Abstract
Microendoscopes allow clinicians to view subcellular features in vivo and in real-time, but their field-of-view is inherently limited by the small size of the probe’s distal end. Video mosaicing has emerged as an effective technique to increase the acquired image size. Current implementations are performed post-procedure, which removes the benefits of live imaging. In this manuscript we present an algorithm for real-time video mosaicing using a low-cost high-resolution microendoscope. We present algorithm execution times and show image results obtained from in vivo tissue.
OCIS codes: (170.0170) Medical optics and biotechnology, (170.2150) Endoscopic imaging, (170.3010) Image reconstruction techniques, (110.2350) Fiber optics imaging
1. Introduction
Histopathological examination of excised tissue is the standard of care for the diagnosis of epithelial cancers. During this procedure pathologists examine cellular features such as nuclear size, nuclear-to-cytoplasmic ratio, and architectural alterations. Microendoscopy is a type of optical imaging that allows clinicians to view these features in vivo at subcellular resolution, effectively bringing the microscope to the patient. These non-invasive devices can be used to (1) screen biopsy sites in order to reduce the number of biopsies taken, (2) function as point-of-care diagnostic tools during surgical procedures, or (3) monitor disease progression throughout longitudinal in vivo studies [1]. Although microendoscopes can have excellent resolution and light efficiency, their field-of-view (FOV) is inherently limited by the size of the probe’s distal end. In most microendoscopes the FOV is <1 mm2, which is comparable to the field of a 40X microscope objective. This “tunnel vision” can make it difficult for users to obtain a broad sense of tissue morphology [2] and result in sampling error when imaging heterogeneous biological tissue [3].
Video mosaicing has been demonstrated as an effective tool for increasing the FOV acquired by microendoscopy. This technique stitches together consecutive video frames acquired as the probe is scanned over the tissue, thus creating a 2 – 30 times larger high-resolution image. Several papers have discussed development and clinical application of off-line video mosaicing using commercial microendoscopes [4–6]. These methods combine the frames of a saved video using non-rigid deformation algorithms and template matching. Although these approaches can account for tissue stretching and motion artifacts, they are too computationally intensive for video-rate implementation and are carried out post-procedure. Therefore the clinician does not have the benefit of live evaluation of a large FOV. Another approach, called real-time video mosaicing, computes a simpler rigid registration in order to build the mosaic as each frame is acquired (see Fig. 1 ). This provides the user immediate feedback about broad tissue morphology, aids in maintaining probe stability, and helps create a better video for additional off-line processing. The concept was discussed for a commercial confocal system in proceedings [7], mentioned in [8], and performed on a larger-FOV fluorescence endoscope in [9]. Unfortunately, these publications do not provide a thorough description of a real-time video mosaicing algorithm for microendoscopy.
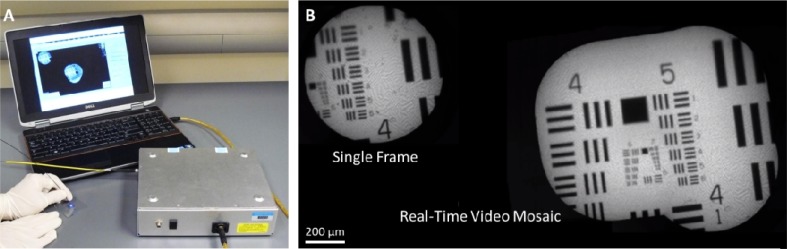
Fig. 1.
The high-resolution microendoscope (HRME) is a portable fluorescence imaging device that provides in vivo subcellular detail with a field-of-view of <1 mm. (B) Real-time video mosaicing increases the field-of-view (Media 1 (697.5KB, AVI) ).
In this manuscript we present an algorithm and initial results for real-time in vivo video mosaicing. The technique is implemented on an inexpensive (<$5,000) high-resolution microendoscope (HRME). The algorithm is a three step process that includes: fiber bundle pattern removal, image registration, and insertion into a mosaic. Mosaic results are compared to benchtop confocal imaging as well as histopathology. To the best of our knowledge, this is the first publication to provide a complete description and histopathology comparison of real-time video mosaicing using a microendoscope.
2. Methods
2.1. High-resolution microendoscope (HRME)
The system that we modified for real-time video mosaicing was previously used in several in vivo tissue studies [10,11]. It is well-suited for animal experiments and clinical applications because it is small, battery-powered, and interfaces with a standard laptop computer (see Fig. 1A). Although the system is thoroughly described in [12], we provide a brief description here.
The HRME is a wide-field epi-fluorescence fiber bundle microendoscope. It uses a 5W 455 nm light emitting diode (LED) and lenses to provide Koehler illumination at the proximal face of a 30,000-element coherent fiber bundle. It has a circular FOV with 700 μm diameter. Tissue at the fiber bundle’s distal end is typically labeled with the fluorescent dye proflavine (0.01% in sterile saline), which binds to DNA and has a peak emission of 515 nm. Emitted light exiting the proximal fiber bundle passes through a dichroic filter and a long-pass filter. It is then imaged onto a CCD using a 10X objective and tube lens. Images are acquired at a rate of 11 frames per second (FPS) at 16-bit digitization. Camera settings, image display, and file saving are controlled with custom LabVIEW software. Figure 2 (left) shows a raw image of non-keratinized stratified squamous epithelium acquired by the device. Nuclei appear as bright circles throughout the image and individual fibers within the fiber bundle can be seen.
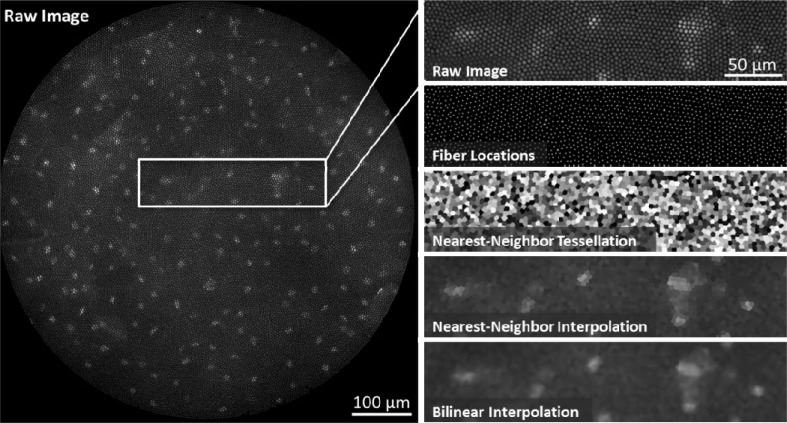
Fig. 2.
Fiber pattern removal. Left, the raw HRME image shows squamous epithelial cells in the oral cavity. Nuclei are labeled with the 0.01% Proflavine. The intensity information from the sample is “pixelated” by the 30,000-element fiber bundle. Right, the fiber pixelation pattern is removed using two-dimensional interpolation. First, the fiber center locations are identified using a regional maxima function. A Voronoi tessellation is then constructed from these points in order to determine the nearest-neighbor fiber pixel for every non-fiber pixel. A lookup table is determined from this tessellation and is used to reconstruct fiber-less HRME images in real-time. Other interpolation methods, such as bilinear interpolation, can be used but have longer computation times.
In comparison to a confocal microendoscope, the HRME has reduced axial response because there is no optical sectioning mechanism. Therefore, the HRME cannot clearly visualize subsurface cellular features or reject out-of-focus background signal. However, we found that the small penetration depth of 455 nm light following proflavine application limits background signal from most tissue sites. The FOV and lateral resolution are similar, because they are both dependent on the size and density of the fiber bundle.
2.2. Fiber pattern rejection
The first step to implement video mosaicing is to remove the fiber bundle pattern from HRME images. This is necessary because the fixed spatial frequencies produced by the bundle structure hinder the subsequent image registration algorithm. For real-time video mosaicing a fast, accurate, and memory efficient method is necessary. Ideally, this step should run much faster than the HRME’s nominal operational speed of 11 FPS. A few publications discuss methods to remove the bundle pattern while preserving image content, including: spectral filtering in the frequency domain [13], median filtering in the spatial domain [14], or spatial interpolation [15]. We developed a custom spatial interpolation algorithm because none of these methods were tailored for real-time execution. Based on the nearest-neighbor lookup table method suggested in [15], our approach (1) preserves all intensity information of the sample image and (2) requires minimal computation time and memory.
The nearest-neighbor interpolation algorithm reconstructs an image based on pixel intensities at regional maxima. As shown in Fig. 2 (right, “Raw Image”), fiber cores appear as regional maxima within the images due to the Gaussian intensity profile that they create on the detector. The pixel values at the center of these regional maxima best represent the intensity of the underlying object; therefore we use only these values to reconstruct a fiber-less image. First, fiber center pixels are determined using the Matlab regional maxima function imregionalmax.m described in [16] (Fig. 2, “Fiber Locations”). Next, the functions griddata.m and TriScatteredInterp.m are used to compute a Voronoi tessellation based on the fiber center coordinates (Fig. 2, “Nearest-Neighbor Tessellation”). The tessellation is then used to assign all non-center pixels the same intensity value as the nearest fiber center pixel.
For real-time implementation, we assume that the fiber bundle pattern remains fixed within a sequence of images. Therefore, the tessellation is computed only once at the beginning of the procedure to determine a lookup table of fiber center reference coordinates. The lookup table is calculated from an image of a uniform fluorescent slide (Fluor-Ref Green, Ted Pella). Subsequent HRME images are then divided by this “calibration” image to normalize the fiber bundle intensity response. In addition, given that fibers are spaced approximately 5 pixels apart, we found that downsampling the lookup table by a factor of 2 – 3 also reduces processing time while maintaining image quality. The interpolated and downsampled result is approximately 375 x 375 pixels. Applying the lookup table to a raw 1.3 megapixel HRME image takes 1.9 ms on our laptop computer (Intel i7-2720QM), as shown in Table 1 . Figure 2 (“Nearest-Neighbor Interpolation”) shows our fast interpolation method removes the fiber pattern while preserving image edges and contrast.
Table 1. Time Requirements for Video Mosaicing Algorithm.
| Mosaic Size (pixels) | Fiber Pattern Rejection (ms) | Registration (ms) | Insertion (ms) | Total, Incl. All GUI Functions (ms) |
|---|---|---|---|---|
| No Mosaicing | - | - | - | 49.5 |
| 1120 x 770 | 1.9 | 18.1 | 9.2 | 64.5 |
| 1600 x 1100 | 1.9 | 18.1 | 10.6 | 65.8 |
| 2080 x 1430 | 1.9 | 18.1 | 12.3 | 66.5 |
| 2400 x 1650 | 1.9 | 18.1 | 13.3 | 67.5 |
| 3200 x 2200 | 1.9 | 18.1 | 17.1 | 71.4 |
2.3. Frame registration
The registration algorithm determines the best translational offset between two consecutive video frames and calculates an error for their fit. Before registration, a rectangular region of interest is cropped from the circular FOV of processed HRME images. This prevents registration errors caused by fiber bundle edges. Time and memory constraints for registration are the same as for the interpolation algorithm. For this reason, we chose to implement the fast registration algorithm presented in [17], which performs an optimized cross correlation using discrete Fourier transforms (DFTs). The cross correlation of two images f(x,y) and g(x,y) is given by:
| (1) |
where N and M are the image dimensions, uppercase letters represent the DFT, and * denotes complex conjugation. The approach to finding the cross-correlation peak is to (1) compute the product F(u,v)G*(u,v), (2) take the inverse FFT to obtain the cross correlation, and (3) locate its peak [17]. Error of registration fit is given by the normalized root-mean-square error (NRMSE). On average the registration is executed in 18.1 ms (see Table 1).
2.4. Frame insertion
Registered HRME frames are inserted into a large zero-value image called a canvas. The canvas size is selected by the user and is typically 1 to 7 MP, which is 7 to 50 times larger than a downsampled HRME frame. The first frame in a video sequence is copied to the center of the canvas. Subsequent frames are inserted into the canvas at an offset determined by the registration algorithm. We tested two real-time insertion strategies: (1) pixel maxima and (2) dead leaves.
The pixel maxima approach inserts the maximum intensity value for each overlapping pixel of the newest frame and current mosaic. This strategy works well for more rigid tissue such as epidermis, but results in a motion-blurred mosaic when used on deformable tissue or targets with free-moving debris. For our application it is important to preserve size and separation of features such as nuclei, so we typically use the dead leaves approach [7], which simply replaces overlapping regions with the new frame. Figure 3 shows mosaicing results from fluorescently-labeled lens paper using each of these techniques. Although individual frame boundaries can become apparent when using the dead leaves approach, spatial features within the image maintain their size. To reduce frame boundary visibility when the probe is moving slowly, we only insert frames that have moved at least one tenth of a FOV from the last inserted frame.
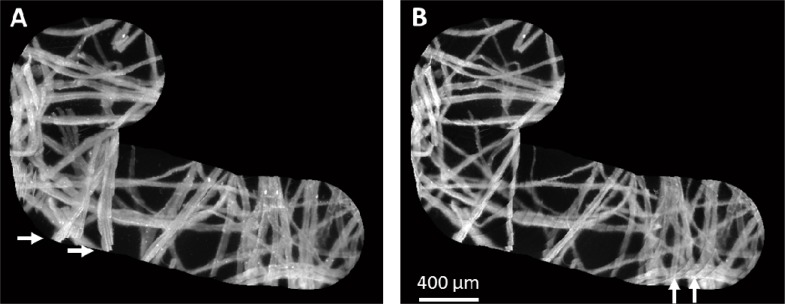
Fig. 3.
The HRME was translated over fluorescently-labeled lens paper to build a mosaic. (A) When video frames are inserted into the mosaic using the pixel maxima approach, frame boundaries are smooth, but disturbed fibers appear blurred as shown by the arrows (Media 2 (1.1MB, AVI) ). (B) When the dead leaves approach is used, blurring does not occur, but frame boundaries can become noticeable (Media 3 (1.2MB, AVI) ). The dead leaves approach is preferred for tissue imaging because probe movement can deform the tissue.
2.5. Error handling and file saving
Registration errors can occur for several reasons. First, registration errors can result from motion blurred images. This is prevented by selecting a low camera exposure (<10 ms) and by translating the probe slowly across the tissue. Slow translation also ensures that consecutive frames have overlapping spatial features to register. We found registration works well when consecutive cropped frames have at least 75 percent overlap, which corresponds to a maximum speed of ~1.375 mm/s for a 700 μm diameter circular HRME probe imaging at 11 FPS. Registration errors can also occur when there is little spatial information (such as in uniform gray images) or when there is loose debris. The normalized root-mean-square error (NRMSE) is used to determine if a registration error has occurred. Frames with NRMSE error above 0.5 were considered misregistered. In general, we often found that small spatial discontinuities from misregistered frames did not significantly degrade the overall quality of the mosaic (see Fig. 4C , white arrows). Therefore, instead of restarting the mosaic after a registration error, the misregistered frames can alternatively be highlighted and inserted into the current mosaic. As an indicator of the probe moving too quickly, we also chose to highlight frames that overlap less than 75% with the previous frame.
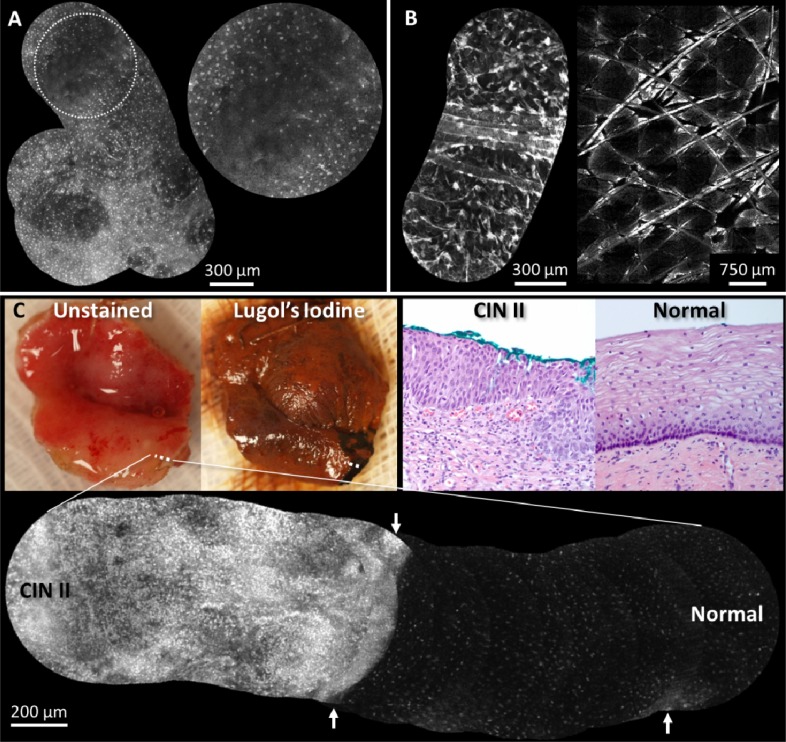
Fig. 4.
Real-time video mosaics (see Media 4 (688.4KB, AVI) ) from (A) normal in vivo oral tissue, (B) normal in vivo epidermis, and (C) pre-cancerous ex vivo cervical tissue. These results show that the algorithm correctly aligns and combines morphological features of several cell types and anatomical locations. In (A) the distribution of normal squamous epithelial cells is seen in the gingiva and in (B, left) features such as hair and keratin sheaths are visible. An image mosaic (B, right) from a benchtop confocal microscope obtained during the same imaging session reveals similar morphologic features as (B, left). The normal/abnormal junction of ex vivo cervical tissue is shown (C), where the white dotted line indicates the path of the mosaic. Lugol’s iodine was used to help visualize the extent of abnormal tissue; dark brown staining indicates normal mucosa. Corresponding histopathology from inside and outside the normal region is shown (images labeled “Normal” and “CIN II,” respectively). The mosaic also shows that it can be useful to display a large mosaic even with misregistered/blurred frames (white arrows).
Although the canvas is displayed in real-time, it is impractical to save every frame of the reconstructed mosaic because of the large canvas size. Instead, we allow the user to save snapshots of the mosaic or save a movie of the raw camera data. Mosaic snapshots are saved as 16-bit PNG images, and movies are saved as two 8-bit AVI videos, which contain the high and low bits of 16-bit frames.
3. Results
All steps of the video mosaicing procedure were implemented and benchmarked in a real-time LabVIEW graphical user interface (GUI) on a standard laptop computer (Intel i7-2720QM). A summary of the computational time requirements for our real-time video mosaicing algorithm is shown in Table 1. The first row shows the execution time with no mosaicing, which represents time to read the image buffer and display the image. Times for fiber rejection and frame registration remain constant because these are only dependent on the raw image size. However, frame insertion time is dependent on the total empty mosaic size; therefore, the results are shown for several canvas sizes. In all cases the execution time of the mosaicing GUI is faster than the frame rate of the camera (which has a maximum speed of 11 FPS or 91 ms/frame).
Real-time video mosaicing was tested on several ex vivo and in vivo tissue specimens. All data collection followed IRB approved protocols (Rice University #11-119E and MD Anderson Cancer Center #PA11-0570). Figure 4A/B shows video mosaics obtained in vivo from the gingiva and skin of a normal volunteer, which demonstrates the algorithm performs well on different anatomical sites. In these examples video mosaicing increased the FOV 3-10 times. Our microendoscopy results reveal similar morphological features, such as hair strands and keratin sheaths, when compared to a skin mosaic obtained from a commercial benchtop confocal microscope (Lucid 2500, Fig. 4B, right). In Fig. 4C we present video mosaicing results from an ex vivo cervical specimen that was excised using electrosurgical techniques due to biopsy proven high grade dysplasia. The HRME probe was scanned from normal squamous epithelium to abnormal squamous epithelium (shown by the dotted white line). Lugol’s iodine was used to aid visualization of the abnormal tissue; dark brown staining indicates normal mucosa. In the high-resolution mosaic, the normal region shows small regularly-spaced nuclei, whereas the dysplastic region shows disorganized nuclear morphology. Determining this boundary is more difficult with the limited FOV of a single HRME frame, but is clearly defined in the mosaic result. Images labeled “Normal” and “CIN II” show histology sections at 20X optical magnification from the same boundary region. Throughout imaging experiments real-time mosaicing provided immediate feedback about probe stability and velocity, which is important when additional offline processing is desired.
4. Discussion and Conclusion
In this paper we describe a complete algorithm for real-time video mosaicing using a portable low-cost high-resolution microendoscope. The algorithm is executed on the laptop computer’s CPU well within the nominal frame rate of the camera. Speed could be further improved with the use of GPU programming, which can take advantage of parallel architecture and massive multithreaded data processing. While our technique provides accurate mosaics in many imaging applications, as shown with ex vivo and in vivo samples, there are several inherent limitations to the technique. There must be (1) spatial features within consecutive images, (2) spatial overlap between consecutive images, (3) minimal free-moving debris, (4) minimal tissue deformation, and (5) no probe rotation. These limitations could be prohibitive for some endoscopic cases, such as deep tissue sites where it may be difficult to slide the probe across the tissue, locations with significant debris, or lower gastro-intestinal sites where rotation may be encountered.
The solution to these limitations could be to use a two-step mosaicing strategy which involves (1) real-time mosaicing for live image acquisition and (2) post-procedural mosaicing for higher accuracy reconstruction. With post-procedural mosaicing there is no strict time requirement, which allows for techniques such as non-rigid image registration and template matching. These algorithms can overcome the limitations of real-time mosaicing because they can selectively ignore potential debris, deform images before registration, register images based on their best fit within an entire mosaic, rotate images, and blend overlapping images. Additionally, without a time limitation, the fiber pattern rejection step could be improved by estimating fiber core intensity values based on all pixels within each fiber core region. This could improve the signal-to-noise ratio of the reconstructed HRME images and mosaics.
Our current work on real-time video mosaicing provides a live picture of broad tissue morphology (>5 mm2) using a <1 mm diameter microendoscope probe. Future work aims to use supplemental post-procedural mosaicing with the HRME and to develop algorithms to automatically quantify features such as nuclear-to-cytoplasmic ratio and nuclei spacing within mosaics. We also plan to implement video mosaicing on a confocal microendoscope in order to combine the benefits of an extended FOV with increased axial response. These new techniques will then be used to test the diagnostic value of microendoscopy video mosaicing for several cancer types in clinical trials.
Acknowledgments
We would like to thank Matthew Kyrish and Benjamin Grant for assistance with image acquisition. This work was supported by NIH grants R01CA124319 and R01EB007594. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health.
References and links
- 1.Flusberg B. A., Cocker E. D., Piyawattanametha W., Jung J. C., Cheung E. L., Schnitzer M. J., “Fiber-optic fluorescence imaging,” Nat. Methods 2(12), 941–950 (2005). 10.1038/nmeth820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loewke K. E., Camarillo D. B., Jobst C. A., Salisbury J. K., “Real-time image mosaicing for medical applications,” Stud. Health Technol. Inform. 125, 304–309 (2007). [PubMed] [Google Scholar]
- 3.Hearp M. L., Locante A. M., Ben-Rubin M., Dietrich R., David O., “Validity of sampling error as a cause of noncorrelation,” Cancer 111(5), 275–279 (2007). 10.1002/cncr.22945 [DOI] [PubMed] [Google Scholar]
- 4.Becker V., Vercauteren T., von Weyhern C. H., Prinz C., Schmid R. M., Meining A., “High-resolution miniprobe-based confocal microscopy in combination with video mosaicing (with video),” Gastrointest. Endosc. 66(5), 1001–1007 (2007). 10.1016/j.gie.2007.04.015 [DOI] [PubMed] [Google Scholar]
- 5.Vercauteren T., Perchant A., Malandain G., Pennec X., Ayache N., “Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy,” Med. Image Anal. 10(5), 673–692 (2006). 10.1016/j.media.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 6.Wu K., Liu J. J., Adams W., Sonn G. A., Mach K. E., Pan Y., Beck A. H., Jensen K. C., Liao J. C., “Dynamic real-time microscopy of the urinary tract using confocal laser endomicroscopy,” Urology 78(1), 225–231 (2011). 10.1016/j.urology.2011.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T. Vercauteren, A. Meining, F. Lacombe, and A. Perchant, “Real time autonomous video image registration for endomicroscopy: fighting the compromises,” in Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing XV (International Society for Optical Engineering, San Jose, CA, USA, 2008). [Google Scholar]
- 8.Loewke K. E., Camarillo D. B., Piyawattanametha W., Mandella M. J., Contag C. H., Thrun S., Salisbury J. K., “In vivo micro-image mosaicing,” IEEE Trans. Biomed. Eng. 58(1), 159–171 (2011). 10.1109/TBME.2010.2085082 [DOI] [PubMed] [Google Scholar]
- 9.Behrens A., Bommes M., Stehle T., Gross S., Leonhardt S., Aach T., “Real-time image composition of bladder mosaics in fluorescence endoscopy,” Comput. Sci. Res. Dev. 26, 51–64 (2011). [Google Scholar]
- 10.Muldoon T. J., Pierce M. C., Nida D. L., Williams M. D., Gillenwater A., Richards-Kortum R., “Subcellular-resolution molecular imaging within living tissue by fiber microendoscopy,” Opt. Express 15(25), 16413–16423 (2007). 10.1364/OE.15.016413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muldoon T. J., Roblyer D., Williams M. D., Stepanek V. M., Richards-Kortum R., Gillenwater A. M., “Noninvasive imaging of oral neoplasia with a high-resolution fiber-optic microendoscope,” Head Neck 34(3), 305–312 (2012). 10.1002/hed.21735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce M., Yu D., Richards-Kortum R., “High-resolution fiber-optic microendoscopy for in situ cellular imaging,” J. Vis. Exp. 47(47), 2306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. Elter, S. Rupp, and C. Winter, “Physically motivated reconstruction of fiberscopic images,” in 18th International Conference on Pattern Recognition (Hong Kong, 2006), pp. 599–602. [Google Scholar]
- 14.Luck B. L., Carlson K. D., Bovik A. C., Richards-Kortum R. R., “An image model and segmentation algorithm for reflectance confocal images of in vivo cervical tissue,” IEEE Trans. Image Process. 14(9), 1265–1276 (2005). 10.1109/TIP.2005.852460 [DOI] [PubMed] [Google Scholar]
- 15.S. Rupp, C. Winter, and M. Elter, “Evaluation of spatial interpolation strategies for the removal of comb-structure in fiber-optic images ” in 31st Annual International Conference of the IEEE EMBS (Minneapolis, Minnesota, USA, 2009), pp. 3677–3680. [DOI] [PubMed] [Google Scholar]
- 16.P. Soille, Morphological Image Analysis: Principles and Applications (Springer, 1999). [Google Scholar]
- 17.Guizar-Sicairos M., Thurman S. T., Fienup J. R., “Efficient subpixel image registration algorithms,” Opt. Lett. 33(2), 156–158 (2008). 10.1364/OL.33.000156 [DOI] [PubMed] [Google Scholar]