Abstract
Pomegranate juice (PJ) is a natural product that inhibits prostate cancer progression. A clinical trial on patients with recurrent prostate cancer resulted in none of the patients progressing to a metastatic stage during the period of the trial. We have previously found that, in addition to causing cell death of hormone-refractory prostate cancer cells, PJ also markedly increases adhesion and decreases migration of the cells that do not die. However, because PJ is a very complex mixture of components and is found in many different formulations, it is important to identify specific components that are effective in inhibiting growth and metastasis. Here, we show that the PJ components luteolin, ellagic acid, and punicic acid together inhibit growth of hormone-dependent and hormone-refractory prostate cancer cells and inhibit their migration and their chemotaxis toward stromal cell-derived factor 1α (SDF1α), a chemokine that is important in prostate cancer metastasis to the bone. These components also increase the expression of cell adhesion genes and decrease expression of genes involved in cell cycle control and cell migration. Furthermore, they increase several well-known tumor-suppression microRNAs (miRNAs), decrease several oncogenic miRNAs, and inhibit the chemokines receptor type 4 (CXCR4)/SDF1α chemotaxis axis. Our results suggest that these components may be more effective in inhibiting prostate cancer growth and metastasis than simply drinking the juice. Chemical modification of these components could further enhance their bioavailability and efficacy of treatment. Moreover, because the mechanisms of metastasis are similar for most cancers, these PJ components may also be effective in the treatment of metastasis of other cancers.
Introduction
Prostate cancer is the second leading cause of death by cancer among men in America. It accounts for approximately 30% of all male malignancies; one in six men will be diagnosed with prostate cancer and 1 in 35 will die of the disease [1]. Early stages of localized prostate cancer can be effectively treated with surgery and radiation, but a majority of patients develop locally advanced or widespread cancer that requires hormone ablation therapy. Moreover, 80% to 90% of patients who receive hormone ablation therapy ultimately develop metastatic castration-resistant prostate cancer (CRPC) 12 to 33 months after initiation of hormone ablation therapy [2,3]. Chemotherapy can be used to treat CRPC, but chemotherapeutic drugs are aggressive and have many side effects [4,5]. Therefore, there is a major need for more effective and less toxic therapies to treat prostate cancer.
Sipuleucel-T (Provenge; Dendreon, Seattle, WA), an autologous cellular immunotherapy, was approved by the Food and Drug Administration (FDA) in 2010 to treat metastatic prostate cancer. The overall survival rate of patients who received Sipuleucel-T was improved, but the median survival rate was only improved by 4.5 months. In addition, the effect on time of progression in patients who were asymptomatic or with limited metastatic disease did not reach statistical significance and treatment is costly [6]. Abiraterone, an inhibitor of androgen biosynthesis, has been shown as very promising antiandrogen therapy to prolong overall survival rate among patients with metastatic prostate cancer [7]. Moreover, novel androgen receptor antagonist MDV3100, which blocks androgen from binding to androgen receptor and prevents nuclear translocation, showed promising antitumor effects in recent clinical trials [8,9]. Another novel drug, Cabozantinib, a potent dual inhibitor of the tyrosine kinases met proto-oncogene (MET) and vascular endothelial growth factor receptor 2, has been shown to reduce or stabilize metastatic bone lesions in CRPC patients [10,11]. However, all of these treatments have adverse side effects. Recently, there has been a renewed push to identify natural remedies to fight prostate cancer. Among the latter is Pomegranate juice (PJ).
Mounting evidence shows that PJ has great potential to inhibit the growth and reduce the invasiveness of prostate cancer cells both in vitro and in vivo [12–14]. In a phase II clinical trial, patients with rising prostate-specific antigen (PSA) were given 8 oz of PJ by mouth daily. PSA doubling time significantly increased with treatment from a mean of 15 months at baseline to 54 months post-treatment (P < .001). This statistically significant prolongation of PSA doubling time and the lack of metastatic progression in any of the patients strongly suggest a potential of PJ for treatment of prostate cancer [15]. As a result of these findings, several studies have shown that PJ affects many of the cellular processes involved in cell death and also affects signaling pathways that could inhibit cell migration and invasion [16,17]. We have shown previously that PJ inhibits the migratory and metastatic properties of hormone refractory prostate cancer cells by stimulating cell adhesion and inhibiting cell migration/chemotaxis [18]. However, the soluble phase of PJ contains many components, and as a whole, it is difficult to determine how to best maximize its use in treating prostate cancer. A way to overcome this challenge is to identify chemical components of PJ that are responsible for the antimetastatic effect of the whole juice.
The fruit of PJ can be divided into several anatomic compartments such as seeds, juice, and peel and all these compartments have been reported to possess antiproliferative and antimetastatic effect against prostate cancer cells [17,19–21]. The juice is a rich source of polyphenolic compounds including anthocyanins such as delphinidin, cyanidin, and pelargonidin, which give the fruit and juice its red color. It is also rich in punicalin, punicalagin, gallagic, quercetin, ferulic acid, caffeic acid, ellagic acid, and luteolin, which largely account for the antioxidant activity of the whole fruit [13,22–24]. The seed oil of PJ, which is comprised of 65% to 80% conjugated fatty acids, also contains many compounds of interest with known anticancer activities. The predominant component among these fatty acids is punicic acid [25,26]. However, the specific components of PJ that have antimetastatic effects against prostate cancer are largely unknown. There is a short one-and-a-half-page communication reporting that ellagic acid, caffeic acid, luteolin, and punicic acid inhibit in vitro invasion of human prostate cancer (PC3) cells across Matrigel (BD Biosciences, San Jose, CA) [27]. Their findings are interesting but very limited. No studies were presented on the effects on other processes involved in metastasis nor was mechanism of action of these PJ components addressed. Given these findings, we hypothesized that these specific components of PJ are able to replace the effects of the full PJ in inhibiting cellular and molecular processes involved in adhesion, migration, and chemotaxis of prostate cancer. Here, we show that luteolin (L), ellagic acid (E), and punicic acid (P), but not caffeic acid (C), stimulate molecule cell adhesion, inhibit molecules involved in cell migration, and inhibit chemotaxis of the cancer cells through CXCR4/stromal cell-derived factor 1α (SDF1α), a chemokine axis that is very important in metastasis of prostate cancer cells to the bone. Our findings strongly suggest that L + E + P can potentially be used to prevent metastasis of prostate cancer.
Materials and Methods
Cell Culture
DU145 and PC3 are hormone-independent prostate cancer epithelial cell lines. LNCaP is an androgen-responsive prostate cancer epithelial cell line. DU145 and LNCaP prostate cancer epithelial cell lines were purchased from American Type Culture Collection (Manassas, VA). PC3 prostate epithelial cell line was a gift from A. Walker (University of California, Riverside).Cells were cultured at 37°C with 5% CO2 in RPMI-1640 supplemented with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 µg/ml) and used at the times indicated in the results.
Adhesion Assay
PC3, DU145, or LNCaP cells (3 x 105) were plated on gelatin-coated six-well plates (BD Biosciences, San Jose, CA), allowed to adhere, and 24 hours later, treated with PJ and/or PJ components for 12 and/or 24 hours. Cells were then trypsinized, and the time required to detach all cells was recorded as an indicator of cell adhesiveness.
Migration Assay
Confluent PC3, DU145, or LNCaP cells were wounded using a rubber scraper to create a scratch, washed, and treated with PJ and/or PJ components at various concentrations. Cell migration was determined by measuring the distance migrated by the cells from the wounded edge to the leading edge of migration at 12, 24, 48, and 72 hours after treatments were initiated. Scraped cells without treatment were used as controls.
Chemotaxis Assay
The upper side of 8-µm pore size polycarbonate membranes of transwells (BD Biosciences) were coated with 50 ng/ml type I collagen (Sigma Chemical Co, St Louis, MO). DU145, PC3, or LNCaP cells (1 x 105) in 100 µl of culture medium were plated on the upper side of transwell membranes and were allowed to adhere for 3 hours. Then, the wells were introduced into 24-well plate and 1000 µl of RPMI-1640 with 10% FBS medium was added to the lower chamber. Cells were treated with PJ and/or PJ components for 12 hours. SDF1α (100 ng/ml) was added to the lower chamber and the cells were allowed to migrate for 4 hours at 37°C. The cells on the side of the membrane facing the upper chamber were removed with a cotton swab, and the membranes were then fixed and stained with 2% toluidine blue in 4% paraformaldehyde. Cells were counted in eight high power fields per filter to obtain the average number of cells per field.
Total RNA Extraction
DU145 and PC3 cells were treated with PJ components for 12 hours, and total RNA was extracted using the RNeasy RNA Isolation Kit according to the manufacturer's protocol. Briefly, cells were washed with ice-cold 1x phosphate-buffered saline (PBS) and lysed on ice with lysis buffer. Cell lysates were then spun at 12,000 rpm for 5 minutes to remove cell debris, followed by organic extraction to remove proteins. Then, lysates were loaded into isolation columns and the final RNA product was dissolved in nuclease-free water. RNA quality was assessed on the Agilent Bioanalyzer 2100 using the Agilent RNA 6000 Nano Assay Kit (Agilent Technologies, Waldbronn, Germany), and the concentration was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc, Wilmington, DE).
Affymetrix Microarray and Data Analysis
Affymetrix Human Genome U133 Plus 2.0 Arrays, which contain more than 54,000 probe sets representing approximately 38,500 genes and gene sequences, were used. Cells were treated with PJ components for 12 hours, and total RNA was extracted and evaluated as described above. Capillary electrophoresis using an Agilent Bioanalyzer 2100 to confirm the RNA quality levels was used before performing the array assays. RNA from cells without treatment was used as control. A single log2 expression measure for each probe set was calculated from image files (CEL format) using the robust multiarray analysis (RMA) procedure using Agilent GeneSpring GX software. The changes of expression level between untreated and specific PJ component-treated samples were compared. Only genes that were overexpressed or underexpressed by more than two-fold were considered. Once we identified genes of potential interest, we verified their increase or decrease in expression by real-time quantitative polymerase chain reaction (qPCR).
Real-Time qPCR
RNA (1 µg) was reverse-transcribed to cDNA by RETROscript Reverse Transcription Kit (Ambion) at 44°C for 1 hour and 92°C for 10 minutes. cDNA (2 µl) from the reverse transcription reaction was added to 23 µl of real-time qPCR mixture containing 12.5 µl of 2x SYBR Green Supermix (Bio-Rad, Hercules, CA) and 200 nM oligonucleotide primers. PCRs were carried out in a Bio-Rad MyiQ5 Real-Time PCR Detection System (Bio-Rad). The thermal profile was 95°C for 3 minutes followed by 40 amplification cycles, consisting of denaturation at 95°C for 10 seconds and annealing at 60°C for 30 seconds. Fluorescence was measured and used for quantitative purposes. At the end of the amplification period, melting curve analysis was done to confirm the specificity of the amplicon. Fold changes of genes after treatment with PJ were calculated by the Pfaffl method to normalize the Ct values to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) internal control. The following primer sequences were designed with IDT PrimerQuest (http://www.idtdna.com/Scitools/Applications/Primerquest/) and used for the reactions:
GAPDH, TCGACAGTCAGCCGCATCTTCTTT and ACCAAATCCGTTGACTCCGACCTT;
BCL2, TTTCTCATGGCTGTCCTTCAGGGT and AGGTCTGGCTTCATACCACAGGTT;
ROCK2, TTCCAGTGGAGCCAGTTGGAGAAA and TACAAGCCTCACAGTTGGTTGGGA;
EZH2, CAGTTTGTTGGCGGAAGCGTGTAA and AGGATGTGCACAGGCTGTATCCTT;
CDK6, ATTCACTGCCTGGGACACAGTCTT and ACAGGCCACTGTGGTAACTCTCAA;
DTL, ATTTGGATCTGTGCTGCCTTGCTG and AGGTAGCGTTCCACAGCTTTCTGA;
CCNE2, ATGACACCACCGAAGAGCACTGAA and TTGGCTAGGGCAATCAATCACAGC;
FSCN1, CAACGATGGCGCCTACAACATCAA and TGGCCACCTTGTTATAGTCGCAGA;
CDC25B, TCAGGTGCTGTCCATGGGAAAGAT and AACTCAACAGACTGGGCTCTTCCA;
TWIST, ACCATCCTCACACCTCTGCATTCT and TTCCTTTCAGTGGCTGATTGGCAC;
CCNB1, TGTGGATGCAGAAGATGGAGCTGA and TTGGTCTGACTGCTTGCTCTTCCT;
PTEN, GGTTGCCACAAAGTGCCTCGTTTA and AACTGGCAGGTAGAAGGCAACTCT;
CDKN1A, TTAGCAGCGGAACAAGGAGTCAGA and ACACTAAGCACTTCAGTGCCTCCA.
MicroRNA PCR Array and Data Analysis
MicroRNAs (miRNAs) were extracted from total RNA using RT2 qPCR-Grade miRNA Isolation Kit from SABiosciences (Frederick, MD). For PCR arrays, miRNA cDNA (100 ng) was amplified using RT2 miRNA First Strand Kit and RT2 miRNA PCR Array (MAH-001A) from SABiosciences following the manufacturer's instructions. The fluorescence threshold value (Ct) was calculated using SABiosciences web-based RT2 profiler PCR array data analysis portal (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php).
siRNA and Vector Transfection
PC3 cells (80–90% confluent) were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer's protocols; 40 nM E-cadherin chimera small-interfering RNA (siRNA) (Abnova, Taiwan, China) was transfected, and 2 µg/ml pcDNA3.1 HMMR vector, pcDNA4.1 TWIST vector, or pcDNA3 CCNE2 vector (Addgene plasmid 19935; Addgene, Cambridge, MA) was transfected. Scratch wound assay, adhesion assay, and/or cell growth assay were performed as described above, 24 hours after transfection.
Statistical Analysis
Data analysis was performed using the one-way analysis of variance on raw data using GraphPad InStat software (GraphPad Software Inc, La Jolla, CA).
Results
Effect of Luteolin, Ellagic Acid, Punicic Acid, and Caffeic Acid on Prostate Cancer Cell Growth, Adhesion, Migration, and Chemotaxis
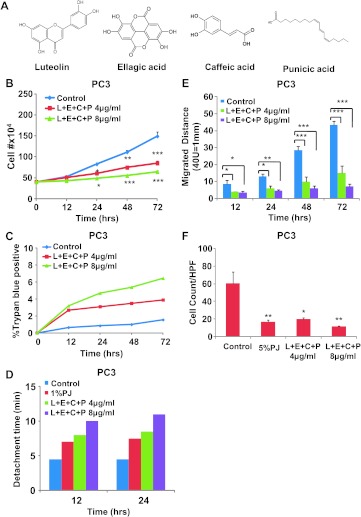
We treated hormone-independent prostate cancer cells, PC3 and DU145, with the four PJ components L, E, P, and C (Figure 1A) at concentrations of both 4 and 8 µg/ml, which are relevant to physiological concentration of these components in the juice [27]. We found that this combination significantly inhibited cell growth of both PC3 (Figure 1B) and DU145 (Figure W1A). When compared to PJ treatment, the number of cells floating in the medium after the treatment with the components was much smaller. Trypan blue staining showed that the majority of the cells remained viable after treatment with the components at either 4 or 8 µg/ml (Figures 1C and W1B). At both 12 and 24 hours, the cells all still looked healthy and no floating cells were seen in any of the cultures. We also found that the cells strongly adhered to gelatin-coated substrate, as indicated by increased time to detach all cells from the culture dish in the presence of treatment with the components; by 12 hours of 4 µg/ml L + E + C + P treatment, the cultures required about double the time to be released by trypsinization (Figures 1D and W1C). Moreover, the effect was dose dependent with 8 µg/ml of the combination of PJ components being more potent than treatment with either 4 µg/ml of the components or 1% PJ. Using the scratch wound assay, we found that the combination of L + E + C + P significantly inhibited the migratory capabilities of these cells. We measured the distance that the cells migrated from the wounded edge to the migration front and found it to be significantly reduced in the treated cells beginning as early as 12 hours after treatment, which continued more than 72 hours, and the effect was dose dependent (Figures 1E and W1D).
Figure 1.
L + E + C + P inhibits growth, stimulate adhesion, inhibits migration, and inhibits chemotaxis toward SDF1α of hormone-independent prostate cancer cells. (A) The chemical structure of L, E, P, and C. (B) PC3 cells were treated with four PJ components L + E + C + P at 4 and 8 µg/ml and counted for increasing times after initiation of treatment. Controls represent no treatment. The medium containing L + E + C + P was changed daily. (C) PC3 cells were treated with L + E + C + P at 4 and 8 µg/ml, and the percentage of dead cells was determined by Trypan blue staining at the indicated time points. (D) PC3 cells were plated on gelatin-coated dishes, and 24 hours later, the medium was changed and the cells were treated with L + E + C + P at 4 and 8 µg/ml. We tested for adhesion to the substrate at 12 and 24 hours after initiation of treatment by recording the time it took for trypsinization to remove all of the cells from the dish. Control represents no treatment. The reason for not presenting statistical significance is because the loss of adhesion is very similar from culture to culture and it occurs rapidly when the cells begin to detach. Within each experiment, the times of trypsinization were the same within 1 minute for each specific treatment. (E) PC3 cells were treated with L + E + C + P at 4 and 8 µg/ml for 72 hours and the distance migrated by the cells from the wounded edge to the leading edge was measured at the indicated time points. Controls represent no treatment. The medium containing the components was changed daily. (F) PC3 cells were allowed to attach to the top of the filter of the chemotaxis assay chambers for 4 hours and then treated with L + E + C + P at 4 and 8 µg/ml for 12 hours. At this time, 100 ng/ml SDF1α was introduced into the lower chamber and the cells found on the bottom of the filter were counted 3.5 hours later. Control had no treatment. The number of cells found on the underside of the filter was counted 3.5 hours later. Bars represent SEM. ***P < .001; **P < .01; *P < .05.
SDF1α is a critical chemokine secreted by bone marrow cells that attracts prostate cancer cells to the bone. Therefore, we tested whether the combination of L + E + C + P also inhibits chemotaxis toward SDF1α. Cells pretreated with L + E + C + P at 4 or 8 µg/ml for 12 hours before initiation of the chemotaxis assay toward SDF1α showed significantly inhibited chemotaxis at both concentrations (Figures 1F and W1E); at 8 µg/ml, the effects were similar to those of 5%PJ. Therefore, L + E + C + P has the potential to inhibit metastasis of these cells to the bone marrow.
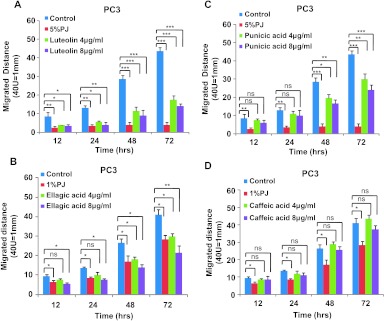
Because the combination of L + E + C + P significantly inhibited processes involved in metastasis, we also tested their effects individually using the scratch migration assay. L, E, and P each significantly inhibited cell migration of PC3 and DU145 cells when individually applied at 4 or 8 µg/ml (Figure 2, A–C, and W2, A–C). However, caffeic acid when applied individually did not show any effects on cell migration of either cell type (Figures 2D and W2D). On the basis of these findings, we eliminated caffeic acid as a potential anticancer component and focused on the effects of the other three components, L, E, and P.
Figure 2.
Luteolin, ellagic acid, and punicic acid but not caffeic acid individually inhibit cell migration of hormone-independent PC3 cells. PC3 cells were treated with individual PJ components (A) luteolin, (B) ellagic acid, (C) punicic acid, and (D) caffeic acid at 4 and 8 µg/ml for 72 hours, and migration assay was performed as described in Figure 1. Bars represent SEM. ***P < .001; **P < .01; *P < .05.
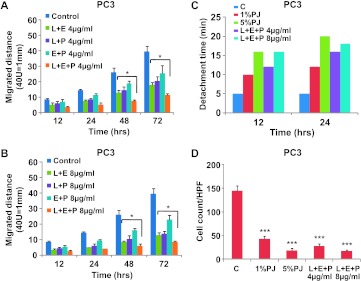
To test whether the combination of L + E + P is more potent than the combination of any two components, we treated PC3 and DU145 cells with two-component combinations (L + E, L + P, and E + P) and three-component combination (L + E + P) at 4 or 8 µg/ml (Figures 3, A and B, and W3, A and B). More than 72 hours of the scratch wound assay, L + E + P was significantly more potent than any dual combinations at 48 and 72 hours. On the basis of these findings, we performed the remainder of the studies using L + E + P.
Figure 3.
L + E + P is the most potent combination for inhibition of cell migration and chemotaxis toward SDF1α as well as increasing cell adhesion of hormone-independent prostate cancer cells. PC3 cells were treated with different combination of PJ components L, E, and P at (A) 4 and (B) 8 µg/ml for up to 72 hours, and migration assay was performed as described in Figure 1. (C) PC3 cells were plated on gelatincoated dishes, and 24 hours later, the medium was changed and the cells were treated with PJ components L + E + P at 4 and 8 µg/ml. Adhesion assay was performed as described in Figure 1. (D) PC3 cells were allowed to attach to the top of the filter of the chemotaxis chamber for 4 hours and then treated with L + E + P at 4 and 8 µg/ml for 12 hours. Chemotaxis assay was performed as described in Figure 1. Bars represent SEM. ***P < .001; **P < .01; *P < .05.
L + E + P significantly increased cell adhesion of PC3 (Figure 3C) and DU145 (Figure W3C) cells. The detachment time was increased by 4 or 8 µg/ml of L + E + P treatment for 12 and 24 hours. When compared to the effect of PJ, L + E + P at 8 µg/ml showed similar effect to 5% PJ. In addition, chemotaxis toward SDF1α was significantly inhibited by pretreatment of PC3 and DU145 cells with L + E + P at 4 or 8 µg/ml (Figures 3D and W3D). The effect of L + E + P at 8 µg/ml mimicked the effect of 5% PJ. We also tested the effects of L + E + P combinations at different ratios on cell migration. However, we found that the ratios of 1:1:1 are the most potent combination.
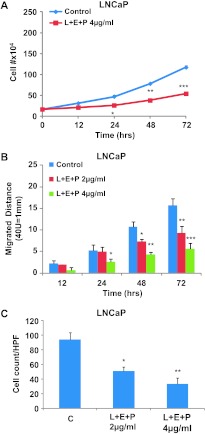
To determine whether these effects of L + E + P are also found in androgen-sensitive cells, we used LNCaP, a prostate cancer cell line that is responsive to androgen, and performed similar studies using growth, migration, and SDF1α chemotaxis assays. L + E + P at 4 µg/ml significantly suppressed cell growth of LNCaP cells (Figure 4A). However, LNCaP cells are considerably more sensitive to these PJ components as we observed that cells were largely killed by L + E + P at 8 µg/ml (data not shown). As a result, we used lower concentrations of L + E + P to treat LNCaP cells in scratch migration assays and chemotaxis assays. L + E + P at 2 and 4 µg/ml significantly inhibited cell migration more than 72 hours of treatment (Figure 4B) and chemotaxis toward SDF1α (Figure 4C). These results suggest that L + E + P are even more effective in androgen-sensitive than androgen-independent prostate cancer cells.
Figure 4.
L + E + P suppresses cell growth, stimulates cell adhesion, and inhibits chemotaxis to SDF1α of LNCaP cell. (A) LNCaP cells were treated with L + E + P at 4 µg/ml and counted for increasing times after initiation of treatment. (B) LNCaP cells were plated on gelatin-coated dishes, and 24 hours later, the medium was changed and the cells were treated with L+ E + P at 2 and 4 µg/ml. Adhesion assay was performed as described in Figure 1. (C) LNCaP cells were allowed to attach to the top of the filter of the chemotaxis chamber for 4 hours and then treated with L + E + P at 2 and 4 µg/ml for 12 hours. Chemotaxis assay was performed as described in Figure 1. Bars represent SEM. ***P < .001; **P < .01; *P < .05.
Effect of L + E + P on the Expression of Genes Involved in Cell Growth, Adhesion, Migration, and Chemotaxis
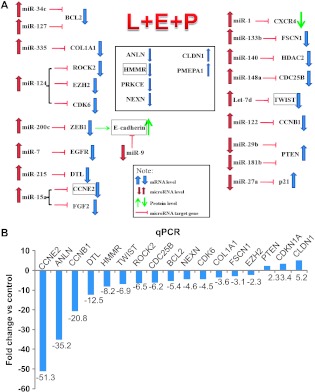
To understand how L + E + P inhibit cell growth, cell migration, and chemotaxis and increase cell adhesion, we performed Affymetrix microarray analysis to examine their effects on the expression of genes involved in these cell functions. For these studies, we used total RNA from PC3 cells treated with L + E + P at 8 µg/ml for 12 hours and analyzed for gene expression using Affymetrix Human U133 Plus 2.0 microarrays. PC3 cells were used because these are the most invasive cells of all of the three types of cells we studied. The data show that L + E + P inhibit the expression of several important genes involved in cell growth and cell migration while stimulating the expression of several genes involved in cell adhesion (Figure 5A, blue arrows and Table 1). The genes we show that are increased all significantly enhance adhesion or are tumor suppressor genes. Those that are decreased are all related to stimulation of migration and cytoskeletal components or extracellular matrix (ECM) molecules that facilitate migration.
Figure 5.
Effect of L + E + P on genes involved in cell growth, cell adhesion, and migration of hormone-independent prostate cancer cells. (A) Schematic summary of the effects of L + E + P on gene and miRNA expression in hormone-independent prostate cancer cells. (B) The mRNA levels of CCNE2, ANLN, CCNB1, DTL, HMMR, TWIST, ROCK2, CDC25B, BCL2, NEXN, CDK6, COL1A1, FSCN1, EZH2, PTEN, CDKN1A, and CLDN1 were determined by using qPCR with RNA extracted from PC3 cells treated with L + E + P at 8 µg/ml for 12 hours.
Table 1.
Gene Analysis of the Effects of L + E + P on PC3 Cells.
| Gene Name | Product | Fold Change | Function |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B; p15 | →4.9 | Cell cycle control |
| CLDN1 | Claudin 1 | →3.6 | Adhesion |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A; p16 | →2.2 | Cell cycle control |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A; p21 | →2 | Cell cycle control |
| PTEN | Pten | →1.6 | Tumor suppressor |
| CCNE2 | Cyclin E2 | ↓18 | Cell cycle control |
| ANLN | Anillin | ↓16 | Migration |
| HMMR | HA-mediated motility receptor | ↓8 | Migration |
| DTL | Denticleless homolog | ↓7 | Cell cycle control |
| CCNB1 | Cyclin B1 | ↓5.1 | Cell cycle control |
| CCNB2 | Cyclin B2 | ↓4.5 | Cell cycle control |
| ROCK2 | Rock2 | ↓4 | Migration |
| ZEB1 | Zinc finger E-box binding homeobox 1 | ↓3 | EMT |
| TWIST | Twist | ↓2.6 | EMT |
| CDC25B | Cell division cycle 25 homolog B | ↓2.5 | Cell cycle control |
| EZH2 | Enhancer of zeste homolog 2 | ↓2.3 | Oncogenic |
| NEXN | Nexilin | ↓2.3 | Migration |
| BCL2 | Bcl2 | ↓2 | Antiapoptotic |
| COL1A1 | Collagen 1 | ↓2 | Migration |
| CDK6 | Cyclin-dependent kinase 6 | ↓1.8 | Cell cycle control |
| FSCN1 | Fascin 1 | ↓1.7 | Migration |
L + E + P changes the expression profile of genes involved in the cell growth, cytoskeleton, and cell adhesion machinery. RNA was extracted from PC3 cells that had been treated with L + E + P at 8 µg/ml for 12 hours, and Affymetrix array analysis was performed as described in Materials and Methods. Relative mRNA levels are presented as fold change compared with untreated controls.
To verify the effects of L + E + P on gene expression revealed by the Affymetrix arrays, we used real-time qPCR to examine mRNA levels of the following specific genes that are upregulated or downregulated in the gene arrays: claudin 1 (CLDN1), p21 (CDKN1A), Pten (PTEN), cyclin E2 (CCNE2), anillin (ANLN), cyclin B1 (CCNB1), denticleless homolog (DTL), hyaluronan (HA)-mediated motility receptor (HMMR), Twist (TWIST), Rock2 (ROCK2), cell division cycle homolog B (CDC25B), Bcl2 (BCL2), nexilin (NEXN), cyclin-dependent kinase 6 (CDK6), collagen I (COL1A1), fascin1 (FSCN1), and enhancer of zeste homolog 2 (EZH2). For the real-time PCR, total RNA was extracted from PC3 cells treated with L + E + P at 8 µg/ml for 12 hours (Figure 5B). The mRNA fold change of these genes was highly consistent with the Affymetrix array results.
Effect of L + E + P on the Level of Cancer-Related miRNAs
miRNAs are naturally occurring small non-coding RNAs that function as negative regulators of gene expression. They regulate important cellular functions such as cell proliferation, apoptosis, differentiation, and development. Mature miRNAs bind to target mRNAs, which subsequently results in either direct cleavage of the targeted mRNAs or inhibition of translation. To determine the effects of L + E + P on cancer-related miRNAs and how they correlate with gene expression, we used miRNA PCR arrays to analyze the RNA obtained from PC3 cells treated with L + E + P at 8 µg/ml for 12 hours. Among 88 well-known cancer-related miRNAs in the array, we found that those that function as tumor suppressors are highly increased, whereas those that function as oncogenic miRNAs are highly decreased (Table 2). Moreover, we found a consistent correlation between the level of miRNAs and the expression levels of their predicted target genes as shown in the Affymetrix mRNA arrays. These correlations are shown in Figure 5A.
Table 2.
Effect of L + E + P on Cancer-Related miRNAs.
| miRNA | Function | Fold Change | Targets |
| miR144 | Tumor-suppressive | →772941-fold | Notch-1 |
| miR-133b | Tumor-suppressive | →177812-fold | c-MET, FSCN1 |
| miR-1 | Tumor-suppressive | →66913-fold | Cyclin D2, CXCR4, SDF1α |
| miR-122 | Proapoptotic | →9741-fold | Cyclin B1 |
| miR-34c | Tumor-suppressive | →6700-fold | E2F3, Bcl2, c-MET |
| miR-200c | Tumor-suppressive | →5077-fold | ZEB1, ZEB2, FN1, MSN |
| miR-127 | Tumor-suppressive | →4067-fold | E2F3, Notch-1, Bcl2 |
| miR-335 | Tumor-suppressive | →3983-fold | COLA1, SOX4, TNC |
| miR-124 | Tumor-suppressive | →1389-fold | ROCK2, EZH2, CDK6 |
| miR-181a | Tumor-suppressive | →962-fold | K-ras, Bcl2 |
| miR-7 | Tumor-suppressive | →849-fold | Bcl2, EGFR, IGF1R |
| miR-215 | Tumor-suppressive | →820-fold | DTL |
| miR-15a | Tumor-suppressive | →786-fold | Fgf2, cyclin E2 |
| Let-7d | Tumor-suppressive | →4-fold | Twist |
| miR-20a | Oncogenic | ↓13587-fold | APP |
| miR-21 | Oncogenic | ↓1260-fold | TMP1, Pdcd4, MARCKS |
| miR-9 | Oncogenic | ↓173-fold | E-cadherin |
| miR-29b | Oncogenic | ↓48-fold | PTEN |
| miR-181b | Oncogenic | ↓39-fold | PTEN, TIMP3 |
Effects of PJ on the levels of metastasis-related miRNAs. RNA from PC3 cells treated with L + E + P at 8 µg/ml for 12 hours was submitted to miRNA PCR array analysis. Relative miRNA levels are shown as fold change compared with untreated control.
Testing Mechanistically Specific Genes Involved in Cell Proliferation, Adhesion, and Migration
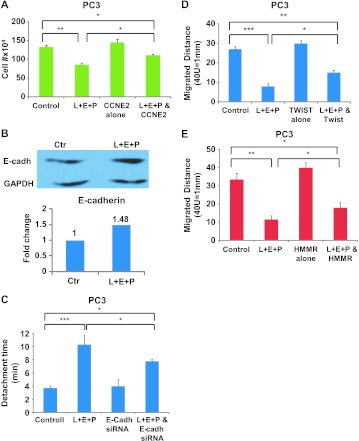
To determine whether the inhibitory effect of L + E + P on cell proliferation can be reversed, we chose to overexpress cyclin E (CCNE2) that is the most decreased gene shown in the gene array and is known to be critical in cell proliferation. We found that the proliferation inhibitory effect of L + E + P was significantly reversed by CCNE2 overexpression (Figure 6A).
Figure 6.
Mechanistic study of the effect of L + E + P on cell growth, cell adhesion, and migration of hormone-independent prostate cancer cells. (A) PC3 cells were transfected with pcDNA3 CCNE2 vector (2 µg/ml) and treated with L + E + P at 8 µg/ml 24 hours after transfection. Cell number was counted at a 48-hour time point. (B) Immunoblot analysis for E-cadherin with protein extracts from PC3 cells treated with L + E + P at 8 µg/ml for 24 hours. (C) PC3 cells were transfected with 40 nM E-cadherin siRNA. Twenty-four hours after transfection, cells were treated with L + E + P at 8 µg/ml for 12 hours and adhesion assay was performed. PC3 cells were transfected with 2 µg/ml (D) pcDNA3.1 HMMR vector or (E) pcDNA4.1 TWIST vector and treated with L + E + P at 8 µg/ml 24 hours after transfection. Migrated distance was determined at a 48-hour time point. Bars represent SEM. **P < .01; *P < .05.
We also found that L + E + P significantly increases the protein levels of E-cadherin (Figure 6B). Loss of this adhesion protein is critical for invasion of epithelial tumor cells. To determine whether the effect of L + E + P on cell adhesion can be reversed, we used siRNA to inhibit E-cadherin and performed adhesion assays 36 hours after PC3 cells were transfected with E-cadherin siRNA. We found that L + E + P treatment significantly increases attachment of the untransfected (control) cells and that the effect is partially reversed by E-cadherin siRNA (Figure 6C). These results indicate that the effects of L + E + P on cell adhesion are mediated significantly by increased E-cadherin.
To determine whether the inhibitory effects of L + E + P on cell migration can be reversed, we chose to overexpress HMMR (a gene that is known to promote cell migration and was highly decreased in the gene array) and TWIST (a gene that is known to play a role in epithelial-to-mesenchymal transitions (EMTs) and was also decreased in the gene array). The inhibitory effects of L + E + P on cell migration are partially reversed by overexpression of HMMR or TWIST. Scratch wounds on PC3 cell cultures were made 24 hours after transfection of HMMR or TWIST and the migrated distances were measured 36 or 48 hours later. We found that L + E + P treatment significantly decreased cell migration of untransfected cells, but the effect was partially reversed by overexpressing HMMR (Figure 6D) and TWIST (Figure 6E). These results indicate that the effects of L + E + P on cell migration are significantly mediated through decreasing HMMR and/or TWIST.
Effect of L + E + P on CXCR4/SDF1α Signaling in Prostate Cancer Cells
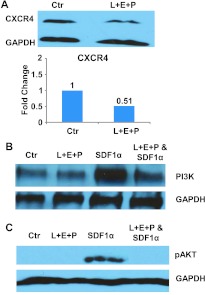
Approximately 80% of patients who have died of advanced hormone refractory prostate cancer have clinical evidence of bone metastases and 100% have histologic bone involvement [28]. Constitutive production of SDF1α by bone marrow stromal cells is a major source of this chemokine. We show here that L + E + P inhibits chemotaxis of hormone-independent prostate cancer cells toward SDF1α (Figures 3D and W3D). Moreover, we found that L + E + P significantly decreased the protein levels of CXCR4, the receptor for SDF1α that is present on the prostate cancer cells, by 50% in PC3 cells (Figure 7A) and that they inhibit the downstream signaling pathway of CXCR4/SDF1α. We found that L + E + P inhibits activation of PI3K (Figure 7B) and abolishes phosphorylation/activation of AKT induced by SDF1α (Figure 7C). These findings show that L + E + P inhibits the CXCR4/SDF1α chemotaxis axis both at the receptor level and the downstream signaling pathways, making these components strong contenders for treatment of prostate cancer metastasis.
Figure 7.
Effect of L + E + P on the SDF1α chemotaxis in prostate cancer cells. (A) Immunoblot analysis for CXCR4 in extracts prepared from PC3 cells treated with L + E + P at 8 µg/ml for 24 hours. (B) PI3K immunoblot analysis with protein extract from PC3 cells treated with 200 ng/ml SDF1α for 8 hours in the presence of L + E + P at 8 µg/ml. (C) Phosphor-Akt immunoblot analysis with protein extract from PC3 cells induced with 200 ng/ml SDF1α for 2 minutes in the presence of L + E + P at 8 µg/ml.
Discussion
The studies presented here identify L, E, and P as potential antimetastatic components in PJ. We delineate cellular and molecular mechanisms involved in inhibition of processes critical for metastasis of prostate cancer cells and show that L + E + P 1) suppresses the growth of two prostate cancer cell lines that are irresponsive to androgen deprivation and one line that is responsive to androgen deprivation, 2) stimulates prostate cancer cells to adhere strongly to the substrate, 3) inhibits the migratory capabilities of prostate cancer cells and their chemotaxis toward SDF1α, 4) stimulates expression of genes involved in cell adhesion while reducing expression of genes involved in cell migration and cell cycle control, and 5) increases the levels of tumor-suppressive miRNAs while reducing the level of oncogenic miRNAs.
During metastasis, cancer cells lose adhesion to each other and become migratory. E-cadherin has been shown as a critical adhesion molecule holding epithelial cells together and preventing them from breaking away. We found that the protein levels of E-cadherin are increased, whereas the expression levels of Zeb1 (ZEB1), a zinc finger transcription repressor of E-cadherin, is inhibited by L + E + P. Furthermore, when the cells are treated with siRNA for E-cadherin, we observe that the effect of the PJ components is reversed, strongly suggesting that E-cadherin is a key target of L + E + P. Moreover, the expression of a tight junction protein claudin 1 is increased by treatment with these components. Therefore, L + E + P increase cell adhesion through up-regulation of these cell adhesion proteins.
Several molecules involved in cell migration are downregulated by L + E + P. For example, HMMR binds HA, a molecule that facilitates cell migration. Binding of HA to HMMR stimulates the RhoA-activated protein kinase (ROCK) signal transduction pathway, leading to tumor cell migration and invasion in various cancers [29]. We found that the expression of HMMR and ROCK2 were downregulated by L + E + P. Other proteins such fascin, which is an actin-bundling protein, that regulates the actin cytoskeleton and the formation of migration initiating filopodia and actin-binding proteins anillin and nexillin, which are involved in the regulation of the structure of the cytoskeleton [30,31], are downregulated by L + E + P. Twist (TWIST) is a basic helix-loop-helix transcription factor that has been implicated in EMT characterized by loss of cell adhesion and increased cell motility [32,33]. Therefore, our results strongly suggest that L + E + P decrease cell migration through down-regulation of genes involved in cell migration and transcriptional factors involved in EMT.
The three PJ components also inhibit cell division. For example, cyclins E2 and B1 and cell cycle regulator CDK6 are highly downregulated by L + E + P treatment. p21, also known as CDK inhibitor 1, binds to and inhibits the activity of cyclin-CDK complex and therefore functions as a key regulator to control cell cycle progression [34]. We found that p21 was upregulated by L + E + P treatment. Denticleless homolog (DTL) is a substrate-specific adaptor of E3 ubiquitin-protein ligase complex required for cell cycle control. DTL mediates the ubiquitination and subsequent degradation of p21 [35]. Down-regulation of DTL together with up-regulation of p21 by L + E + P can potentially lead to suppression of cell cycle progression. Therefore, L + E + P may inhibit cell growth through down-regulation of genes involved in cell cycle control.
miRNAs have been shown to have profound impact on posttranscriptional gene regulation. Aberrant expression of miRNAs occurs in diverse types of human cancer and in different stages of disease progression. Our miRNA PCR array results show that many well-known tumor-suppression miRNAs are highly upregulated, whereas many oncogenic miRNAs are highly downregulated by L + E + P. Among the tumor-suppression miRNAs, miR-144 is known to negatively regulate the Notch-1 signaling pathway that has been suggested to be involved in a wide variety of human cancers [36]. miR-133b is known to negatively target receptor tyrosine kinase c-MET and actin-bundling protein fascin. Aberrantly activated c-MET (also known as hepatocyte growth factor receptor) has been shown to promote tumor growth, angiogenesis, and metastasis in various cancers, and fascin is known as an important regulator of actin cytoskeleton and cell migration [37,38]. miR-1 is known to negatively target cyclin D2, a protein required for cell cycle progression, and also CXCR4 and its ligand SDF1α that are known as important chemotactic proteins in cancer metastasis [39,40]. miR-212 is known to negatively target antiapoptotic protein phosphoprotein enriched in diabetes (PED) and transcriptional factor c-Myc. miR-122 is known to negatively target cyclin B1 [41] and miR-34c to negatively target transcriptional factor E2F3 and apoptosis key regulator Bcl2 [42,43]. miR-200c has been shown to inhibit ZEB1 and ZEB2, which are transcriptional repressors of the E-cadherin gene, whose product is critical in cell adhesion [44]. As a result, L + E + P might stimulate the expression of E-cadherin by inhibiting its transcriptional repression ZEB1 and ZEB through up-regulating miR-200c. miR-335 has been identified as a metastasis-suppressive miRNA in breast cancer by inhibiting type I collagen and tenascin C. Type I collagen is an extracellular matrix molecule involved in cytoskeletal control, and tenascin C is involved in the regulation of cell migration [45]. miR-124 is known to negatively regulate ROCK2, a kinase involved in cell migration, and CDK6, a key regulator of cell cycle [46]. Among the oncogenic miRNAs that are highly downregulated by L + E + P, miR-21 is one of the first discovered and best-studied oncogenic miRNAs that negatively regulate several tumor suppressor genes including tropomyosin 1 (TMP1) and programmed cell death 4 (PDCD4) protein [47]. miR-9 is known to negatively target E-cadherin [48], and miR-29b and miR-181b are oncogenic miRNAs known to negatively target tumor suppressor Pten [49]. Interestingly, the mRNA level of Pten is upregulated by L + E + P treatment that correlates with the down-regulation of miR-29b and miR-181b. These results show that L + E + P significantly up-regulate many tumor-suppression miRNAs and significantly downregulate many oncogenic miRNAs.
It is also known that, with time, prostate cancer cells develop ways to bypass the need for testosterone and then the cancer progresses very rapidly. It has been shown that the CXCR4/SDF1α chemotaxis axis may play a critical role in the metastasis of prostate cancer to bone and constitutive production of SDF1α by bone marrow stromal cells is a major source of this chemokine. We have shown that L + E + P is capable of inhibiting prostate cancer migration toward SDF1α as shown in our chemotaxis experiments. In addition, we found that L + E + P significantly reduced the level of CXCR4 in prostate cancer cells and inhibited the phosphorylation/activation of AKT. Therefore, L + E + P may act by suppressing the levels of CXCR4 and inhibiting the downstream signaling pathway of CXCR4/SDF1α.
Conclusion and Future Prospects
To date, there is no cure for prostate cancer when recurrence occurs after surgery and/or radiation. In particular, when it recurs after hormone ablation therapy, there are no other effective treatments for deterrence of cancer progression. Here, we show that luteolin, ellagic acid, and punicic acid, components of PJ, can potentially be used as antimetastatic treatments to deter prostate cancer metastasis. L + E + P interfere with multiple biologic processes involved in metastasis of cancer cells such as suppression of cell growth, increase in cell adhesion, inhibition of cell migration, and inhibition of chemotaxis toward proteins that are important in prostate cancer metastasis to the bone.
Supplementary Materials and Methods
Materials
RPMI-1640 medium was acquired from Mediatech (Manassas, VA) and FBS from Sigma-Aldrich (St Louis, MO). SDF1a was obtained from ProSpec (Boca Raton, FL) and the RNeasy RNA Isolation Kit for RNA preparation from Qiagen Inc (Valencia, CA). Secondary antibody (Ab) was from Thermo Scientific (Rockford, IL). Detection was done using Supersignal West Dura Kit from Thermo Scientific. Transfection reagents Lipofectamin 2000 were purchased from Invitrogen. All oligonucleotide primers for qPCR were obtained from IDT (Coralville, IA) and the qPCR iQ SYBR Green Supermix Kit from Bio-Rad. RT2 qPCR-Grade miRNA Isolation Kit and RT2 miRNA PCR Array (MAH-001A) were obtained from SABiosciences. pcDNA 3.1HMMR vector was a gift from Dr E. Turley (University of Western Ontario). pcDNA4 TWIST vector was a gift from Dr C. Glackin (City of Hope); pcDNA3 CCNE2 vector (Plasmid 19935) was purchased from Addgene; E-cadherin siRNA was purchased from Abnova. CXCR4 Ab was from Abcam (Cambridge, MA); E-cadherin Ab, phosphor-Akt Ab, and PI3K Ab were from Cell Signaling (Danvers, MA). PJ was purchased from POMx Wonderful (Los Angeles, CA).
Immunoblot Analysis
DU145 and PC3 cells were treated with PJ components for 12 hours, washed with ice cold 1x PBS, and lysed on ice with lysis buffer containing 0.5% Triton X-100, 0.5% NP-40, 10 mM Tris (pH 7.5), 2.5 mM KCl, 150 mM NaCl, 30 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 0.1% sodium dodecyl sulfate, and additional protease inhibitor cocktails (Sigma-Aldrich). Protein concentrations were measured using the DC Protein Assay Kit (Bio-Rad). Equal amounts of protein in the cell extracts were mixed with sample buffer, boiled, and analyzed using 10% acrylamide sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblot analysis was performed with the HRPconjugated secondary Ab (Thermo Scientific), followed by incubation with West Dura extended duration substrate (Thermo Scientific). Blots were then reprobed for histone 2A or GAPDH antibody to show equal loading of proteins.
Acknowledgments
We would like to thank Barbara Walter for help with Affymetrix microarray. We thank Dr E. Turley (University of Western Ontario) for the HMMR vector. We thank Mike Franco and Sandeep Dhall for helpful discussion on the manuscript.
Footnotes
None of the authors have conflict of interest related to this work.
This article refers to supplementary materials, which are designated by Figures W1 to W3 and are available online at www.transonc.com.
References
- 1.Stavridi F, Karapanagiotou EM, Syrigos KN. Targeted therapeutic approaches for hormone-refractory prostate cancer. Cancer Treat Rev. 2010;36:122–130. doi: 10.1016/j.ctrv.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Coffey DS, Pienta KJ. New concepts in studying the control of normal and cancer growth of the prostate. Prog Clin Biol Res. 1987:1–73. [PubMed] [Google Scholar]
- 3.Chuu CP, Kokontis JM, Hiipakka RA, Fukuchi J, Lin HP, Lin CY, Huo C, Su LC. Androgens as therapy for androgen receptor-positive castrationresistant prostate cancer. J Biomed Sci. 2011;18:63. doi: 10.1186/1423-0127-18-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrylak DP. Chemotherapy for advanced hormone refractory prostate cancer. Urology. 1999;54:30–35. doi: 10.1016/s0090-4295(99)00452-5. [DOI] [PubMed] [Google Scholar]
- 5.Harzstark AL, Small EJ. Castrate-resistant prostate cancer: therapeutic strategies. Expert Opin Pharmacother. 2010;11:937–945. doi: 10.1517/14656561003677382. [DOI] [PubMed] [Google Scholar]
- 6.Higano CS, Small EJ, Schellhammer P, Yasothan U, Gubernick S, Kirkpatrick P, Kantoff PW. Sipuleucel-T. Nat Rev Drug Discov. 2010;9:513–514. doi: 10.1038/nrd3220. [DOI] [PubMed] [Google Scholar]
- 7.Shah S, Ryan CJ. Abiraterone acetate CYP17 inhibitor oncolytic. Drugs Fut. 2009;34:873–880. [Google Scholar]
- 8.Scher HI, Beer TM, Higano CS, Anand A, Taplin M-E, Efstathiou E, Rathkopf D, Shelkey J, Yu EY, Alumkal J, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1-2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrijvers D, Van Erps P, Cortvriend J. Castration-refractory prostate cancer: new drugs in the pipeline. Adv Ther. 2010;27:285–296. doi: 10.1007/s12325-010-0038-1. [DOI] [PubMed] [Google Scholar]
- 10.Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P, Qian F, Chu F, Bentzien F, Cancilla B, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- 11.Smith DC, Smith MR, Small EJ, Sweeney C, Kurzrock R, Gordon MS, Vogelzang NJ, Scheffold C, Ballinger MD, Hussain M. Phase II study of XL184 in a cohort of patients (pts) with castration-resistant prostate cancer (CRPC) and measurable soft tissue disease. J Clin Oncol. 2011;29(suppl 7) Abstract 127. [Google Scholar]
- 12.Albrecht M, Jiang W, Kumi-Diaka J, Lansky EP, Gommersall LM, Patel A, Mansel RE, Neeman I, Geldof AA, Campbell MJ. Pomegranate extracts potently suppress proliferation, xenograft growth, and invasion of human prostate cancer cells. J Med Food. 2004;7:274–283. doi: 10.1089/jmf.2004.7.274. [DOI] [PubMed] [Google Scholar]
- 13.Syed DN, Suh Y, Afaq F, Mukhtar H. Dietary agents for chemoprevention of prostate cancer. Cancer Lett. 2008;265:167–176. doi: 10.1016/j.canlet.2008.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rettig MB, Heber D, An J, Seeram NP, Rao JY, Liu H, Klatte T, Belldegrun A, Moro A, Henning SM, et al. Pomegranate extract inhibits androgenindependent prostate cancer growth through a nuclear factor-κβ-dependent mechanism. Mol Cancer Ther. 2008;7:2662–2671. doi: 10.1158/1535-7163.MCT-08-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantuck AJ, Leppert JT, Zomorodian N, Seeram N, Seiler D, Liker H, Wang H-J, Elashoff R, Heber D, Belldegrun AS. Phase II study of pomegranate juice for men with rising PSA following surgery or radiation for prostate cancer. J Urol. 2005;173:225–226. [Google Scholar]
- 16.Malik A, Afaq F, Sarfaraz S, Adhami VM, Syed DN, Mukhtar H. Pomegranate fruit juice for chemoprevention and chemotherapy of prostate cancer. Proc Natl Acad Sci USA. 2005;102:14813–14818. doi: 10.1073/pnas.0505870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lansky EP, Jiang W, Mo H, Bravo L, Froom P, Yu W, Harris NM, Neeman I, Campbell MJ. Possible synergistic prostate cancer suppression by anatomically discrete pomegranate fractions. Invest New Drugs. 2005;23:11–20. doi: 10.1023/B:DRUG.0000047101.02178.07. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Alcon A, Yuan H, Ho J, Li QJ, Martins-Green M. Cellular and molecular mechanisms of pomegranate juice-induced anti-metastatic effect on prostate cancer cells. Integr Biol (Camb) 2011;3:742–754. doi: 10.1039/c0ib00122h. [DOI] [PubMed] [Google Scholar]
- 19.Kim ND, Mehta R, Yu W, Neeman I, Livney T, Amichay A, Poirier D, Nicholls P, Kirby A, Jiang W, et al. Chemopreventive and adjuvant therapeutic potential of pomegranate (Punica granatum) for human breast cancer. Breast Cancer Res Treat. 2002;71:203–217. doi: 10.1023/a:1014405730585. [DOI] [PubMed] [Google Scholar]
- 20.Syed DN, Afaq F, Mukhtar H. Pomegranate derived products for cancer chemoprevention. Semin Cancer Biol. 2007;17:377–385. doi: 10.1016/j.semcancer.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Adhami VM, Khan N, Mukhtar H. Cancer chemoprevention by pomegranate: laboratory and clinical evidence. Nutr Cancer. 2009;61:811–815. doi: 10.1080/01635580903285064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil MI, Tomas-Barberan FA, Hess-Pierce B, Holcroft DM, Kader AA. Antioxidant activity of pomegranate juice and its relationship with phenolic composition and processing. J Agric Food Chem. 2000;48:4581–4589. doi: 10.1021/jf000404a. [DOI] [PubMed] [Google Scholar]
- 23.El Kar C, Ferchichi A, Attia F, Bouajila J. Pomegranate (Punica granatum) juices: chemical composition, micronutrient cations, and antioxidant capacity. J Food Sci. 2011:C795–C800. doi: 10.1111/j.1750-3841.2011.02211.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q, Yan B, Hu X, Li XB, Zhang J, Fang J. Luteolin inhibits invasion of prostate cancer PC3 cells through E-cadherin. Mol Cancer Ther. 2009;8:1684–1691. doi: 10.1158/1535-7163.MCT-09-0191. [DOI] [PubMed] [Google Scholar]
- 25.Grossmann ME, Mizuno NK, Schuster T, Cleary MP. Punicic acid is an ω-5 fatty acid capable of inhibiting breast cancer proliferation. Int J Oncol. 2010;36:421–426. [PubMed] [Google Scholar]
- 26.Gasmi J, Sanderson JT. Growth inhibitory, antiandrogenic, and proapoptotic effects of punicic acid in LNCaP human prostate cancer cells. J Agric Food Chem. 2010;58:12149–12156. doi: 10.1021/jf103306k. [DOI] [PubMed] [Google Scholar]
- 27.Lansky EP, Harrison G, Froom P, Jiang WG. Pomegranate (Punica granatum) pure chemicals show possible synergistic inhibition of human PC-3 prostate cancer cell invasion across Matrigel. Invest New Drugs. 2005;23:379. doi: 10.1007/s10637-005-5856-7. [DOI] [PubMed] [Google Scholar]
- 28.Roudier MP, Vesselle H, True LD, Higano CS, Ott SM, King SH, Vessella RL. Bone histology at autopsy and matched bone scintigraphy findings in patients with hormone refractory prostate cancer: the effect of bisphosphonate therapy on bone scintigraphy results. Clin Exp Metastasis. 2003;20:171–180. doi: 10.1023/a:1022627421000. [DOI] [PubMed] [Google Scholar]
- 29.Lin S-L, Chang D, Ying S-Y. Hyaluronan stimulates transformation of androgen-independent prostate cancer. Carcinogenesis. 2007;28:310–320. doi: 10.1093/carcin/bgl134. [DOI] [PubMed] [Google Scholar]
- 30.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 31.Ohtsuka T, Nakanishi H, Ikeda W, Satoh A, Momose Y, Nishioka H, Takai Y. Nexilin: a novel actin filament-binding protein localized at cellmatrix adherens junction. J Cell Biol. 1998;143:1227–1238. doi: 10.1083/jcb.143.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 35.Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- 36.Sureban SM, May R, Mondalek FG, Qu D, Ponnurangam S, Pantazis P, Anant S, Ramanujam RP, Houchen CW. Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J Nanobiotechnology. 2011;9:40. doi: 10.1186/1477-3155-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akcakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39:311–318. doi: 10.3892/ijo.2011.1043. [DOI] [PubMed] [Google Scholar]
- 38.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 39.Leone V, D'Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1α. J Clin Endocrinol Metab. 2011;96:E1388–E1398. doi: 10.1210/jc.2011-0345. [DOI] [PubMed] [Google Scholar]
- 40.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Sasaki K, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, et al. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int J Oncol. 2011;39:1099–1107. doi: 10.3892/ijo.2011.1096. [DOI] [PubMed] [Google Scholar]
- 41.Incoronato M, Garofalo M, Urso L, Romano G, Quintavalle C, Zanca C, Iaboni M, Nuovo G, Croce CM, Condorelli G. miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70:3638–3646. doi: 10.1158/0008-5472.CAN-09-3341. [DOI] [PubMed] [Google Scholar]
- 42.Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]
- 43.Cai KM, Bao XL, Kong XH, Jinag W, Mao MR, Chu JS, Huang YJ, Zhao XJ. Hsa-miR-34c suppresses growth and invasion of human laryngeal carcinoma cells via targeting c-Met. Int J Mol Med. 2010;25:565–571. doi: 10.3892/ijmm_00000378. [DOI] [PubMed] [Google Scholar]
- 44.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2011:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 47.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 48.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Bian Z, Wei D, Zhang JG. miR-29b regulates migration of human breast cancer cells. Mol Cell Biochem. 2011;352:197–207. doi: 10.1007/s11010-011-0755-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.