Abstract
The phytohormone auxin contributes to virtually every aspect of the plant development. The spatiotemporal distribution of auxin depends on a complex interplay between auxin metabolism and intercellular auxin transport. Intracellular auxin compartmentalization provides another link between auxin transport processes and auxin metabolism. The PIN-LIKES (PILS) putative auxin carriers localize to the endoplasmic reticulum (ER) and contribute to cellular auxin homeostasis. PILS proteins regulate intracellular auxin accumulation, the rate of auxin conjugation and, subsequently, affect nuclear auxin signaling. Here, we investigate sequence diversification of the PILS family in Arabidopsis thaliana and provide insights into the evolution of these novel putative auxin carriers in plants. Our data suggest that PILS proteins are conserved throughout the plant lineage and expanded during higher plant evolution. PILS proteins diversified early during plant evolution into three clades. Besides the ancient Clade I encompassing non-land plant species, PILS proteins evolved into two clades. The diversification of Clade II and Clade III occurred already at the level of non-vascular plant evolution and, hence, both clades contain vascular and non-vascular plant species. Nevertheless, Clade III contains fewer non- and increased numbers of vascular plants, indicating higher importance of Clade III for vascular plant evolution. Notably, PILS proteins are distinct and appear evolutionarily older than the prominent PIN-FORMED auxin carriers. Moreover, we revealed particular PILS sequence divergence in Arabidopsis and assume that these alterations could contribute to distinct gene regulations and protein functions.
Keywords: PILS proteins, auxin, evolution, phylogeny, auxin metabolism, auxin homeostasis
Introduction
Plant development is particularly flexible due to its postembryonic growth behavior, allowing individual adjustment of the body plan according to the environment (Finet and Jaillais, 2012). The phytohormone auxin is crucial for these adaptive responses and, hence, has drawn enormous research attention (Teale et al., 2008). The importance of auxin for plant development seems to be also reflected in the complex regulation of auxin perception and its spatiotemporal distribution (Vanneste and Friml, 2009). Up to date three auxin receptor classes have been suggested to jointly regulate auxin-signaling output. Most auxin responses have been assigned to the F-box proteins TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX (TIR1/AFB). Auxin binding to the co-receptors TIR1/AFB and the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) will initiate the proteasome-dependent degradation of the transcriptional repressors Aux/IAAs. The subsequent release of AUXIN RESPONSE FACTOR (ARF) transcription factors eventually leads to the transcriptional reprogramming of the respective cell (Leyser, 2006; Chapman and Estelle, 2009). Another F-box protein S-PHASE KINASE-ASSOCIATED PROTEIN 2A (SKP2A) also binds to auxin and might contribute to auxin-dependent modulation of the cell cycle (Jurado et al., 2010). Rapid and non-genomic auxin effects appear to be mainly perceived by the AUXIN BINDING PROTEIN1 (ABP1; Jones and Venis, 1989; Robert et al., 2010; Xu et al., 2010). However, ABP1 action might also affect auxin-dependent gene transcription and cell cycle regulation (Braun et al., 2008; Tromas et al., 2009).
Beside the complex cell type-dependent regulation of auxin signaling, also auxin metabolism is multifaceted. Several redundant auxin biosynthesis pathways determine auxin levels in various tissues and the decay/inactivation of auxin is regulated via oxidation or mostly reversible conjugation (Woodward and Bartel, 2005; Ruiz Rosquete et al., 2012; Zhao, 2012). Auxin metabolism is highly dynamic and has pronounced importance for the spatiotemporal regulation of auxin.
Intercellular (polar) auxin transport also determines cellular auxin levels (Zazímalová et al., 2010). The most prominent auxin carriers are AUXIN-RESISTANT1/LIKE AUX1 (AUX/LAX) influx carriers, ATP BINDING CASSETTE (ABC) auxin transporters of a MULTIDRUG RESISTANCE (MDR) subfamily, and the PIN-FORMED (PIN) auxin carriers (Bennett et al., 1996; Chen et al., 1998; Gälweiler et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998; Geisler et al., 2005). PIN proteins have a particular developmental importance as their polar localization at a given cell side determines the direction of the intercellular auxin flow (Wisniewska et al., 2006). PIN proteins can be grouped into two subclasses according to the length of the central hydrophilic loop. Canonical PIN1-type auxin efflux carriers have a long loop, localize to the plasma membrane (PM) and perform a rate-limiting function in cellular auxin efflux (Petrásek et al., 2006). In contrast, PIN5 and PIN8 have a dramatically reduced central hydrophilic loop, localize to the endoplasmic reticulum (ER) and regulate intracellular auxin compartmentalization and homeostasis (Mravec et al., 2009; Bosco et al., 2012; Ding et al., 2012).
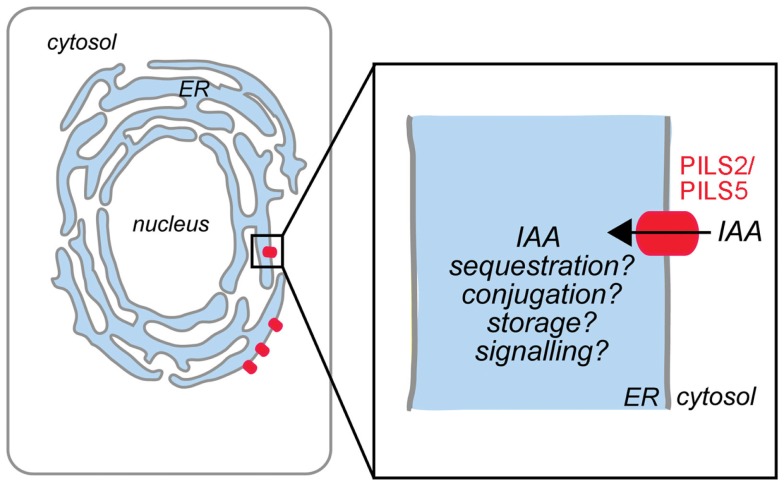
We have recently discovered a novel putative auxin carrier family of seven members in Arabidopsis thaliana (Barbez et al., 2012) and designated them as PIN-LIKES (PILS), because their predicted protein topology is highly similar to the topology of the PIN proteins. Similar to PIN proteins, PILS contain the so-called Interpro auxin carrier domain, an in silico defined domain to predict auxin transport function. Functional PILS5-GFP fusion proteins localize to the ER and stimulate intracellular auxin accumulation in plant and yeast cells (Barbez et al., 2012). PILS2 and PILS5 activity increases amide auxin conjugates, thereby reducing the free auxin levels, and negatively affecting nuclear auxin signaling (Barbez et al., 2012). Our current working model proposes that PILS2 and PILS5 proteins regulate auxin compartmentalization into the ER lumen, where auxin might be the substrate for compartmentalized auxin metabolism (Figure 1). It needs to be experimentally tested whether PILS proteins affect nuclear auxin signaling mainly by limiting the excess of auxin to diffuse into the nucleus or by the effect on presumably compartmentalized auxin conjugation. This mode of action is reminiscent to auxin carrier PIN5 that has been shown to regulate intracellular auxin homeostasis by modulating auxin compartmentalization and metabolism at the ER (Mravec et al., 2009). Further research will address whether the distinct protein families have redundant and interchangeable function at the ER.
Figure 1.
Model of PILS protein function in Arabidopsis thaliana. PILS2 and PILS5 proteins localize to the ER and mediate intracellular auxin accumulation. We hypothesize that PILS proteins are putative auxin carriers that regulate the auxin transport from the cytosol into the lumen of the ER (black arrow). PILS activity affects auxin metabolism and might control the cytoplasmic availability of auxin (adapted from Barbez et al., 2012).
PILS overexpression strongly distorts plant patterning and development, while pils2 and pils5 loss of function mutants show comparably weaker defects in plant growth regulation. Moderate PILS5 gain and pils2pils5 loss of function phenotypes can be largely explained by low and high auxin content, respectively. For example, PILS5 overexpressors have reduced free auxin levels/signaling, shorter hypocotyls and fewer lateral roots, while pils2pils5 double mutants display higher free auxin levels, enhanced hypocotyl growth and lateral rooting. In contrast, PILS5 gain andpils2pils5 loss of function leads to reduced and enhanced root growth, which might be not related to the overall changes in auxin content, but could indicate a more specific PILS2 and PILS5 function in the cellular regulation of root growth (Barbez et al., 2012).
The identification of PILS proteins and their role in auxin homeostasis at the ER reveal the molecular complexity of intracellular auxin compartmentalization and its eminent importance for the plant development. Here we present in silico analysis to further reveal insight into the organization and regulation of this novel family of putative auxin transport facilitators.
Material and Methods
Sequence information
Sequences were downloaded from PLAZA1, NCBI2 by using tblastx program (Altschul et al., 1997; nr/nt database, PILS and PIN sequences from A. thaliana as queries) or Phytozome3 servers. The information and the ID of the presented sequences can be found in the Supplementary Data.
Online servers
The online available servers used to perform in silico analyses of PILSes are found at: http://www.arabidopsis.org (chromosome localization; alternative splicing), http://bioinformatics.psb.ugent.be/plaza/ (intron/exon organization; sequence information), www.genevestigator.com (expression), http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi (expression), http://www.enzim.hu/hmmtop/index.php (prediction of protein topology), http://biophysics.biol.uoa.gr/TMRPres2D (visual representation of transmembrane protein models), http://weblogo.berkeley.edu/logo.cgi (sequence logo), http://www.cbs.dtu.dk/services/NetPhos/ (prediction of phosphorylation sites), http://www.cbs.dtu.dk/services/NetPhosK (prediction of phosphorylation sites), http://www.ebi.ac.uk/Tools/msa/clustalw2/ (multiple amino acid sequences alignment), http://blast.ncbi.nlm.nih.gov/ (sequence information), http://www.phytozome.net/ (sequence information), http://www.r-project.org/ (analysis of collinearity).
Analysis of collinearity
We investigated possible collinearity among A. thaliana PILS genes by comparing 200 surrounding translated genes for each PILS. The comparison was performed for pairs of PILS genes by using blastp program (Altschul et al., 1997). The homology was determined according to E-value from blast results. The analysis was performed in R environment4.
Phylogenetic analysis
A multiple alignment was built by using Muscle in MEGA5 software (Tamura et al., 2011). Only the conserved domains were used and all positions with less than 80% site coverage were eliminated. The evolutionary history was inferred by using the Maximum Likelihood method based on the Whelan and Goldman (2001) + Freq. model with discrete Gamma distribution (five categories, G parameter = 3.0640) for analysis of PILS amino acid sequences or on the Whelan and Goldman model with discrete Gamma distribution (five categories, G parameter = 2.9920) for analysis of PIN-PILS dataset. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The PILS analysis involved 42 amino acid sequences and 322 positions in the final dataset. The PIN-PILS analysis involved 67 amino acid sequences and 354 positions. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). For the sequence alignments see Figures S2 and S3 in Supplementary Material.
Results
Phylogeny of PILS proteins
Using available online tools, we previously showed that PILS proteins are highly conserved among plant species (Barbez et al., 2012). To further investigate the evolution of PILS protein diversification, we analyzed PILS protein sequences from all sequenced taxa of Viridiplantae. The PILS family is present in all the 26 available sequenced genomes and is represented by 202 genes (Table 1; Van Bel et al., 2012; confirmed by reciprocal blast, Altschul et al., 1997). PILS family obviously diversified in the different plant lineages (Table 1). Ancient species, such as algae (1–2), mosses (5), and spike mosses (8), have 1–8 PILS genes, while seed plants, such as Oryza (6), Zea (10), Medicago (13), or Populus (18), have 6 to 18 PILS genes (Table 1). The steadily increasing number in seed plants suggests that PILS genes have duplicated independently in several plant lineages and indicate a more diversified function of PILS proteins in higher plants.
Table 1.
Size of PILS gene families in different plant species.
| Lineage | Species | Number of genes* |
|---|---|---|
| Eukaryota | ||
| Viridiplantae | ||
| Chlorophyta | Micromonas sp. | 1 |
| Micromonas pusilla | 1 | |
| Ostreococcus lucimarinus | 1 | |
| Ostreococcus tauri | 1 | |
| Chlamydomonas reinhardtii | 2 | |
| Volvox carteri | 2 | |
| Bryophyta | Physcomitrella patens | 5 |
| Lycopodiophyta | Selaginella moellendorffii | 8 |
| Euphyllophyta | ||
| Monocots | Oryza sativa japonica | 6 |
| Oryza sativa indica | 6 | |
| Sorghum bicolor | 7 | |
| Brachypodium distachyon | 8 | |
| Zea mays | 10 | |
| Dicots | Carica papaya | 4 |
| Arabidopsis lyrata | 6 | |
| Arabidopsis thaliana | 7 | |
| Ricinus communis | 7 | |
| Fragaria vesca | 8 | |
| Manihot esculenta | 9 | |
| Lotus japonicus | 12 | |
| Medicago truncatula | 13 | |
| Vitis vinifera | 13 | |
| Malus domestica | 14 | |
| Theobroma cacao | 16 | |
| Glycine max | 17 | |
| Populus trichocarpa | 18 |
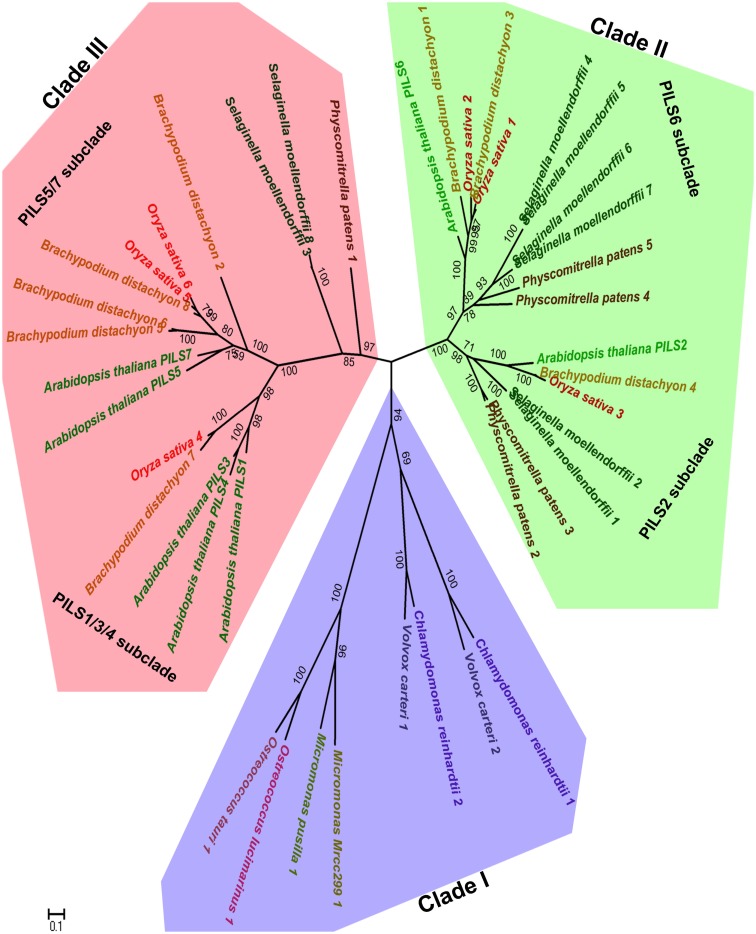
To assess the evolutionary relationship among PILS proteins, we constructed phylogenetic trees with PILS sequences from selected model organisms such as available green algae, Physcomitrella, Selaginella, Picea, Brachypodium, Oryza, Medicago, Arabidopsis, and Populus sequences (Figure 2; Figure S1 in Supplementary Material; for sequence alignment see Figure S2 in Supplementary Material). The phylogenetic tree presented in Figure 2 shows that PILSes from Viridiplantae can be grouped into three evolutionary clades: Clade I, Clade II, and Clade III.
Figure 2.
Phylogeny of PILS proteins. The phylogenetic tree of PILS proteins can be divided into three clades: Clade I (blue), Clade II (green), and Clade III (red). The Arabidopsis PILSes are found in the Clade II and Clade III, while Clade I is represented only by PILS proteins from algae. The Maximum Likelihood molecular phylogenetic analysis was performed in MEGA5 (Tamura et al., 2011) by using 42 amino acid PILS sequences from algae, Physcomitrella, Selaginella, Brachypodium, Oryza, and Arabidopsis as explained in the Materials and Methods.
The available green algae genomes from the lineage Chlorophyta have a relatively low number of only one or two PILS genes per species. All these PILS algae orthologs cluster together and define the Clade I that contains the so far oldest known PILS genes of the Viridiplantae (Figure 2). We could also identify putative PILS genes in the genomes of sequenced algae from lineage Streptophyta from which the land plants evolved. However, we did not include these sequences in the phylogenetic analysis because they were incomplete (only EST fragments are currently available).
The evolutionary Clade II and III already emerged early during non-vascular plant evolution and both contain PILS sequences from Embryophytes (land plants; Figure 2). The main lineages of land plants are mosses, liverworts, hornworts, lycophytes, ferns, gymnosperms, and angiosperms. Clade II includes the well-conserved PILS2- and PILS6-like subclades, including orthologs of PILS2 and PILS6 from Physcomitrella, Selaginella, Brachypodium, or Oryza (Figure 2).
Clade III encompasses the PILS1/PILS3/PILS4- and PILS5/PILS7-like subclades and displays particular expansion in higher seed plants (Figure 2; Figure S1 in Supplementary Material). Accordingly, this clade encompasses also most Brachypodium and Oryza orthologs (Figure 2; Figure S1 in Supplementary Material). Interestingly, one Physcomitrella and two Selaginella PILS sequences are present at the root of the Clade III (Figure 2). The relatively low number of moss and the relative over amount of higher plant sequences in Clade III may suggest particular importance of this clade in vascular plant evolution.
Our analysis reveals that PILS proteins are evolutionarily conserved throughout plant evolution and might uncover the versatile importance of compartmentalized auxin homeostasis throughout the plant kingdom.
PILS proteins are evolutionarily distinct of PIN proteins
The canonical PIN proteins act in the cellular efflux of auxin at the plasma membrane, but the most ancient members of PIN proteins (PIN5-type) localize to the ER and regulate the subcellular compartmentalization of auxin and auxin metabolism (Mravec et al., 2009). Hence, both PILS and PIN5-like proteins localize to the ER and regulate auxin homeostasis, presumably by mediating auxin transport at the ER (Mravec et al., 2009; Barbez et al., 2012; Bosco et al., 2012; Ding et al., 2012).
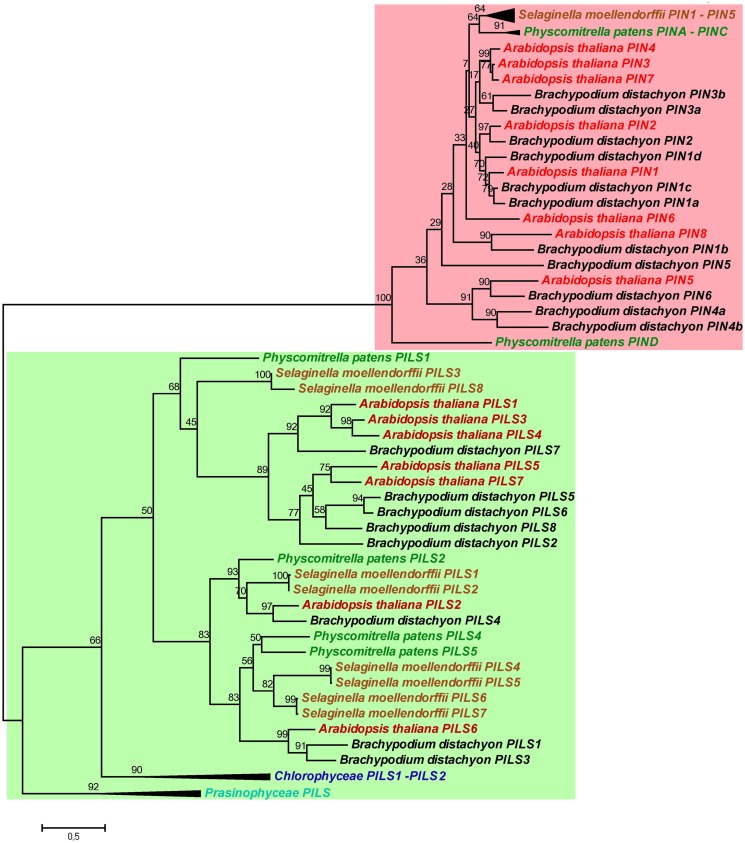
Next, we investigated the evolutionary relationship between PILS and PIN proteins (Figure 3; for sequence alignment see Figure S3 in Supplementary Material). The phylogenetic analysis of PILS and PIN sequences from algae, moss, spikemoss, and several Angiosperms revealed that PILSes and PINs form two distinct phylogenetic clades (Figure 3). Although having a similar predicted protein structure and possibly similar function at the ER, PIN and PILS proteins are evolutionarily distinct in plants. In contrast to PILSes, we could not find any PIN sequence in the genomes of Chlorophyta algae, such as Chlamydomonas, Micromonas, Ostreococcus, or Volvox. Notably, a truncated PIN sequence has been found in the genome of Spirogyra (De Smet et al., 2011). These findings indicate that PILS proteins are more conserved during plant evolution and seem evolutionarily older than PIN proteins. Therefore, we assume that the PILS proteins are central to the evolution of intracellular auxin transport, which presumably has preceded the evolution of PIN-dependent intercellular and intracellular auxin transport.
Figure 3.
Phylogeny of PILS and PIN proteins. PILS proteins are evolutionarily distinct of PIN proteins. Note the two separated subtrees. The Maximum Likelihood phylogenetic analysis was performed in MEGA5 (Tamura et al., 2011) by using PILS and PIN amino acid sequences from algae, Physcomitrella, Selaginella, Brachypodium, and Arabidopsis. For a better visualization the algae and sometimes the Physcomitrella and Selaginella branches were collapsed.
PILS diversification in Arabidopsis thaliana
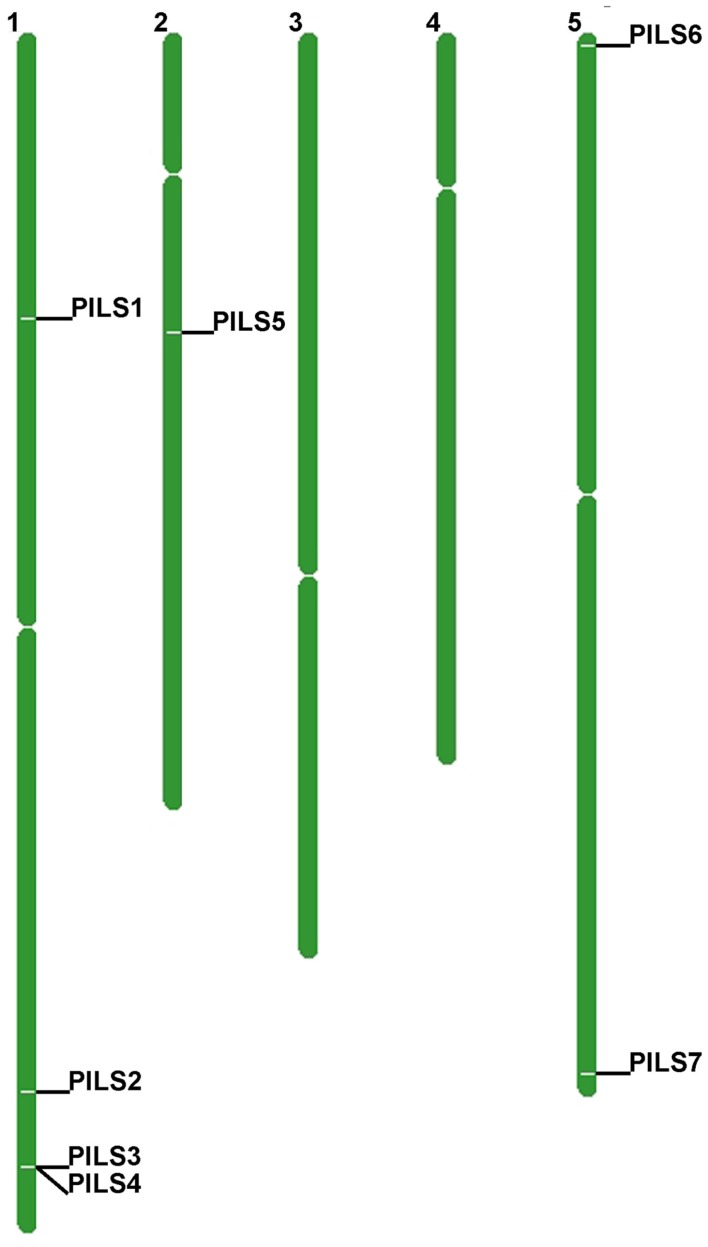
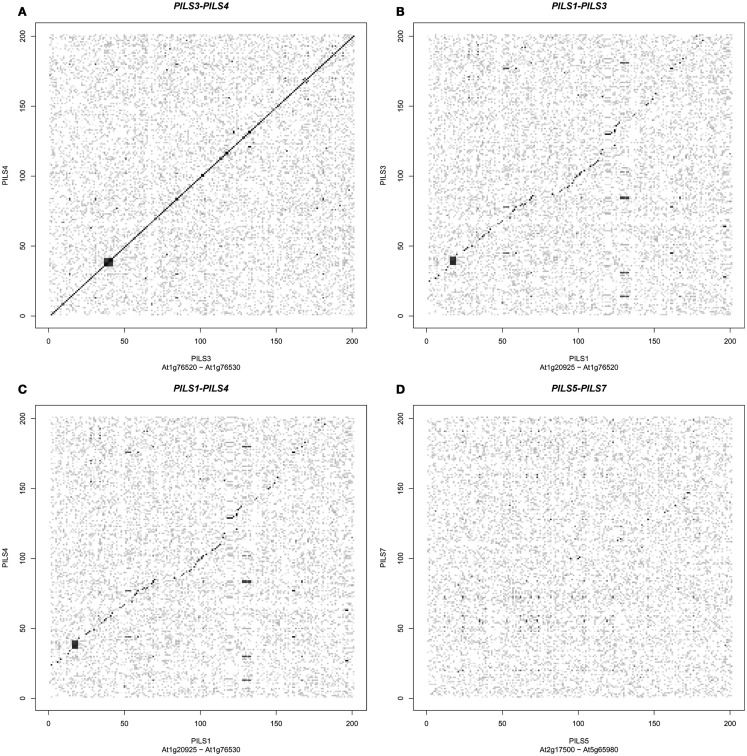
The seven Arabidopsis PILS genes are placed on chromosome 1, 2, and 5 (Figure 4). PILS1 to PILS4 are found on chromosome 1, PILS5 on chromosome 2, while PILS6 and PILS7 are both placed at the ends of the chromosome 5 (Figure 4). PILS3 and PILS4 are neighboring genes at the bottom arm of the chromosome 1 (Figure 4), indicating that PILS3 and PILS4 may resulted from a gene duplication event. To investigate PILS paralogs in A. thaliana we performed comparative sequence analysis of genes that surround the seven PILS genes (Figure 5). Rows of 200 translated genes surrounding each of the seven PILS genes were analyzed in pairs by blastp program (Altschul et al., 1997) and homology between all genes in all unique pairs of gene rows were determined according to E-value from blast results. Pairs of gene rows with high diagonal homology were assigned as collinearity. In the PILS1/PILS3/PILS4 group we found very high collinearity between PILS3 and PILS4 (Figure 5A). These genes appear to be products of very recent gene duplication. Between PILS1 and the PILS3/PILS4 pair we also found high collinearity (Figures 5B,C) and assume that these genes arose during full-genome duplication at Brassicaceae family level (20 million years ago; Mya). Only very weak or no collinearity was detectable between PILS5 and PILS7 (Figure 5D).
Figure 4.
Chromosomal distribution of Arabidopsis thaliana PILS genes. A. thaliana PILS genes are found on chromosome 1, 2, and 5.
Figure 5.
Origin of PILS paralogs in Arabidopsis thaliana. (A–D) PILS genes show high collinearity between gene pairs PILS3/PILS4 (A), PILS1/PILS3 (B), and PILS1/PILS4 (C). No or only very weak collinearity could be detected between PILS5 and PILS7 (D).
To further elaborate on the recent duplication of PILS3 and PILS4, we analyzed the microevolutionary relationship between PILS sequences of A. thaliana and A. lyrata (Figure S1 in Supplementary Material). A. lyrata is the closest known relative of A. thaliana and has a genome of eight chromosomes and six PILS proteins (Van Bel et al., 2012). In contrast, A. thaliana has five chromosomes and seven PILS proteins (Barbez et al., 2012; Van Bel et al., 2012). It has been shown that the reduction of genome size in A. thaliana is the result of chromosomes fusion that presumably occurred about 5 Mya (Yogeeswaran et al., 2005). The phylogenetic analysis revealed that all six A. lyrata PILSes have highly similar orthologs in A. thaliana, while AtPILS4 is a lineage-specific gene (Figure S1 in Supplementary Material). This indicates that AtPILS4 is a duplicated gene that has arisen after the separation of A. thaliana from A. lyrata 5 Mya.
Next we addressed the sequence diversifications among the A. thaliana PILS proteins and performed a ClustalW Multiple sequence alignment (Larkin et al., 2007; Table 2; for sequence alignment see Figure S4 in Supplementary Material). PILS3/PILS4 showed the highest identity (82%), followed by PILS1/PILS3 (69%), PILS5/PILS7 (64%), and PILS1/PILS4 (61%; Table 2). Interestingly, PILS2 and PILS6 showed sequence identity of only 39% (Table 2). Taking together, the amino acid identity proposes that A. thaliana PILS proteins belong to three subgroups: (i) PILS1/PILS3/PILS4, (ii) PILS5/PILS7, and (iii) PILS2/PIL6.
Table 2.
Percentages* of Arabidopsis thaliana PILS amino acid sequence identity calculated by ClustalW multiple sequence alignment (Larkin et al., 2007).
| PILS7 | |||||||
| 31 | PILS6 | ||||||
| 31 | 64 | PILS5 | |||||
| 39 | 28 | 43 | PILS4 | ||||
| 82 | 41 | 31 | 43 | PILS3 | |||
| 32 | 28 | 29 | 39 | 30 | PILS2 | ||
| 28 | 69 | 61 | 40 | 29 | 42 | PILS1 | |
| PILS1 | PILS2 | PILS3 | PILS4 | PILS5 | PILS6 | PILS7 | |
*The identity percentages were calculated as the identities between two PILS sequences, divided by the length of the alignment.
PILS gene regulation and organization in Arabidopsis thaliana
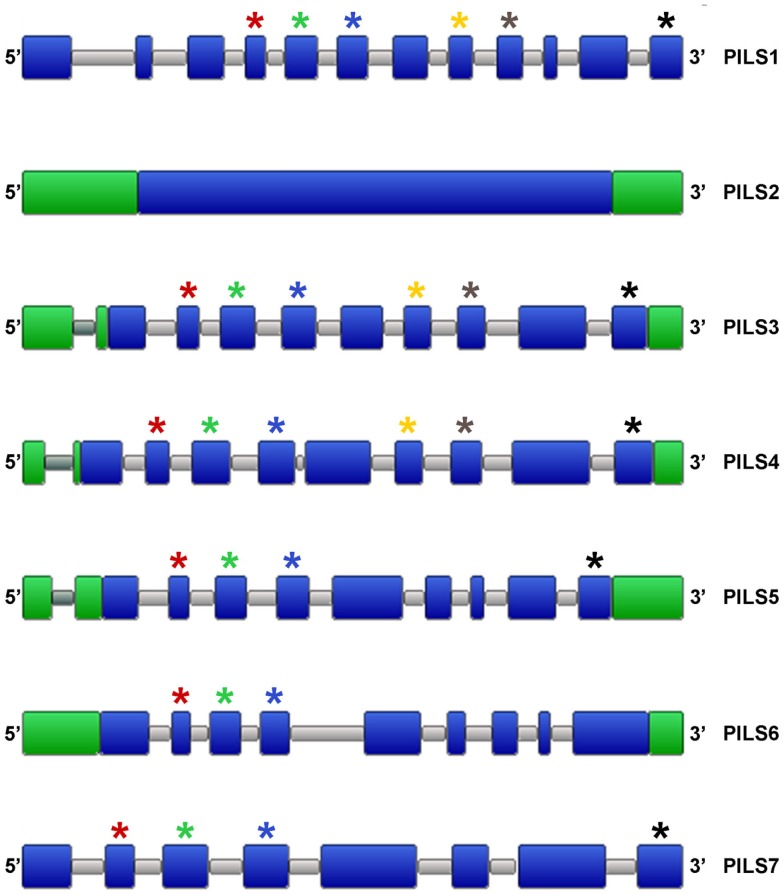
To get further insight into the regulation of PILS activity, we analyzed in silico PILS gene organization and expression. A. thaliana PILS gene transcripts organization is pretty well-conserved regarding the number and size of the exons (Figure 6). PILS3 to PILS6 genes contain nine exons with more or less conserved size and placements (Figure 6). In contrast, PILS1, PILS2, and PILS7 have a divergent exon/intron structure. PILS1 has 12 exons, PILS7 bares eight exons and PILS2 is even intron less. The size of exon number 2 (80 nucleotides), 3 (125 nucleotides), and 4 (122 nucleotides) is largely kept in AtPILS genes and encode for a highly conserved region of the predicted transmembrane helices 2–4 (109 aa in total). Also a C-terminal transmembrane domain seems to be encoded by the last exon (125 nucleotides) in almost all AtPILS genes (Figure 6).
Figure 6.
Organization of Arabidopsis thaliana PILS genes. Schematic intron/exon representation of A. thaliana PILS genes (Van Bel et al., 2012). Exons and introns are depicted in blue and gray, respectively. 3′ UTR and 5′ UTR are in green. Stars are showing exons of similar sizes (nucleotides): red (80), green (125), blue (122), yellow (93), gray (101), and black (125).
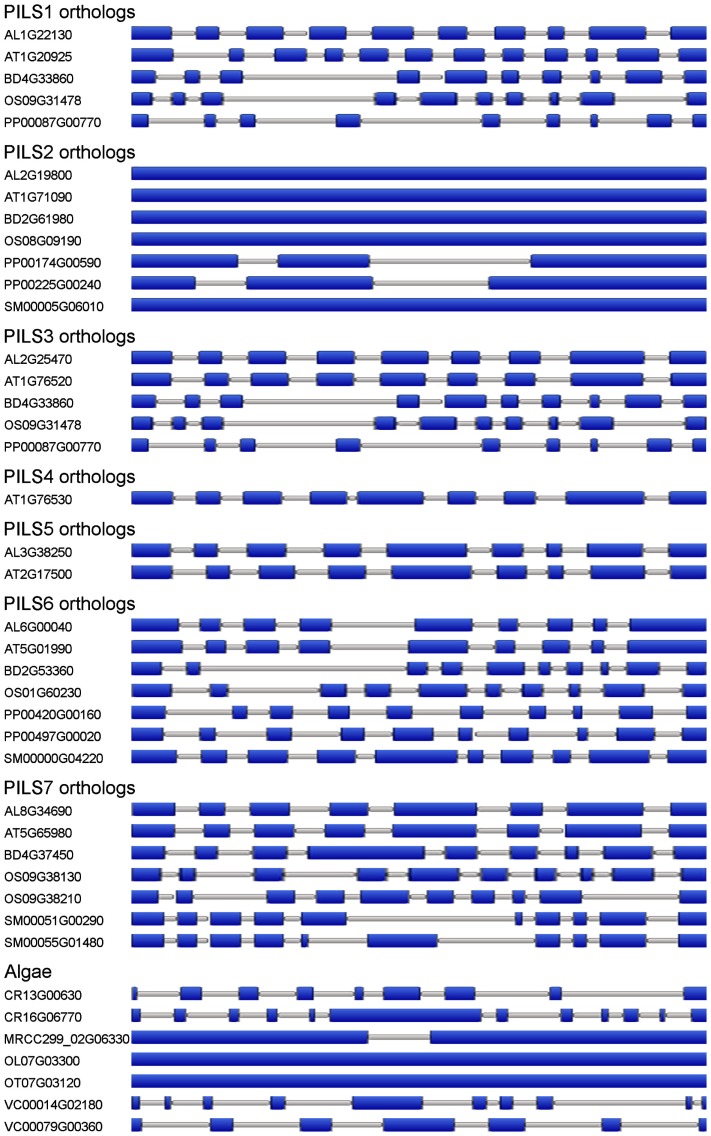
Next, we analyzed the intron/exon organization of PILS genes from algae, Physcomitrella, Selaginella, and several Angiosperms. Our results show that PILS intron/exon organization is largely conserved among PILS orthologous (Figure 7). The variations of 1–2 more or less exons may be the result of insertions, deletions, or both processes along the lineage evolution. The subfamily of PILS2 genes is most particular, because they display single-exon genes in Angiosperms and Selaginella and 3-exons genes in Physcomitrella (Figure 7). Thus, PILS genes belong to two structural groups with 1–3 exons (PILS2 orthologs and PILS genes from Ostreococcus and Micromonas) and 7–12 exons (all the other PILSes; Figure 7).
Figure 7.
Organization of PILS orthologs. Schematic intron/exon representation of PILS genes from Arabidopsis lyrata (AL), Arabidopsis thaliana (AT), Brachypodium distachyon (BD), Chlamydomonas reinhardtii (CR), Micromonas (MRCC299), Oryza sativa (OS), Ostreococcus lucimarinus (OL), Ostreococcus tauri (OT), Physcomitrella patens (PP), Selaginella moellendorffii (SM), and Volvox carteri (VC; Van Bel et al., 2012). Exons are depicted in blue boxes, introns in gray lines.
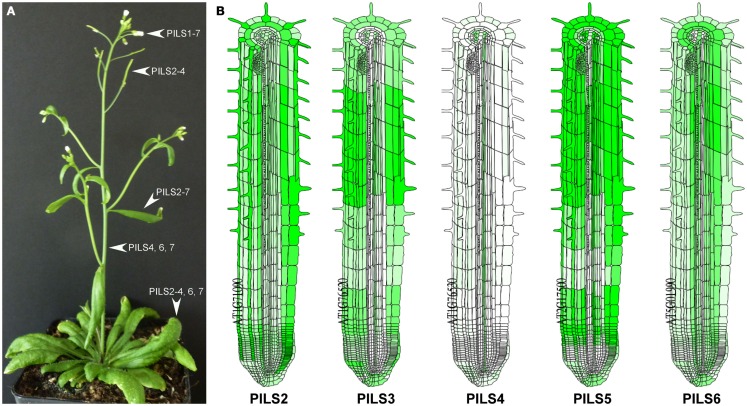
PILS gene activity can be detected in all tissues of A. thaliana as shown by RT-PCR (Barbez et al., 2012) or by micro array-based online tools such as Genevestigator5. PILS genes display either relatively low (PILS1, PILS4, PILS7), medium (PILS6) or high (PILS2, PILS3, and PILS5) expression levels (see text footnote 5). PILS2-to-AtPILS6 are expressed in seedlings, leaves, and flowers (Figure 8; Barbez et al., 2012; see text footnote 5). PILS4 displays the strongest expression in the rosette leaves (Barbez et al., 2012). PILS6 transcripts are particularly abundant in the stem and together with PILS5 in the cauline leaves and flowers, while PILS2 is highest in siliques (Barbez et al., 2012). Interestingly, some PILS gene products were excluded from certain tissues. PILS1 was found to be expressed only in flowers, PILS2 and PILS3 are not expressed in the stem, PILS5 is absent in the rosette leaves, stem, and siliques, while PILS6 and PILS7 were present in all plant organs but not in siliques (Barbez et al., 2012). Except PILS1, all the other PILSes were expressed in seedlings, with PILS5 and PILS2 having the highest expression (Barbez et al., 2012). PILS2-to-PILS6 showed expression in pollen with PILS5 being the most abundant (see text footnote 5). Based on these evidences it seems that PILS genes show specific and partially overlapping expression patterns in all plant tissues.
Figure 8.
Transcription pattern of Arabidopsis thaliana PILS genes. (A,B) Transcription of PILS genes in A. thaliana plant (A) and root (B). Based on the RT-PCR data from individual organs published in Barbez et al. (2012). (A) As predicted by online server Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi); (B).
Alternative splicing might furthermore contribute to the regulatory complexity and diversity for PILS gene activity. PILS3 and PILS5 appear to bear two and four alternative transcripts, respectively6. In both cases the alternative gene splicing seems to occur in the 5′ region and may modulate PILS3 and PILS5 function. However, the importance of PILS transcript splicing remains to be demonstrated.
The pronounced differences in the expression levels and tissue distributions might indicate that PILS-mediated regulation of plant growth and development may be largely determined by gene regulation.
PILS protein organization in Arabidopsis thaliana
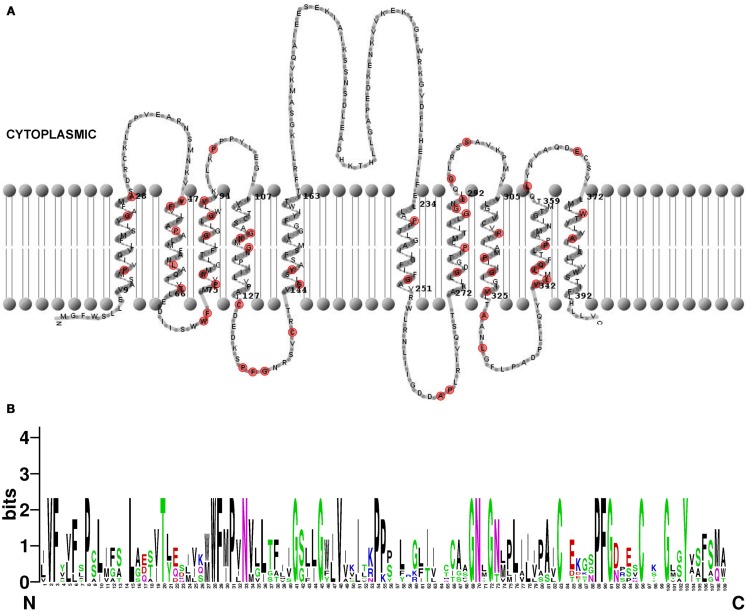
The temporal and spatial regulation of PILS genes will give rise to tissue specific distribution of distinct PILS proteins. Next we analyzed predicted PILS protein organization and searched for domains to speculate on PILS function. PILS proteins range in size from 390 (43 kDa; PILS3) to 472 (52 kDa; PILS1) amino acids. However, the predicted protein topology is highly similar for all PILS proteins. PILS proteins are presumably characterized by two hydrophobic transmembrane regions found at N- and C-termini (Figure 9A; Tusnády and Simon, 1998, 2001; Spyropoulos et al., 2004). The two transmembrane regions flank a short hydrophilic region (loop) with a presumable cytosolic orientation (Figure 9A). Each hydrophobic region appears to be organized in five transmembrane helices that are very similar and highly conserved among the PILS proteins (Figure 9). In contrast, the loop is less conserved and is the most divergent part of the PILS sequences. We assume that the transmembrane domains have central roles in the putative carrier function, while the presumably cytosolic loop might have rather regulatory functions.
Figure 9.
Structure of Arabidopsis thaliana PILS proteins. (A) Predicted topology of A. thaliana PILS5 protein. The prediction was done by HMMTOP 2.0 (Tusnády and Simon, 1998, 2001) and visualized by TMRPres2D (Spyropoulos et al., 2004). Conserved amino acids in all seven PILS proteins are marked in red. (B) Sequence logos generated by WebLogo (Schneider and Stephens, 1990) representing a ClustalW multiple sequence alignment (Larkin et al., 2007) of 109 amino acids from N-terminal region of A. thaliana PILS proteins (exons 2–4). Note the PILS sequence conservation at the highest, single symbol positions.
PILS and PIN proteins share only 10–18% sequence identity and belong to distinct protein families (Figure 3; Barbez et al., 2012). However, the predicted topology of PILS proteins is reminiscent to the predicted topology of PIN proteins (Krecek et al., 2009) and allowed the identification of this novel putative auxin carrier family (Barbez et al., 2012). Based on the hydrophilic loop size, PIN proteins are sub-grouped into two subfamilies. The subfamily of PIN1-type encompasses the PIN members with a long hydrophilic loop and PM localization (PIN1–PIN4, PIN7), while the subfamily of PIN5-type encompasses PIN5 and PIN8 that have very short hydrophilic loops and ER localization. Although PIN6 shows a reduction of the loop size, PIN6 is often included in the PIN1-type subfamily due to high sequence similarity in the transmembrane regions (Krecek et al., 2009). However, it is also localized to the ER in transient localization studies (Mravec et al., 2009).
Similarly to PIN proteins, PILS family members are characterized by the presence of the Interpro auxin carrier domain. This Interpro domain is relatively long and spans almost the whole length of the PILS protein and, hence, it is difficult to ascertain functional residues within the “domain”.
Nothing is yet known about the post-translational modification of PILS proteins but generic phosphorylation sites (non-kinase-specific, such as serine, threonine, and tyrosine), kinase-specific phosphorylation sites and isoform variations could be predicted for PILS proteins by available online servers such as NetPhos (Blom et al., 1999) and NetPhosK (Blom et al., 2004). Interestingly, according with the number of the predicted serine, threonine, and tyrosine phosphorylation sites, PILS proteins can be grouped into three classes: (i) less than 10 (PILS5 and PILS7), (ii) between 10 and 15 (PILS2 and PILS6), and (iii) more than 15 (PILS1, PILS3, and PILS4). This finding may indicate the functional diversification among the PILS members and may suggest that different phosphorylation-based mechanisms are required for the regulation of PILS activity.
Discussion
Auxin has pronounced importance for the plant development. Recent research shed light on a particular link between intracellular auxin transport processes and auxin metabolism (Mravec et al., 2009; Barbez et al., 2012; Bosco et al., 2012; Ding et al., 2012). Here, we report in silico analyses of PILS putative auxin flux facilitator sequences from A. thaliana and revealed certain features that might be functionally important for PILS activity.
The phylogenetic analysis of PILS sequences revealed that four Physcomitrella PILSes are found in Clade II, while only one is found in Clade III (Figure 2). Moreover, two Selaginella PILSes are found in each, Clade III and Clade II-PILS2 subclade, while four paralogs are found in the Clade II-PILS6 subclade (Figure 2). This, together with the distribution of the Brachypodium, Oryza, and Arabidopsis PILS sequences, indicates that the initial PILS divergence occurred in two separate clades already at the level of Bryophytes. We do not know if PILSes are present in the genome of Rhodophytes, but we can speculate that Clade II- and Clade III-PILSes may have originated before land plant evolution at the level of Streptophytes, as these algae are direct ancestors of land plants. Moreover, Clade II presumably diverged before or during the origin of Embryophytes, because this clade is already diversified in PILS2- and PILS6-like subclades in mosses (Figure 2). Clade III particularly expanded during higher plant evolution (Figure 2; Figure S1 in Supplementary Material). This clade is divided in PILS1/PILS3/PILS4 and PILS5/PILS7 subclades (Figure 2; Figure S1 in Supplementary Material). We could not estimate when these subclades emerged because PILS sequences from conifers and ferns are either incomplete (only ESTs available) or not available. More than 30000 ESTs derived from gametophyte of fern Adiantum can be found on NCBI but we could not identify any PILS sequence which indicates that PILSes might be not transcribed in gametophyte.
Combining the gene and protein analyses, AtPILS4 is likely to be a recent duplication of AtPILS3, because they show very high amino acid identity (Table 2), strong gene collinearity (Figure 5A), and no particular PILS4 orthologs could be identified in the genomes of the other sequenced species. PILS3/PILS4 seem to be originally derived from PILS1 (69% amino acid identity; Table 2). Accordingly, by analyzing the amino acid sequence similarities and the PILS phylogeny, we can conclude that from seven PILSes in A. thaliana genome, six in Oryza sativa ssp japonica and eight in Brachypodium distachyon genome only four are true orthologs. The other PILS members presumably represent lineage-specific duplications that occurred after the separation of the dicots and monocots about 200–250 Mya.
The existence of PILS2 as a single-exon gene in most species is intriguing since single-exon genes are rather typical for prokaryotes. However, single-exon or intronless genes are present in eukaryotic genomes (Sakharkar et al., 2004) and can have many origins, but could pinpoint the relatedness to a prokaryotic gene (Zou et al., 2011). However, moss PILS2 orthologs display intronexon structure (Figure 7) and might suggest that PILS2 genes lost the intron structure during evolution.
Our findings might highlight certain functional diversifications among PILS proteins. Notably, PILS2 and PILS5 have only 29% amino acid sequence identities (Table 2), display very diverged gene organization (Figure 6), and belong to diverse evolutionary sub clades (Figure 2). However, their gene regulation and function seem to be highly similar in Arabidopsis, because PILS2 and PILS5 have overlapping expression pattern in the root transition zone and redundantly control seedling growth and development (Barbez et al., 2012). Therefore, defined research is needed to evaluate the functional importance of the distinct features of the respective PILS genes and PILS proteins.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Traffic_and_Transport/10.3389/fpls.2012.00227/abstract
Molecular phylogenetic analysis of PILS proteins. The diagram shows an extended phylogentic tree of PILS proteins with collapsed branches for algae, Physcomitrella, and Selaginella. Note the high diversification of PILSes in Medicago and Populus. Because of incomplete sequences some of the PILSes were eliminated. The evolutionary history was inferred by using the Maximum Likelihood method based on the Data specific model (Nei and Kumar, 2000). The tree with the highest log likelihood (−55875.7936) is shown. The percentage of trees in which the associated taxa clustered together is shown above the branches. Initial tree(s) for the heuristic search were obtained automatically as follows. When the number of common sites was <100 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise BIONJ method with MCL distance matrix was used. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 2.6899)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 3.7299% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 75 nucleotide sequences. All positions with less than 0% site coverage were eliminated. That is, fewer than 100% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 1113 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Alignment of PILS amino acid sequences. The multiple amino acid alignment of PILSes was generated by using Muscle in MEGA5 software (Tamura et al., 2011). This alignment was generated for the phylogenetic analysis presented in the Supplementary Figure 1. The alignment for the smaller tree presented in the Figure 2 is similar but with less sequences.
Alignment of PILS and PIN amino acid sequences. The multiple alignment was generated by using Muscle in MEGA5 software (Tamura et al., 2011).
Alignment of Arabidopsis thaliana PILS amino acid sequences. A multiple sequence alignment generated by ClustalW server (Larkin et al., 2007) of the seven PILSes is shown. Amino acids are color coded: red (small, hydrophobic, aromatic, not Y), blue (acidic); magenta (basic), green (hydroxyl, amine, amide, basic), gray (others). “*,” Identical amino acids; “:,” conserved substitutions (same color group); “.,” semi-conserved substitution (similar shapes).
Sequence information.
Acknowledgments
We are indebted to the Vienna Science and Technology Fund (WWTF; to Elena Feraru, Mugurel I. Feraru, Jürgen Kleine-Vehn), Czech Science Foundation Grant GAP305/11/2476 (to Jan Petrášek) and Charles University in Prague, SVV 265203/2012 (to Jan Petrášek and Stanislav Vosolsobě). We thank Elke Barbez for helpful discussions.
Footnotes
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E., Kubeš M., Rolcík J., Béziat C., Pencík A., Wang B., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485, 119–122 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Bennett M. J., Marchant A., Green H. G., May S. T., Ward S. P., Millner P. A., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950 10.1126/science.273.5277.948 [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. (1999). Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 10.1006/jmbi.1999.3310 [DOI] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- Bosco C. D., Dovzhenko A., Liu X., Woerner N., Rensch T., Eismann M., et al. (2012). The endoplasmic reticulum localized PIN8 is a pollen specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 71, 860–870 10.1111/j.1365-313X.2012.05037.x [DOI] [PubMed] [Google Scholar]
- Braun N., Wyrzykowska J., Muller P., David K., Couch D., Perrot-Rechenmann C., et al. (2008). Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20, 2746–2762 10.1105/tpc.108.059048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E. J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43, 265–285 10.1146/annurev-genet-102108-134148 [DOI] [PubMed] [Google Scholar]
- Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., Masson P. H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. U.S.A. 95, 15112–15117 10.1073/pnas.95.12.6919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Voss U., Lau S., Wilson M., Shao N., Timme R. E., et al. (2011). Unraveling the evolution of auxin signaling. Plant Physiol. 155, 209–221 10.1104/pp.110.168161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Wang B., Moreno I., Dupláková N., Simon S., Carraro N., et al. (2012). ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3, 941. 10.1038/ncomms1941 [DOI] [PubMed] [Google Scholar]
- Finet C., Jaillais Y. (2012). AUXOLOGY: when auxin meets plant evo-devo. Dev. Biol. 369, 19–31 10.1016/j.ydbio.2012.05.039 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., et al. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–22230 10.1126/science.282.5397.2226 [DOI] [PubMed] [Google Scholar]
- Geisler M., Blakeslee J. J., Bouchard R., Lee O. R., Vincenzetti V., Bandyopadhyay A., et al. (2005). Cellular efflux of auxincatalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44, 179–194 10.1111/j.1365-313X.2005.02519.x [DOI] [PubMed] [Google Scholar]
- Jones A. M., Venis M. A. (1989). Photoaffinitylabeling of indole-3-acetic acid-binding proteins in maize. Proc. Natl. Acad. Sci. U.S.A. 86, 6153–6156 10.1073/pnas.86.16.6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S., Abraham Z., Manzano C., López-Torrejón G., Pacios L. F., Del Pozo J. C. (2010). The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22, 3891–3904 10.1105/tpc.110.078972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecek P., Skupa P., Libus J., Naramoto S., Tejos R., Friml J., et al. (2009). The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 10, 249. 10.1186/gb-2009-10-12-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). ClustalW and clustalX version 2 (2007). Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2006). Dynamic integration of auxin transport and signalling. Curr. Biol. 16, R424–33 10.1016/j.cub.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R. A., Grisafi P., Fink G. R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187 10.1101/gad.12.14.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J., Skupa P., Bailly A., Hoyerová K., Krecek P., Bielach A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., et al. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911 10.1093/emboj/17.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S. (2000). Molecular Evolution and Phylogenetics. New York: Oxford University Press [Google Scholar]
- Petrásek J., Mravec J., Bouchard R., Blakeslee J. J., Abas M., Seifertová D., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Robert S., Kleine-Vehn J., Barbez E., Sauer M., Paciorek T., Baster P., et al. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143, 111–121 10.1016/j.cell.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Rosquete M., Barbez E., Kleine-Vehn J. (2012). Cellular auxin homeostasis: gatekeeping is housekeeping. Mol. Plant 5, 772–786 10.1093/mp/ssr109 [DOI] [PubMed] [Google Scholar]
- Sakharkar M. K., Chow V. T. K., Chaturvedi I., Mathura V. S., Shapshak P., Kangueane P. (2004). A report on single exon genes (seg) in eukaryotes. Front. Biosci. 9, 3262–3267 10.2741/1478 [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stephens R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 10.1093/nar/18.20.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos I. C., Liakopoulos T. D., Bagos P. G., Hamodrakas S. J. (2004). TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics 20, 3258–3260 10.1093/bioinformatics/bth358 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W. D., Ditengou F. A., Dovzhenko A. D., Li X., Molendijk A. M., Ruperti B., et al. (2008). Auxin as a model for the integration of hormonal signal processing and transduction. Mol. Plant 1, 229–237 10.1093/mp/ssn006 [DOI] [PubMed] [Google Scholar]
- Tromas A., Braun N., Muller P., Khodus T., Paponov I. A., Palme K., et al. (2009). The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS ONE 4, e6648. 10.1371/journal.pone.0006648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády G. E., Simon I. (1998). Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J. Mol. Biol. 283, 489–506 10.1006/jmbi.1998.2107 [DOI] [PubMed] [Google Scholar]
- Tusnády G. E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- Utsuno K., Shikanai T., Yamada Y., Hashimoto T. (1998). Agr, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 39, 1111–1118 10.1093/oxfordjournals.pcp.a029310 [DOI] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., et al. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158, 590–600 10.1104/pp.111.189514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: a trigger for change in plant development. Cell 136, 1005–1016 10.1016/j.cell.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 10.1093/oxfordjournals.molbev.a003851 [DOI] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P. B., Ruzicka K., Blilou I., et al. (2006). Polar PIN localization directs auxin flow in plants. Science 312, 883. 10.1126/science.1121356 [DOI] [PubMed] [Google Scholar]
- Woodward A. W., Bartel B. (2005). A receptor for auxin. Plant Cell 17, 2425–2429 10.1105/tpc.105.036236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J.-G., Wu M.-J., et al. (2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110 10.1016/j.cell.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogeeswaran K., Frary A., York T. L., Amenta A., Lesser A. H., Nasrallah J. B., et al. (2005). Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15, 505–515 10.1101/gr.3436305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazímalová E., Murphy A. S., Yang H., Hoyerová K., Hosek P. (2010). Auxin transporters – why so many? Cold Spring Harb. Perspect. Biol. 2, a001552. 10.1101/cshperspect.a001552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338 10.1093/mp/ssr104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Guo B., He S. (2011). The roles and evolutionary patterns of intronless genes in deuterostomes. Comp. Funct. Genomics 2011, 1–8 10.1155/2011/463645 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular phylogenetic analysis of PILS proteins. The diagram shows an extended phylogentic tree of PILS proteins with collapsed branches for algae, Physcomitrella, and Selaginella. Note the high diversification of PILSes in Medicago and Populus. Because of incomplete sequences some of the PILSes were eliminated. The evolutionary history was inferred by using the Maximum Likelihood method based on the Data specific model (Nei and Kumar, 2000). The tree with the highest log likelihood (−55875.7936) is shown. The percentage of trees in which the associated taxa clustered together is shown above the branches. Initial tree(s) for the heuristic search were obtained automatically as follows. When the number of common sites was <100 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise BIONJ method with MCL distance matrix was used. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 2.6899)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 3.7299% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 75 nucleotide sequences. All positions with less than 0% site coverage were eliminated. That is, fewer than 100% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 1113 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Alignment of PILS amino acid sequences. The multiple amino acid alignment of PILSes was generated by using Muscle in MEGA5 software (Tamura et al., 2011). This alignment was generated for the phylogenetic analysis presented in the Supplementary Figure 1. The alignment for the smaller tree presented in the Figure 2 is similar but with less sequences.
Alignment of PILS and PIN amino acid sequences. The multiple alignment was generated by using Muscle in MEGA5 software (Tamura et al., 2011).
Alignment of Arabidopsis thaliana PILS amino acid sequences. A multiple sequence alignment generated by ClustalW server (Larkin et al., 2007) of the seven PILSes is shown. Amino acids are color coded: red (small, hydrophobic, aromatic, not Y), blue (acidic); magenta (basic), green (hydroxyl, amine, amide, basic), gray (others). “*,” Identical amino acids; “:,” conserved substitutions (same color group); “.,” semi-conserved substitution (similar shapes).
Sequence information.