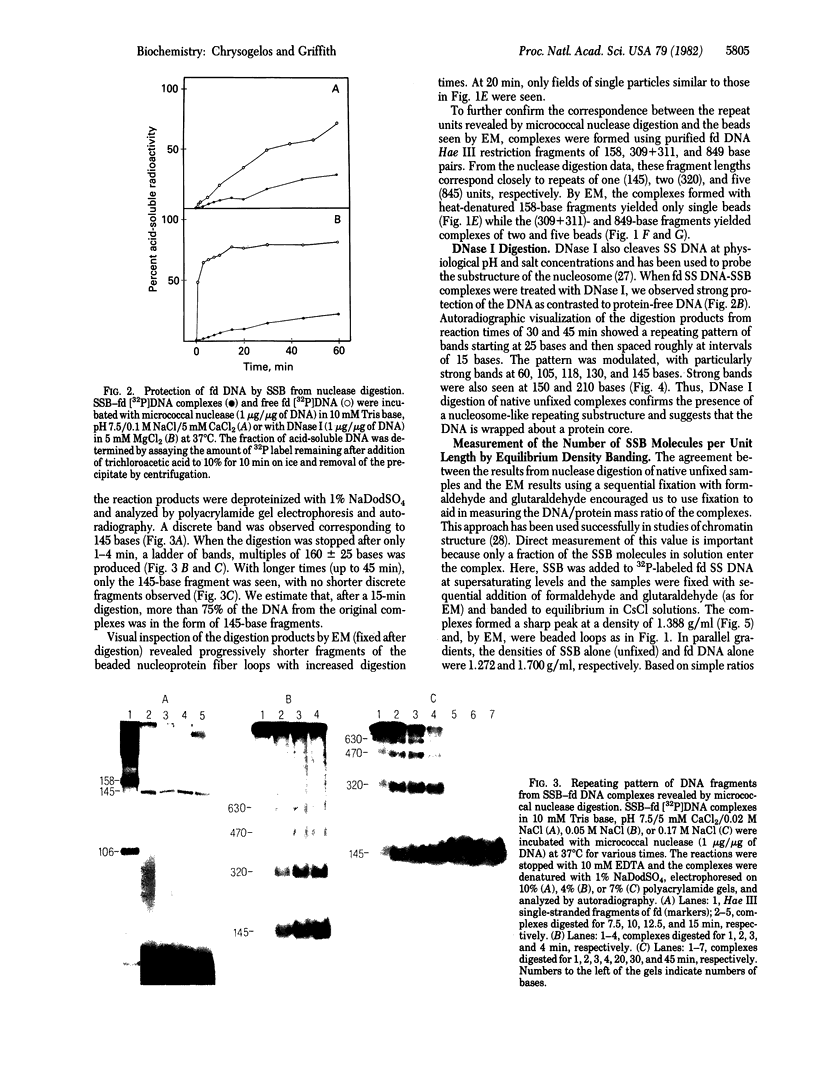
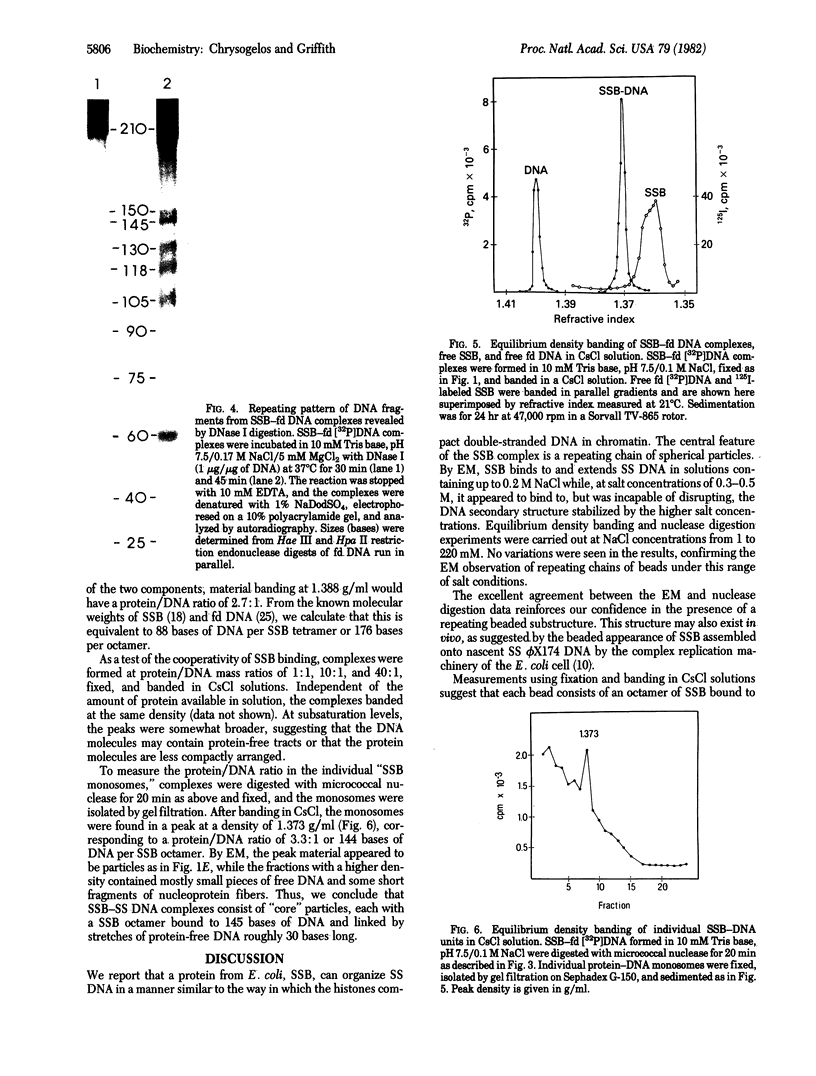
Electron microscopy shows that complexes of the single-strand DNA binding protein (SSB) of Escherichia coli and phage fd DNA appear as beaded fiber loops containing an average of 38 beads, 1 per 170 bases of DNA. Extensive digestion of native unfixed SSB-fd DNA complexes with micrococcal nuclease reveals a protected DNA fragment of 145 bases, while shorter digestion periods result in a sequence of fragments in multiples of 160 +/- 25 bases. Digestion of these complexes with DNase I produces a repeating pattern of bands, multiples of approximately 15 bases with strong bands at 60, 105, 118, 130, 145, 150, and 210 bases. Isopycnic banding in CsCl solution yields densities of 1.272 and 1.700 g/ml, respectively, for SSB alone and for fd DNA and, after fixation, of 1.388 g/ml for fd DNA-SSB beaded fibers and 1.373 g/ml for the individual protein-DNA beads. Based on these data and the molecular weights of SSB and fd DNA, we suggest that the nucleoprotein chain consists of eight molecules of SSB bound to 145 bases of DNA, with these units linked by roughly 30 bases of protein-free DNA. The excellent concord between results obtained by enzyme digestion of unfixed native samples and, after fixation, by electron microscopy and density banding supports the conclusion that SSB organizes single-stranded DNA in a manner similar to the organization of duplex DNA by histones.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brison O., Cambon P. A simple and efficient method to remove ribonuclease contamination from pancreatic deoxyribonuclease preparations. Anal Biochem. 1976 Oct;75(2):402–409. doi: 10.1016/0003-2697(76)90094-4. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Whittier R. F., Auerbach J., Sancar A., Rupp W. D. Amplification of single-strand DNA binding protein in Escherichia coli. Nucleic Acids Res. 1980 Jul 25;8(14):3215–3227. doi: 10.1093/nar/8.14.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of recA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982 Apr;28(4):757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- Dunn K., Griffith J. D. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980 Feb 11;8(3):555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory J., Radding C. M. Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell. 1982 Apr;28(4):747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassberg J., Meyer R. R., Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979 Oct;140(1):14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Gudas L. J., Mount D. W. Identification of the recA (tif) gene product of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5280–5284. doi: 10.1073/pnas.74.12.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. Separation of newly synthesized nucleohistone by equilibrium centrifugation in cesium chloride. Biochemistry. 1974 Sep 10;13(19):3952–3956. doi: 10.1021/bi00716a021. [DOI] [PubMed] [Google Scholar]
- Johnson B. F. Genetic mapping of the lexC-113 mutation. Mol Gen Genet. 1977 Nov 29;157(1):91–97. doi: 10.1007/BF00268691. [DOI] [PubMed] [Google Scholar]
- Krauss G., Sindermann H., Schomburg U., Maass G. Escherichia coli single-strand deoxyribonucleic acid binding protein: stability, specificity, and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry. 1981 Sep 1;20(18):5346–5352. doi: 10.1021/bi00521a040. [DOI] [PubMed] [Google Scholar]
- Makino S., Woolford J. L., Jr, Tanford C., Webster R. E. Interaction of deoxycholate and of detergents with the coat protein of bacteriophage f1. J Biol Chem. 1975 Jun 10;250(11):4327–4332. [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci U S A. 1980 Feb;77(2):857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. R., Glassberg J., Kornberg A. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1702–1705. doi: 10.1073/pnas.76.4.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. R., Glassberg J., Scott J. V., Kornberg A. A temperature-sensitive single-stranded DNA-binding protein from Escherichia coli. J Biol Chem. 1980 Apr 10;255(7):2897–2901. [PubMed] [Google Scholar]
- Molineux I. J., Pauli A., Gefter M. L. Physical studies of the interaction between the Escherichia coli DNA binding protein and nucleic acids. Nucleic Acids Res. 1975 Oct;2(10):1821–1837. doi: 10.1093/nar/2.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Noll H. Structural dynamics of bacterial ribosomes. V. Magnesium-dependent dissociation of tight couples into subunits: measurements of dissociation constants and exchange rates. J Mol Biol. 1976 Jul 25;105(1):111–130. doi: 10.1016/0022-2836(76)90197-2. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Ruyechan W. T., Wetmur J. G. Studies on the cooperative binding of the Escherichia coli DNA unwinding protein to single-stranded DNA. Biochemistry. 1975 Dec 16;14(25):5529–5534. doi: 10.1021/bi00696a023. [DOI] [PubMed] [Google Scholar]
- Schaller H. The intergenic region and the origins for filamentous phage DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):401–408. doi: 10.1101/sqb.1979.043.01.046. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Eisenberg S., Bertsch L. L., Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977 Jan;74(1):193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal N., Delius H., Kornberg T., Gefter M. L., Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. H., Bertsch L. L., Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and functions in replication. J Biol Chem. 1975 Mar 25;250(6):1972–1980. [PubMed] [Google Scholar]
- Wickner S., Hurwitz J. Conversion of phiX174 viral DNA to double-stranded form by purified Escherichia coli proteins. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4120–4124. doi: 10.1073/pnas.71.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]