Abstract
Targeting heat shock protein 90 (Hsp90) provides a promising therapeutic approach to enhance the sensitivity of tumor cells to ionizing radiation (IR). To explore the impact of scheduling drug-IR administration, in the present study, we analyzed the response of lung carcinoma A549 and glioblastoma SNB19 cells to simultaneous drug-IR treatment followed by a long-term drug administration. Cellular response was evaluated at different time intervals after IR-alone, drug-alone, or combined drug-IR treatments by colony counts and expression profiles of Hsp90 and its clients, along with several apoptotic markers and cell cycle-related proteins, as well as by IR-drug-induced cell cycle arrest, DNA damage, and repair. A short 30-minute exposure to either Hsp90 inhibitor did not affect the radiosensitivity of both tumor cell lines. Increasing the duration of post-IR-drug treatment progressively enhanced the sensitivity of SNB19 cells to IR. In contrast, the response of A549 cells to drug-IR combination was largely determined by the cytotoxic effects of both drugs without radiosensitization. Combined drug-IR treatment induced more severe DNA damage in both tumor cell lines than each treatment alone and also protracted the kinetics of DNA damage repair in SNB19 cells. In addition to large cell cycle disturbances, drug-IR treatment also caused depletion of the antiapoptotic proteins Akt and Raf-1 in both cell lines, along with a decrease of survivin in A549 cells in case of NVP-AUY922. The data show that simultaneous Hsp90 inhibition and irradiation may induce cell type-specific radiosensitization as well as cytotoxicity against tumor cells.
Introduction
Heat shock protein 90 (Hsp90) is a molecular chaperone essential for the conformation and function of numerous proteins, termed Hsp90 clients [1]. Hsp90 clients are involved in processes characteristic for the malignant phenotype, such as angiogenesis, invasion, and metastasis [1]. Moreover, Hsp90 stabilizes Raf-1, Akt, and ErbB2 proteins, which are known to be associated with protection against radiation-induced cell [2–4].
Several studies highlighted the Hsp90 inhibitor geldanamycin and its derivatives to be potent radiosensitizers of tumor cells, both in vitro [3–6] and in animal models [7]. However, the excessive toxicity of geldanamycins in vivo [8] has prompted considerable efforts to design small synthetic inhibitors of Hsp90 with low toxicity and better bioavailability. A series of new pyrazole resorcinol compounds, most notably NVP-AUY922 and NVP-BEP800, fulfill the above criteria. Beside their ability to interfere with the Hsp90 function, both compounds show favorable pharmaceutical and pharmacological properties, along with strong antiproliferative potential against various tumor cell lines and primary tumors in vitro and in vivo at well-tolerated doses [7].
Most radiobiologic studies with Hsp90 inhibitors have used a schedule where drug exposure precedes irradiation [3,5,6,9–12]. Studies dealing with the optimum time schedule of drug-ionizing radiation (IR) application led to controversial results. Enmon et al. [13] found that the sequence irradiation first followed by the addition of the geldanamycin derivative 17AAG is superior to that of the reverse order in delaying the spheroid formation of prostate cancer cells. In contrast, another geldanamycin derivative (17-DMAG) has shown the maximum synergy with IR in killing lung cancer cell lines when added 16 hours before radiation [14].
In our hands, the novel inhibitors of Hsp90, NVP-AUY922 and NVP-BEP800, exhibit strong radiosensitizing activity in several tumor cell lines (including the lung carcinoma A549 and glioblastoma SNB19), which were pretreated with the drugs for 24 hours before irradiation [12]. Besides this, we found that NVP-AUY can radiosensitize tumor cells under hypoxic conditions [15]. To prove whether the time schedule of drug-IR administration is critical for radiosensitization, we explore in the present study the response of A549 and SNB19 cells to simultaneous drug-IR treatment followed by a long-term drug exposure for up to 48 hours. To discriminate between the radiosensitization and cytotoxic effects, we thoroughly analyzed both cell lines for colony-forming ability, cell cycle distribution, and expression of Hsp90 and its clients, along with several apoptotic markers, cell cycle-related proteins, and radiation-induced DNA damage and repair, at different time intervals after drug and/or IR treatment.
Materials and Methods
Cells
The lung carcinoma A549 and SNB19 cell lines were obtained from the American Type Culture Collection (Manassas, VA) and cultured under standard conditions in complete growth medium (CGM) containing Dulbecco's modified Eagle's medium supplemented with 10% FBS. The A549 cell line bears mutation in k-Ras, SMARCA4, and STK12, whereas the SNB19 is mutated for p53 and phosphatase and tensin homolog (PTEN), and both cell lines are mutated for CDKN2A and CDKN2a(p14) (http://www.sanger.ac.uk/genetics/CGP/cosmic/; Catalogue of Somatic Mutations In Cancer).
Drug Treatment
The Hsp90 inhibitors NVP-AUY922 and NVP-BEP800 were kindly provided by Novartis Institutes for Biomedical Research (Basel, Switzerland). Exponentially growing cell cultures were treated either with 200 nM 1 hour before irradiation or with different concentrations (up to 5 µM) of inhibitors for the cell viability test. Control cells were treated in parallel with respective amounts of DMSO as a vehicle control.
Antibodies
The primary and secondary antibodies are specified in the Supplementary Materials.
X-ray Irradiation
Irradiation was performed at room temperature using a 6-MV Siemens linear accelerator (Siemens, Concord, CA) at a dose rate of 2 Gy/min. After irradiation, cells were kept in drug-containing CGM for the indicated time until harvest.
Colony Survival Assay
Cell survival curves were generated by a standard colony formation assay as previously described [12]. Subconfluent monolayers of drug-treated and nontreated control cells were irradiated in culture flasks filled with CGM at room temperature by graded single doses (0–10 Gy), seeded in Petri dishes and then cultivated in CGM for the next 2 weeks. At least two quadruplicate experiments were carried out for each radiation dose. After 2 weeks, the cells were fixed and stained with crystal violet (0.6%). Colonies of at least 50 cells were scored as survivors. The mean survival data for each individual cell line were fitted to the linear quadratic (LQ) model, using Origin software (Microcal, Northampton, MA):
| (1) |
where SF is the survival fraction, X is the irradiation dose, and α and β are the fitted parameters.
Cell Viability Assay
The intracellular ATP level, as an indicator of cell viability, was determined by means of the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) according to the manufacturer's instructions. Cell culture aliquots were treated for 24 and 48 hours with serial ∼1/3 log dilutions of Hsp90 inhibitors (10 nM to 5 µM) in CGM. The cytotoxicity of each drug was determined for both incubation times in at least two quadruplicate experiments. Control samples contained the respective concentrations of DMSO. The mean ATP content data (±SE) from three independent experiments were normalized against DMSO-treated controls to generate dose-response curves and then analyzed with the standard four-parameter logistic model (4PLM):
| (2) |
where Y is the cell viability; the abscissa c is drug concentration given in nanomolars; A1 and A2 are the upper and lower asymptotes, respectively; IC50 is the 50% inhibitory concentration (nM); and p is the Hill slope.
Western Blot
For immunoblot analysis, whole-cell lysates were prepared according standard procedures. Samples equivalent to 10 to 50 µg of protein were separated using 4 to 12% sodium dodecyl sulfate (SDS)-polyacrylamide precast gels (Invitrogen, Karlsruhe, Germany) and transferred to nitrocellulose membranes. For protein detection, membranes were incubated with respective primary and species-specific peroxidase-labeled secondary antibodies according to standard protocols. The levels of protein expression were quantified using the Kodak 1D Image analyzing software (Scientific Imaging Systems, Eastman Kodak Company, Rochester, NY) and normalized to the β-actin levels.
Immunocytochemical Detection of Histone γH2AX and Cell Cycle Measurements
Nontreated and drug-treated cell cultures were irradiated as subconfluent monolayers in CGM at room temperature. The cells were then incubated in the same medium under standard conditions and analyzed by flow cytometry 30 minutes, 1 day, and 2 days after IR exposure. For analysis, cells were trypsinized, washed twice in phosphate-buffered saline, fixed, and stained for γH2AX according to a protocol described elsewhere [16]. The cells were then counterstained with propidium iodide (PI, Sigma P-4170, 10 µg/ml) in the presence of RNase A (Sigma R-5250, 25 µg/ml) as described elsewhere [17]. At least 15,000 cells were assayed for either histone γH2AX or DNA distribution using a flow cytometer (FACSCalibur, Becton Dickinson, San Jose, CA) equipped with a 15-mW argon-ion laser. Cellular green (anti-γH2AX antibody coupled with fluorescein isothiocyanate) or red fluorescence (DNA staining with PI) was acquired in logarithmic or linear mode. The output data presented as one-dimensional histograms, that is, the distributions of histone γH2AX or PI-DNA signals within cell samples, were analyzed using the WinMDI program obtained from J. Trotter (The Scripps Research Institute, La Jolla, CA) and the ModFit LT program (Verity Software House, Topsham, ME).
Statistics
Data are presented as means (±SD or ±SE). Mean values were compared by the Student's t test. The threshold of statistical significance was set at P < .05. Statistics and fitting of experimental curves were performed with the program Origin (Microcal).
Results
Effects of Hsp90 Inhibitors on Cellular Radiosensitivity
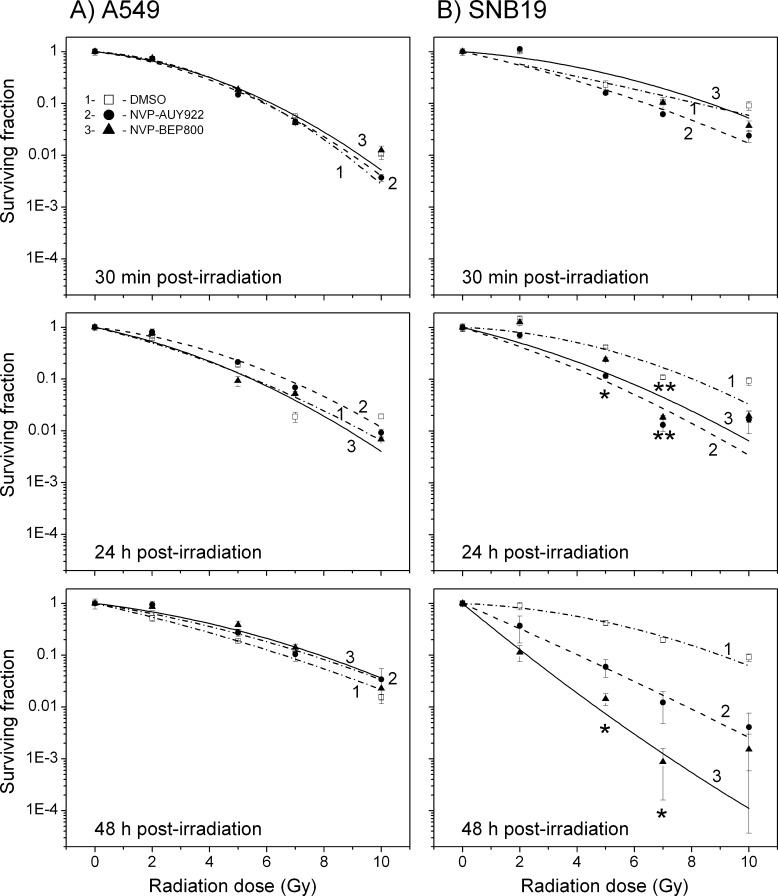
On the basis of the dose-response data reported previously [12], both Hsp90 inhibitors, NVP-AUY922 and NVP-BEP800, were used at a concentration of 200 nM. Drugs were added to cells 1 hour before irradiation. The cells were exposed to an X-ray dose of up to 10 Gy, kept in culture for 30 minutes, 24 hours, or 48 hours, and then seeded for the colony survival test. Figure 1 shows the normalized cell survival responses (symbols) plotted versus the X-ray dose. Table W1 summarizes several parameters calculated with the LQ model, including the surviving cell fractions at 2 Gy (SF2), the radiation doses (D10) resulting in 10% survival, and the growth inhibition factor (IF10), allowing for the radiosensitizing effect. The table also includes the data for the plating efficiencies (PEs) of nonirradiated cell samples, which reflect the cytotoxicity of Hsp90 inhibitors alone.
Figure 1.
Clonogenic survival of A549 (A) and SNB19 (B) cell lines as functions of the radiation dose and the duration of postradiation exposure to an Hsp90 inhibitor. DMSO-treated controls (empty squares), NVP-AUY922-treated (filled circles) cells, and NVP-BEP800-treated cells (filled triangles) were irradiated with single IR doses of up to 10 Gy. At the indicated time after irradiation, cells were plated for the colony survival tests. Survival curves were fitted by an LQ equation (Equation 1). The experiment was repeated at least three times. Significant differences between control and cells treated with NVP-AUY922 or NVP-BEP800 are indicated (*P < .05, **P < .01).
As seen in Figure 1 (left-hand panel), neither NVP-AUY922 nor NVP-BEP800 had any significant effects on the IR dose-response curves of A549 cells over the whole range of postradiation time intervals (0.5–48 hours). Accordingly, the calculated SF2 and D10 values of both drug-treated samples are very similar to the corresponding DMSO controls (Table W1; SF2 ≈ 0.65 ± 0.2 and D10 ≈ 6.6 ± 1.0).
In sharp contrast to A549 cells, the colony-forming ability of SNB19 cells was subject to strong radiosensitization by both Hsp90 inhibitors (Figure 1, right-hand panel). Increasing incubation time from 24 to 48 hours after irradiation further enhanced the radiosensitivity of SNB19 cells (Figure 1, right-hand bottom graphs). Accordingly, the SF2 values decreased two-fold to three-fold, as a result of a 48-hour postirradiation drug treatment, i.e., from 0.7 in the DMSO control to 0.3 for NVP-AUY922 and to 0.23 for NVP-BEP800 (Table W1).
The normalized survival data shown in Figure 1 do not allow for possible cytotoxic and/or antiproliferative effects of the Hsp90 inhibitors alone, which were present in cell culture for up to 48 hours after irradiation. Therefore, Table W1 also shows the data for the PE of nonirradiated cell samples. Even without irradiation, the viability of both cell lines decreased steadily with increasing incubation time in drug-containing medium. The PE of the lung carcinoma A549 cells diminished nearly seven-fold (from 0.4 to 0.06) after a 48-hour post-treatment with NVP-AUY922 and to a lesser extent (about four-fold) in the presence of NVP-BEP800, compared to DMSO-treated control cells (PE ≈ 0.4). Both drugs were less toxic to SNB19 cells, whose PE was reduced only ∼1.5-fold and ∼3-fold, respectively, after a 48-hour treatment at 200 nM with NVP-BEP800 and NVP-AUY922 alone.
Cytotoxicity of Hsp90 Inhibitors to Tumor Cell Lines
Dose-dependent cytotoxic potentials of NVP-AUY922 and NVP-BEP800 against nonirradiated cells were studied over a concentration range from 10 nM to 5 µM. Cell viability was quantified by an ATP-based assay. The cellular ATP levels were normalized against DMSO-treated controls, plotted versus the drug concentration (Figure W1), and fitted to the 4PLM (Equation 2).
Figure W1 shows that the ATP content in both cell lines depends on the drug concentration and on the duration of drug treatment. Following a 24-hour treatment with either drug, the viability gradually decreased with increasing drug concentration. Increasing incubation time from 24 to 48 hours shifted the dose-response curves of both tumor cell lines toward lower drug concentrations (Figure W1, filled symbols). This is particularly evident from a nearly 10-fold decrease of IC50 value of NVP-BEP800 after a 48-hour drug treatment in both cell lines (Table W2). In accordance with the colony formation assays, intermediate concentrations of NVP-AUY922 (also including the 200 nM used in irradiation experiments) were more toxic to A549 cells (the ATP level ≈ 0.2 a.u.) than to SNB19 cells (ATP ≈ 0.45 a.u.).
Expression of Hsp90 Client and (Anti-) Apoptotic Marker Proteins
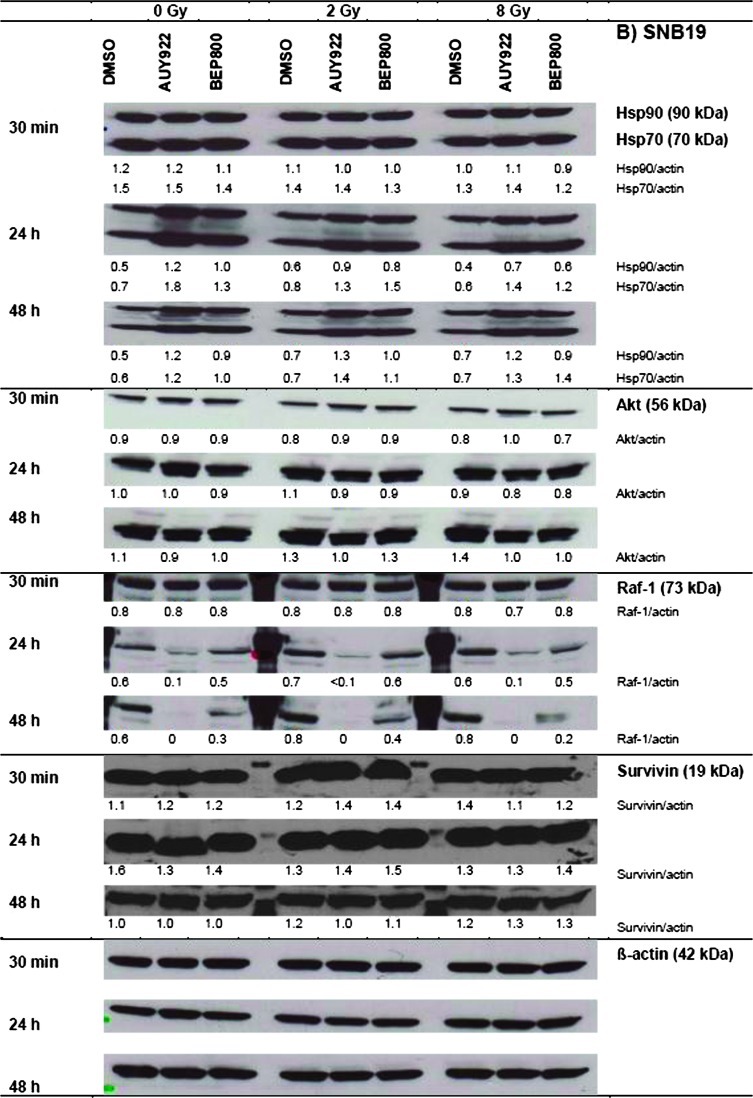
Figure 2 shows representative Western blots of A549 and SNB19 cells probed for Hsp90, Hsp70, Akt, Raf-1, and survivin, at different time intervals (0.5, 24, and 48 hours) after drug-alone, IR-alone, or combined drug-IR treatments. Protein expression was quantified by the protein/actin ratios given in a.u.
Figure 2.
Western blot analysis of expression levels and migration patterns of Hsp90, Hsp70, Akt, Raf-1, and survivin, in DMSO-treated controls, drug-treated and/or irradiated (2 and 8 Gy) A549 (A) and SNB19 (B) cells. Each protein band was normalized to the intensity of β-actin used as loading control, and the ratios are given by numbers. The experiment was repeated three times.
In nonirradiated cell samples of both cell lines (Figure 2, A and B, left-hand columns; 0 Gy), a short exposure (30 minutes) to either Hsp90 inhibitor did not cause any significant changes in the expression of all tested proteins, compared to DMSO controls. A 24-hour drug exposure increased the expression of Hsp90 and Hsp70 in A549 cells, respectively, from 0.5 to 0.6 a.u. and from 0.7 to ∼0.9 a.u. At the same time, the Akt level decreased by half (0.8 to 0.4) and Raf-1 nearly vanished (0.8 to ∼0). The depletion of both antiapoptotic proteins (Akt and Raf-1) might be considered as a sign of ongoing apoptosis, which was also confirmed by an ∼25% reduction in the expression of survivin (1.6 to 1.2) after Hsp90 inhibition with NVP-AUY922.
Irradiation of A549 cells with 2 Gy in the absence of Hsp90 inhibitors (DMSO controls) did not significantly affect the expression of four of five marker proteins, including Hsp90, Hsp70, Akt, and Raf-1. At the same time, the amount of survivin decreased by 30 to 40%. Upon increasing the radiation dose to 8 Gy, Hsp90, Hsp70, Akt, and survivin showed no or little changes, whereas Raf-1 decreased at 24 hours by half (0.8 to 0.4).
Compared to drug alone, irradiation of drug-treated A549 cells with either 2 or 8 Gy produced only little, if any, additional changes in the expression of four of five marker proteins studied here (Hsp90, Hsp70, Akt, and Raf-1). Only in case of survivin, whose expression was markedly reduced or even fully abolished in A549 cells by the combined drug-IR treatment (e.g., NVP-AUY922, 8 Gy, 48 hours), irradiation appeared to add to the effect of the Hsp90 inhibitors.
In SNB19 cells, the kinetics and relative changes in the expression of most marker proteins were qualitatively similar to those in A549 cells but differed in quantitative details. Thus, the NVP-AUY922-IR-treated SNB19 cells (i.e., 8 Gy, 48 hours) exhibited a much greater increase in the expression of chaperones Hsp90 (0.7 to 1.2) and Hsp70 (0.7 to 1.3) over their levels in irradiated DMSO control. On the opposite, the same treatment (i.e., NVP-AUY922, 8 Gy, 48 hours) decreased Akt expression in SNB19 cells only by ∼30% (1.4 to 1.0), as contrasted with a more than 50% reduction of Akt in A549 cells (0.7 to 0.3). While both drugs completely abolished Raf-1 in nonirradiated A549 cells (0.6 to 0, 48 hours), only NVP-AUY922-treated SNB19 cells were totally depleted of Raf-1 (0.6 to 0), whereas NVP-BEP800 decreased only by half the expression of Raf-1 in nonirradiated SNB19 cells (0.6 to 0.3). The expression of survivin in SNB19 was rather insensitive to the different treatment modalities studied here, which contrasted sharply with the complete depletion of survivin in A549 cells in case of NVP-AUY922.
The observed changes in the expression of antiapoptotic proteins (i.e., Akt, Raf-1, and survivin) do not explain the strong radiosensitizing effects of NVP-AUY922 and NVP-BEP800 observed in SNB19 cells (Figure 1B). The depletion of Akt, Raf-1, and survivin in NVP-AUY922-treated and irradiated A549 cells rather correlates with apoptosis and the observed reduction in PE (Table W1) and ATP levels (Figure W1A).
Induction and Restoration of Histone γH2AX, a Marker of DNA Damage
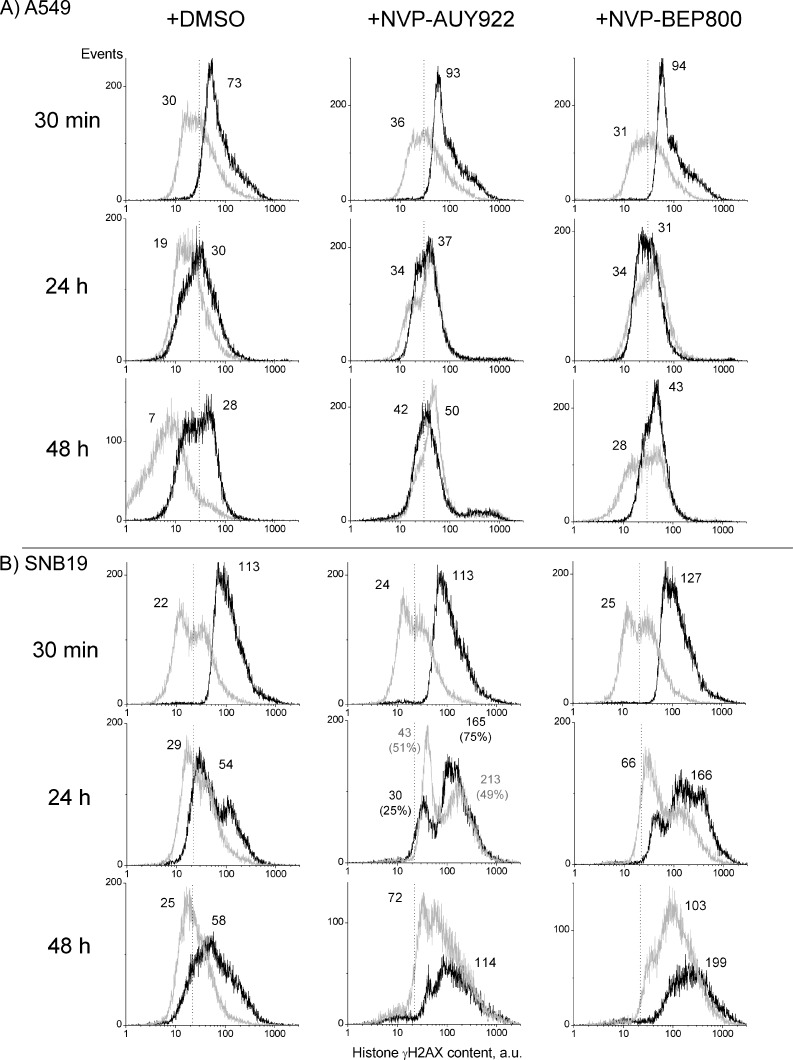
In addition, increased DNA damage leading to proliferative death might be responsible for the cell type-specific radiosensitizing and cytotoxic effects of the Hsp90 inhibitors reported here (Figure 1). The induction and restoration kinetics of DNA damage were studied by phosphorylated histone H2AX (γH2AX), an established marker of DNA double-strand breaks (DSBs) [18]. Cells were exposed to drug and irradiated with either 2 Gy (Figure W2) or 8 Gy (Figure 3) and kept for 30 minutes, 24 hours, or 48 hours. After that, the cells were stained with anti-γH2AX antibodies coupled with fluorescein isothiocyanate and analyzed by flow cytometry. Nonirradiated and DMSO-treated cells served as controls.
Figure 3.
Typical distributions of histone γH2AX, as a marker of DNA DSBs, in A549 (A) and SNB19 (B) cell lines. Black and light gray histograms represent irradiated (8 Gy) and nonirradiated cells, respectively. Numbers denote the mean γH2AX expression for the respective cell subpopulation and the percentage of cells in the subpopulation. To facilitate visual comparison, the modal γH2AX values of DMSO-treated and irradiated samples are indicated by dashed vertical lines. The experiment was repeated at least three times.
As seen in Figure 3 (A and B, upper panels, light gray histograms), the DMSO-treated control A549 and SNB19 cells show almost similar background expression of histone γH2AX. Thirty minutes after irradiation alone with 8 Gy (black histograms), the expression of γH2AX increased more than two-fold in A549 cells (from 30 to 73 a.u.) and about five-fold in SNB19 cells (22 to 113 a.u.). Upon incubation for 24 hours after IR alone, A549 cells completely restored their background γH2AX expression (73 to 30 a.u.), whereas SNB19 accomplished only a partial restoration of the histone (113 to 54 a.u.). Increasing incubation time to 48 hours gave only little further changes of γH2AX expression in both cell lines.
A short-term exposure (30 minutes) to either Hsp90 inhibitor without irradiation caused no or little DNA damage in both cell lines (Figure 3, A and B, second and third columns, top panels, light gray histograms). Upon longer incubation, i.e., for 24 or 48 hours, both drugs induced a large time-dependent increase of histone γH2AX in SNB19 cells, without affecting significantly the amount of γH2AX in A549 cells. Thus, judging from the histograms of SNB19 cells treated for 48 hours, the amount of γH2AX increased three-fold by NVP-AUY922 (72 a.u.) and even four-fold by NVP-BEP800 (103 a.u.) over that of the DMSO control (25 a.u.). Apparently, NVP-BEP800 induced greater DNA damage in this particular cell line than did NVP-AUY922.
Unlike drug-alone regimen, the combined drug-IR treatment of A549 cells initially increased about three-fold the expression of γH2AX (93–94 a.u.) above nonirradiated drug-treated samples (31–36 a.u.) and, to a lesser extent, above the irradiated control (73 a.u.; Figure 3A). Twenty-four or forty-eight hours later, the expression of γH2AX returned nearly to the level of control cells (∼30–40 a.u.), indicating efficient repair of DNA damage in this cell line.
Thirty minutes after a combined drug-IR treatment, SNB19 cells exhibited (similar to A549 cells) high initial γH2AX levels of 113 to 127 a.u., which were four to five times as large as those induced by drug-alone treatments (24–25 a.u.). As with A549 cells, the combined drug-IR treatment, however, caused only little—if any—additional γH2AX increase above that produced by IR alone (113 a.u.).
Unlike A549 cells, SNB19 cells were apparently incapable of restoring the DNA damage induced by IR or drug-IR treatment. This is particularly evident from the finding that the initially elevated γH2AX expression in SNB19 cells did not return to the background level during the following 24 to 48 hours after IR or drug-IR treatment. In SNB19 cells that received a combined NVP-BEP800-IR treatment, the γH2AX level rose steadily with time (127 to 166 to 199 a.u.).
Effects of Hsp90 Inhibitors on the Cell Cycle and Related Proteins
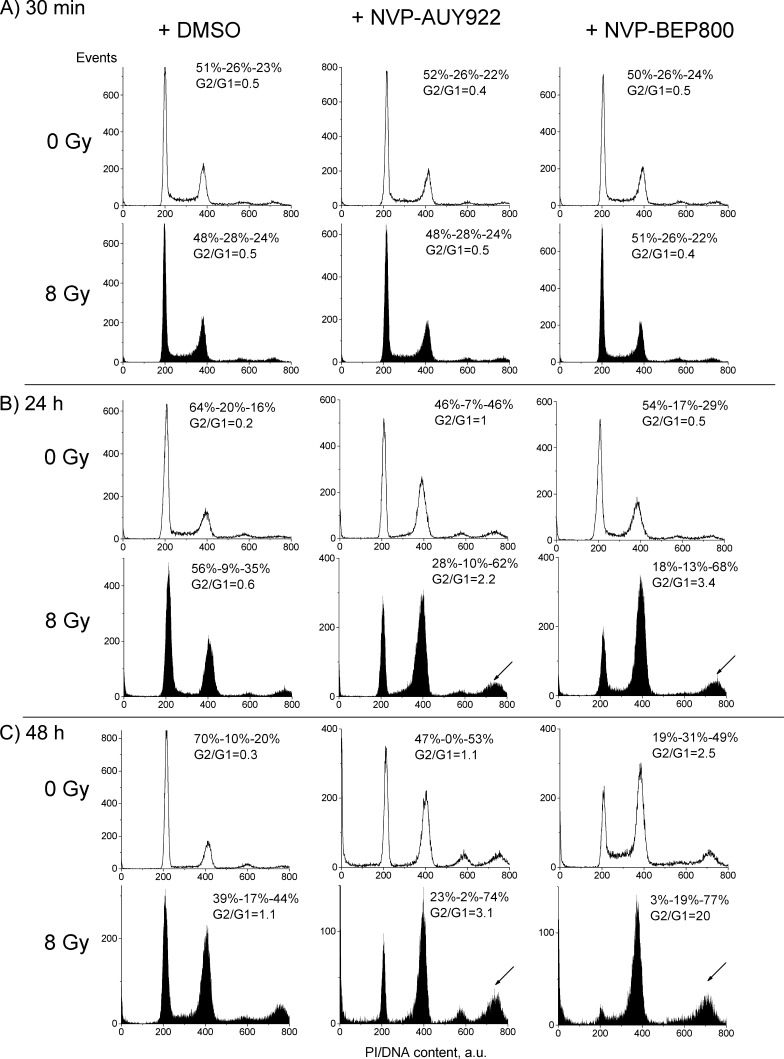
The cell samples probed for DNA damage by histone γH2AX expression (Figure 3) were also analyzed for possible alterations in the cell cycle progression (Figures 4, W3, W4, and W5 and Tables W3 and W4) induced by Hsp90 inhibitors, IR or combination of both treatments.
Figure 4.
Impact of Hsp90 inhibitors, IR, and combined drug-IR treatment on the cell cycle-phase distribution in the SNB19 cell line 30 minutes (A), 24 hours (B), 48 hours (C) after drug and/or IR treatments (8 Gy). Numbers denoted the percentage of cells in G1, S, and G2/M phases and G2/G1 ratios in each cell sample. Filled and unfilled histograms represent irradiated and nonirradiated cells, respectively. Measurements of cell cycle progression were repeated at least three times.
Thirty minutes after irradiation, the cell cycle distribution remained nearly unchanged in drug-free cultures of both cell lines. During the following 24 hours after irradiation, both cell lines showed a strong depletion of S phase and an increase in the G2/M peak. These changes progressively increased in SNB19 cells (Figure 4) as the incubation time was extended from 24 to 48 hours, whereas A549 cells showed no or little further changes in the cell cycle (Figure W3). Interestingly, in both cell lines, the changes in cell cycle distributions induced by an Hsp90 inhibitor without IR (Figures 4 and W3, second and third columns, unfilled histograms) were qualitatively similar to the cell cycle disturbances induced by irradiation alone, after corresponding long-term incubation times (i.e., 24 and 48 hours after irradiation or drug addition).
Combining IR with either Hsp90 inhibitor did not cause any additional changes in the cell cycle distribution of A549 cells (Figure W3), compared to drug-alone or IR-alone protocols. In sharp contrast, the combined drug-IR treatment of SNB19 cells (Figure 4) gave rise to a strong additive effect on the cell cycle distribution, which is evident from the increase of the portion of G2/M cells, most probably through mitotic arrest. Note that the G2/M-to-G1 ratio (G2/G1 = 20) of NVP-BEP800-treated and irradiated SNB19 cells (48 hours) exceeds by far the corresponding data observed after drug-alone (G2/G1 = 2.5) and IR-alone treatment (G2/G1 = 1.1). In addition, the combined drug-IR treatment induced a significant fraction of SNB19 cells with hyperdiploid DNA content (Figure 4, arrows).
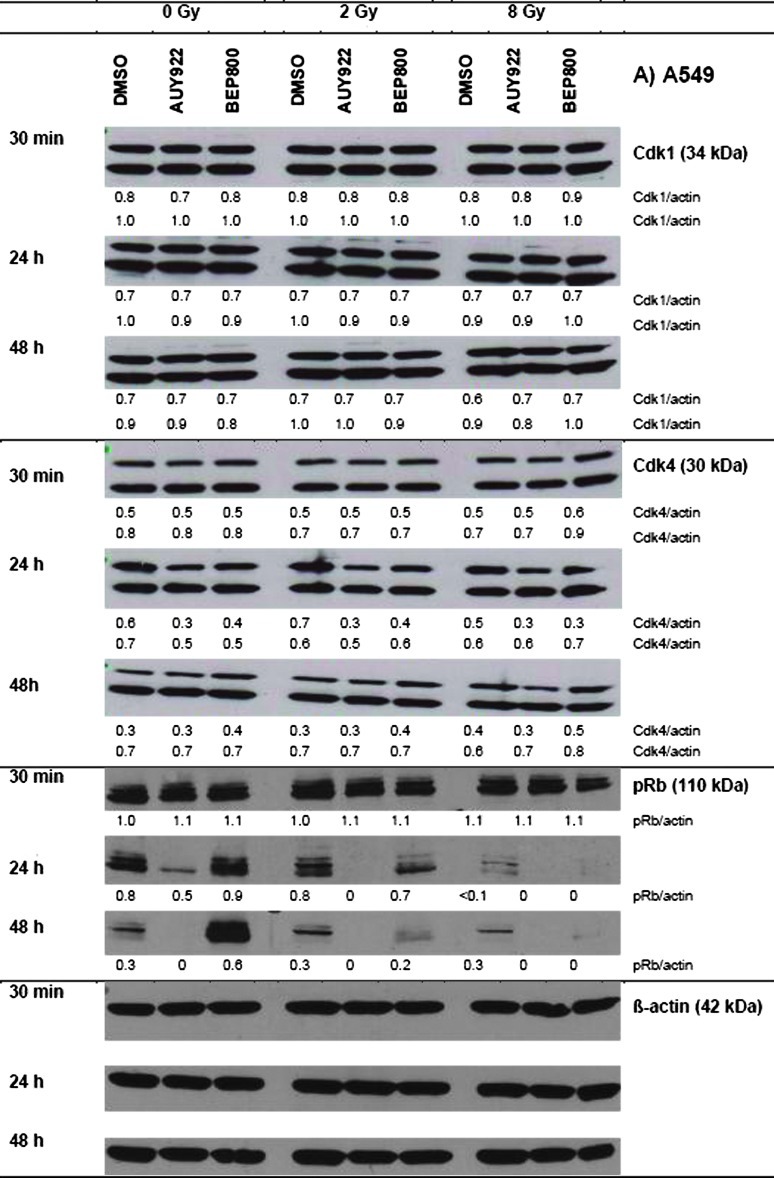
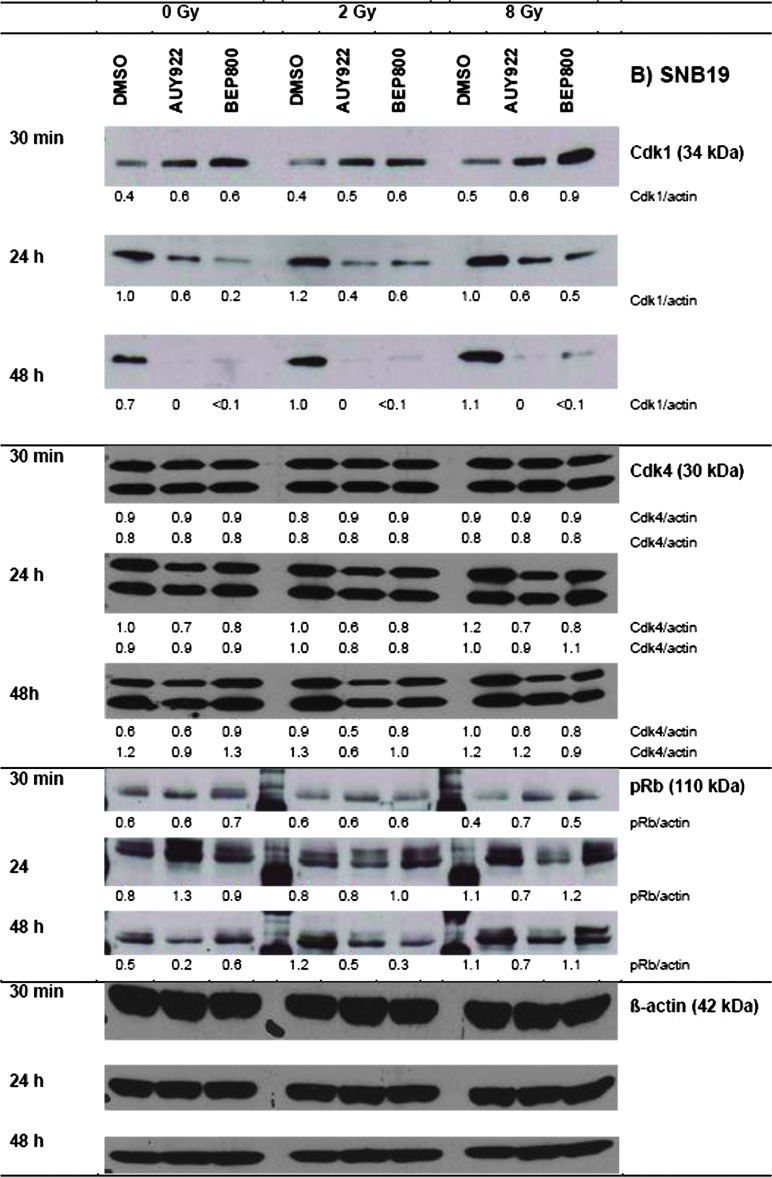
The observed alterations in the cell cycle distributions (Figures 4 and W3 summarized in Tables W3 and W4) prompted us to analyze the expression of several cell cycle regulatory factors, including cyclin-dependent kinases (Cdk1 and Cdk4) and phospho-Retinoblastom protein (pRb). The representative Western blots shown in Figure 5 illustrate cell type and time-dependent changes induced by drug and/or IR treatment.
Figure 5.
Effects of Hsp90 inhibitors on the expression levels of cell cycle regulatory proteins in the A549 (A) and SNB19 (B) tumor cell lines. In case of Cdk1 and Cdk4, which exhibited double isoform bands, the protein/actin ratios are given for both isoforms. Each protein band was normalized to the intensity of β-actin used as loading control, and the ratios are given by numbers. The experiment was repeated three times.
In the absence of Hsp90 inhibitors, irradiation of A549 cells (Figure 5A) with 2 or 8 Gy resulted in a significant down-regulation of pRb at 24 hours, without affecting the expression of Cdk1 and Cdk4 proteins. As evident from Figure 5A, neither NVP-AUY922 nor NVP-BEP800 alone altered the expression of Cdk1 protein in A549 cells. At the same time, nonirradiated A549 cells exhibited down-regulation of Cdk4 and pRb protein after a 24-hour or 48-hour exposure to both Hsp90 inhibitors. In general, changes induced by combined drug-IR treatments were similar to those caused by drug treatment alone, except that drug-IR resulted in a lasting complete depletion of pRb. Downregulation of pRb and the G1/S regulatory protein Cdk4 was less pronounced in SNB19 cells (Figure 5B). In contrast, Cdk1 protein was completely abolished in SNB19 cells after drug alone or after combined drug-IR application.
Discussion
We recently reported strong radiosensitizing effects of NVP-AUY922 and NVP-BEP800 in several tumor cell lines, including A549 and SNB19 cells at normoxic [12] as well as hypoxic [15] conditions. In those studies, we analyzed the impact of a drug-first treatment, i.e., tumor cells were incubated with an Hsp90 inhibitor for 24 hours before irradiation [12]. In contrast, the experiments presented here address the mechanisms of simultaneous drug-IR treatment followed by a prolonged postradiation exposure of tumor cells to an Hsp90 inhibitor.
Unexpectedly, the Hsp90 inhibitors used under this treatment schedule showed considerable differences between the two cell lines, which are summarized in Figure W6. As with drug-first regimen [12], both NVPAUY922 and NVP-BEP800 present during and after IR strongly enhance the radiosensitivity of glioblastoma-derived SNB19 cells (Figure 1). Duration of postirradiation drug exposure appears to be crucial. Moreover, judging by the SF2 values measured for a 48-hour exposure (Table W1), both drugs exerted much stronger radiosensitization compared to the drug-first combination [12]. In A549 cells, there was a pattern of strong cytotoxicity by the drugs itself induced by the apoptotic pathway, G1 arrest, and negligible effects on DNA damage induction and repair.
Consistent with the survival and viability data discussed above, drug-treated A549 cells exhibited strong depletion of ATP and of the antiapoptotic proteins Akt and survivin indicative of apoptotic events. In view of the upregulated expression of the molecular chaperones Hsp90 and Hsp70 (Figure 2), however, the depletion of antiapoptotic proteins is not necessary in conflict with the lack of radiosensitization in drug-treated A549 cells. Given that cell survival could benefit from the drug-mediated induction of molecular chaperones, the above findings suggest that the Hsp90 inhibitors were apparently capable of inducing cell type-dependent antiapoptotic and proapoptotic effects.
We found marked changes in the expression of the cell cycle- associated proteins, Cdk1, Cdk4, and pRb (Figure 5), in drug-treated cells. Cdk1 and Cdk4 are well-known clients of Hsp90. In SNB19 cells, Hsp90 inhibition led to significant down-regulation of the checkpoint protein Cdk1, whereas in A549 cells its expression was mostly unchanged (Figure 5). Given that Cdk1 governs the G2/M transition and mitotic progression, its depletion might have been responsible for the G2/M arrest observed in SNB19 cells (Figure 4). Both Hsp90 inhibitors caused marked down-regulation of Cdk4 in A549 cells. In accord with these data, drug-treated A549 cells exhibited significant hypophosphorylation of Rb protein, which acts as a blocker of cell cycle progression at the G1/S checkpoint in p53wt cells [19]. Drug-treated SNB19 cells were also depleted of both Cdk4 and pRb proteins but to a much lesser extent than A549 cells.
Taken together, our data are consistent with the findings of Enmon et al. [13] who found that the sequence irradiation first followed by the addition of 17AAG is superior to that of the reverse order in delaying the spheroid formation of prostate cancer cells; however, it depends strongly on the cell type.
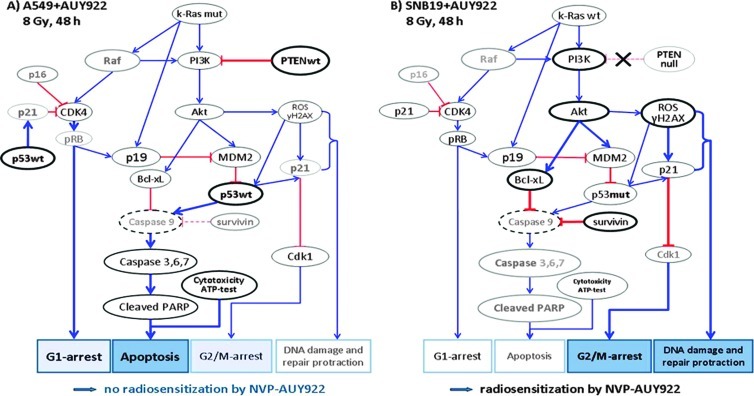
The observed differences between A549 and SNB19 cell lines in their responses to combined drug-IR treatment can be explained by a simplified model illustrated in Figure 6. The model takes account of the different genetic status of the two cell lines, i.e., a k-Ras mutation in A549 cells versus mutations in p53 and pten in SNB19 cells. The model also includes additional data on the expression of several marker proteins: k-Ras, PTEN, phosphoinositid 3-kinase (PI3K), p19ARF, p16, p21WAF1, murine double minute 2 (MDM2), cleaved poly (ADP-ribose) polymerase (PARP), and Bcl-xL, given in Figure W7 (A and B).
Figure 6.
Simplified schema of putative signaling pathways accountable for the differential responses of A549 (A) and SNB19 (B) cell lines to Hsp90 inhibition and IR. In A549 cells (A), the wild-type PTEN along with Hsp90 inhibition apparently suppress the PI3K/Akt pathway. As a consequence, the cells produce less the antiapoptotic protein Bcl-xL. In parallel, IR might activate the wild-type p53 in A549 cells, which in turn triggers the caspase-dependent apoptosis. As a result of Hsp90 inhibition, the expression of the Hsp90 client protein Cdk4 decreases in A549 cells, and through depleted pRB, it cause a G1-phase arrest in the drug-treated and irradiated A549 cells. In contrast to A549 cells, the PTEN-null SNB19 cells (B) express constitutively elevated amounts of the antiapoptotic PI3K and Akt, which favor the survival and growth of this cell line. Besides this, p53 protein is mutated in SNB19 cells, which might be the reason for the lack of activation of caspase 9 (not detected) and subsequently of caspase 3. Moreover, the activated Akt in SNB19 cells, together with IR, induces almost twice as much ROS, which is indicative of DNA damage (about histone γH2AX), compared to that in A549 cells. In addition, SNB19 cells repair the IR-induced DNA damage much more slowly than do A549 cells. Finally, ROS also activate p21, which in turn inhibits Cdk1 in SNB19 cells, thus leading to a strong G2/M arrest (for more details, see text). The schema was proposed using the data presented in Figures 1 to 5, W1 to W3, and W7 and Table W1. Note the size of the letters/symbols and the thickness of the lines; gray means strong reduction or depletion of proteins.
It is well known that cells expressing mutant k-Ras (such as A549 cells) produce elevated levels of p19ARF mRNA, a tumor suppressor protein that links the k-Ras status with p53 expression and hence susceptibility to DNA damage-induced apoptosis and reduced G2 arrest [20]. Besides this, the oncogenic k-Ras results in elevated p53, p21, and p16 levels [21]. The activation of p53 by oncogenic k-Ras with p19ARF is thought to be through a separate pathway to p53 activation from DNA damage [22]. The protein p19ARF, one of the two tumor suppressors encoded by the INK4α-ARF locus, activates p53 both by neutralizing MDM2, which destabilizes p53, and by interacting directly with p53 [22]. The authors have shown that the combination of even mild DNA damage and oncogenic k-Ras signal led to enhanced apoptosis and reduced G2 arrest. The findings of Brooks et al. [20] and Palmero et al. [22] corroborate our results on A549 cell line, which showed an increased apoptosis and G1 arrest after IR and prolonged incubation with the Hsp90 inhibitors thereafter (Figures 2A, 5A, W1, W3, and W7A).
In contrast, cells lacking oncogenic k-Ras tend to respond to DNA damage by entering G2 arrest, observed as an accumulation of cells in the G2/M phases [20]. This finding is consistent with our data for SNB19 cells, which have no mutation in k-Ras and are mostly blocked in G2/M phase upon Hsp90 inhibition and irradiation (Figures 4, 5B, and 6B).
According to the Catalogue of Somatic Mutations In Cancer, the A549 cell line has no mutation in pten gene. Lee et al. [23] have shown that PTEN proficiency sensitizes tumor cells to apoptosis, whereas PTEN-deficient glioma cells undergo senescence through the Akt/reactive oxygen species (ROS)/p53/p21 pathway.
As mentioned above, the SNB19 cell line displays, among others, mutations in p53 and pten. Moreover, SNB19 cells do not express PTEN protein at all [24], thus being a pten-null line, which is in accordance with our data (Figure W7B). The loss of PTEN leads to a constitutive activation in the PI3K/Akt pathway [25], which in turn can enhance tumor cell growth and proliferation. Our Western blot data show an increased expression of PI3K (Figure W7B) and Akt (Figure 2B, 48 hours) in SNB19 cells, compared to A549 cells (Figure 2A, 48 hours, and W7A). Akt promotes the activity of the E3 ubiquitin ligase MDM2, an important negative regulator of p53 protein [26,27]. Furthermore, Akt enhances cell survival by intervening in the apoptosis cascade upstream of cytochrome c release and caspase activation [28]. Accordingly, the activated Akt in SNB19 cells can stimulate, through Bcl-2-associated death promoter (BAD) inhibition (not detected), the antiapoptotic protein Bcl-xL, and thereby inhibiting apoptosis. Besides this, Akt can reduce cell death [29,30] by inhibiting caspase 9 (not detected) and caspase 3 activation, possibly by direct phosphorylation of caspase 9. In agreement with the published data, we observed neither alteration in the expression of caspase 3 nor induction of the cleaved PARP in SNB19 cells (Figures 6B and W7B). In contrast, drug-treated and irradiated A549 cells expressed less antiapoptotic proteins Akt (by a factor of 2, Figure 2A) and Bcl-xL (by a factor of ∼3, Figure W7A) than did control cells, which is indicative of apoptosis (Figure 6A).
In contrast to SNB19 cells, in A549 cells, which are p53wt, an IR-mediated p53 protein can induce activation of caspase 3 through caspase 9 (not detected) and consequently the induction of cleaved PARP (Figures 6A and W7A). Another important protein is survivin, an inhibitor of apoptosis, which acts by repressing caspase 9 and thereafter caspase 3 (Figure 6A and W7A). In our hands, survivin was strongly depleted after Hsp90 inhibition by NVP-AUY922 and IR in A549 cells (Figure 2A, 48 hours), while remaining almost unchanged in the SNB19 cell line (Figure 2B, 48 hours) or after NVP-BEP800 treatment in both cell lines.
IR also induces ROS, which are potent activators of p53 and its downstream target—Cdk inhibitor p21 [31]. Although we did not detect ROS directly, we quantified the induced DNA damage by means of histone γH2AX, a sensitive marker of DNA DSBs [18], which are known to correlate with ROS [32]. We found that Hsp90 inhibition and IR induced more DNA damage, persisting longer in SNB19 cells treated with NVP-AUY922 (Figure 3B). The protracted DNA damage repair along with the Cdk inhibitor p21 is in line with the stronger depletion of Cdk1 in SNB19 (Figure 5B), compared to A549 (Figure 5A), and with the stronger G2/M arrest (Figures 4 and 6B).
In case of NVP-BEP800, in the A549 cells, this inhibitor might operate through a cell cycle arrest and diminished (with respect to NVP-AUY922) DNA damage rather than through apoptosis. In addition, as seen in Figure W1, a 48-hour incubation of the A549 cells with NVP-BEP800 caused much lower (by a factor of ∼2.5) cytotoxicity than did NVP-AUY922. Accordingly, the expressions of survivin (Figure 2A), k-Ras, PI3K, and Bcl-xL (Figure W7A) were not depleted, and the cleaved PARP (Figure W7A) was not expressed after single treatment with NVP-BEP800 (or in combination with IR) in the A549 cells, as it was the case with the NVP-AUY922. In SNB19 cells, NVP-BEP800 also caused a much stronger (by a factor of ∼3) G2/M arrest than did NVP-AUY922.
In summary, our data suggest that, depending on the tumor cell line used, Hsp90 inhibition and radiation may direct damage toward different mode of cell death, i.e., either apoptosis or radiosensitization. Future work on the extended cell sample should therefore try to identify more precisely the mechanism(s) of the observed differences in cell type-specific response of tumor cells to combined Hsp90 inhibition and radiation treatments.
Supplementary Material
Antibodies
The primary antibodies used were mouse monoclonal anti-phospho-histone H2AX FITC conjugate and mouse polyclonal anti-p53 (Millipore, Schwalbach, Germany); rabbit polyclonal antisurvivin (R&D Systems, Minneapolis, MN); mouse monoclonal anti-Hsp70 and mouse monoclonal anti-Hsp90 (BD Pharmingen, Heidelberg, Germany); rabbit polyclonal anti-Raf-1, mouse polyclonal anti-MDM2, mouse polyclonal anti-k-Ras, rabbit polyclonal anti-p19, mouse polyclonal anti-p16, and rabbit polyclonal anti-Cdk4 (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-actin (Sigma, Deisenhofen, Germany); mouse polyclonal anti-p21WAF1 (Thermo Fisher Scientific, Fremont, CA); rabbit polyclonal anti-Akt, rabbit polyclonal anti-PARP, mouse polyclonal anti-caspase 3, mouse monoclonal anti-Cdk1, rabbit polyclonal anti-phospho-Rb, rabbit polyclonal anti-Ku80, rabbit polyclonal anti-Ku70, rabbit polyclonal anti-PI3K p85, rabbit polyclonal anti-PTEN, rabbit polyclonal anti-PI3Kα, rabbit polyclonal anti-Bcl-xL were purchased from Cell Signaling (Danvers, MA). Secondary species-specific antibodies for Western blot were labeled with HRP (DAKO, Hamburg, Germany).
Footnotes
This work was supported by grant B-159 of the Interdisziplinäres Zentrumfür Klinische Forschung (IZKF), University of Würzburg, Germany. Potential conflicts do not exist. Presented in part at the 17th Annual Meeting of the German Society of Radiation Oncology (DEGRO), Wiesbaden, June 2011, and the 14th International Congress of Radiation Research, Warsaw, Poland, August 2011.
This article refers to supplementary materials, which are designated by Figures W1 to W7 and Tables W1 to W4 and are available online at www.transonc.com.
References
- 1.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- 3.Bull EE, Dote H, Brady KJ, Burgan WE, Carter DJ, Cerra MA, Oswald KA, Hollingshead MG, Camphausen K, Tofilon PJ. Enhanced tumor cell radiosensitivity and abrogation of G2 and S phase arrest by the Hsp90 inhibitor 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin. Clin Cancer Res. 2004;10:8077–8084. doi: 10.1158/1078-0432.CCR-04-1212. [DOI] [PubMed] [Google Scholar]
- 4.Camphausen K, Tofilon PJ. Inhibition of Hsp90: a multitarget approach to radiosensitization. Clin Cancer Res. 2007;13:4326–4330. doi: 10.1158/1078-0432.CCR-07-0632. [DOI] [PubMed] [Google Scholar]
- 5.Machida H, Matsumoto Y, Shirai M, Kubota N. Geldanamycin, an inhibitor of Hsp90, sensitizes human tumour cells to radiation. Int J Radiat Biol. 2003;79:973–980. doi: 10.1080/09553000310001626135. [DOI] [PubMed] [Google Scholar]
- 6.Bisht KS, Bradbury CM, Mattson D, Kaushal A, Sowers A, Markovina S, Ortiz KL, Sieck LK, Isaacs JS, Brechbiel MW, et al. Geldanamycin and 17-allylamino-17-demethoxygeldanamycin potentiate the in vitro and in vivo radiation response of cervical tumor cells via the heat shock protein 90-mediated intracellular signaling and cytotoxicity. Cancer Res. 2003;63:8984–8995. [PubMed] [Google Scholar]
- 7.Massey AJ, Schoepfer J, Brough PA, Brueggen J, Chene P, Drysdale MJ, Pfaar U, Radimerski T, Ruetz S, Schweitzer A, et al. Preclinical antitumor activity of the orally available heat shock protein 90 inhibitor NVP-BEP800. Mol Cancer Ther. 2010;9:906–919. doi: 10.1158/1535-7163.MCT-10-0055. [DOI] [PubMed] [Google Scholar]
- 8.Sausville EA, Tomaszewski JE, Ivy P. Clinical development of 17-allylamino, 17-demethoxygeldanamycin. Curr Cancer Drug Targets. 2003;3:377–383. doi: 10.2174/1568009033481831. [DOI] [PubMed] [Google Scholar]
- 9.Russell JS, Burgan W, Oswald KA, Camphausen K, Tofilon PJ. Enhanced cell killing induced by the combination of radiation and the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin: a multitarget approach to radiosensitization. Clin Cancer Res. 2003;9:3749–3755. [PubMed] [Google Scholar]
- 10.Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of Hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 11.Kabakov AE, Makarova YM, Mayutina YV. Radiosensitization of human vascular endothelial cells through Hsp90 inhibition with 17-N-allilamino-17-demethoxygeldanamycin. Int J Radiat Oncol Biol Phys. 2008;71:858–865. doi: 10.1016/j.ijrobp.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 12.Stingl L, Stühmer T, Chatterjee M, Jensen MR, Flentje M, Djuzenova CS. Novel HSP90 inhibitors, NVP-AUY922 and NVP-BEP800, radiosensitise tumour cells through cell-cycle impairment, increased DNA damage and repair protraction. Br J Cancer. 2010;102:1578–1591. doi: 10.1038/sj.bjc.6605683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enmon R, Yang WH, Ballangrud AM, Solit DB, Heller G, Rosen N, Scher HI, Sgouros G. Combination treatment with 17-N-allylamino-17-demethoxy geldanamycin and acute irradiation produces supra-additive growth suppression in human prostate carcinoma spheroids. Cancer Res. 2003;63:8393–8399. [PubMed] [Google Scholar]
- 14.Koll TT, Feis SS, Wright MH, Teniola MM, Richardson MM, Robles A, Bradsher J, Capala J, Varticovski L. HSP90 inhibitor, DMAG, synergizes with radiation of lung cancer cells by interfering with base excision and ATM-mediated DNA repair. Mol Cancer Ther. 2008;7:1985–1992. doi: 10.1158/1535-7163.MCT-07-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djuzenova CS, Blassl C, Roloff K, Kuger S, Katzer A, Niewidok N, Günther N, Polat B, Sukhorukov VL, Flentje M. Hsp90 inhibitor NVP-AUY922 enhances radiation sensitivity of tumor cell lines under hypoxia. Cancer Biol Ther. 2012;13:425–434. doi: 10.4161/cbt.19294. [DOI] [PubMed] [Google Scholar]
- 16.Muslimovic A, Ismail IH, Gao Y, Hammarsten O. An optimized method for measurement of gamma-H2AX on blood mononuclear and cultured cells. Nat Protoc. 2008;3:1187–1193. doi: 10.1038/nprot.2008.93. [DOI] [PubMed] [Google Scholar]
- 17.Djuzenova CS, Sukhorukov VL, Klock G, Arnold WM, Zimmermann U. Effect of electric field pulses on the viability and on the membrane-bound immunoglobulins of LPS-activated murine B-lymphocytes: correlation with the cell cycle. Cytometry. 1994;15:35–45. doi: 10.1002/cyto.990150107. [DOI] [PubMed] [Google Scholar]
- 18.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 19.Stewart ZA, Westfall MD, Pietenpol JA. Cell-cycle dysregulation and anticancer therapy. Trends Pharmacol Sci. 2003;24:139–145. doi: 10.1016/S0165-6147(03)00026-9. [DOI] [PubMed] [Google Scholar]
- 20.Brooks DG, James RM, Patek CE, Williamson J, Arends MJ. Mutant K-ras enhances apoptosis in embryonic stem cells in combination with DNA damage and is associated with increased levels of p19ARF. Oncogene. 2001;20:2144–2152. doi: 10.1038/sj.onc.1204309. [DOI] [PubMed] [Google Scholar]
- 21.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 22.Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 23.Lee JJ, Kim BC, Park MJ, Lee YS, Kim YN, Lee BL, Lee BS. PTEN status switches cell fate between premature senescence and apoptosis in glioma exposed to ionizing radiation. Cell Death Differ. 2011;18:666–677. doi: 10.1038/cdd.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George J, Banik NL, Ray SK. Combination of hTERT knockdown and IFN-γ treatment inhibited angiogenesis and tumor progression in glioblastoma. Clin Cancer Res. 2009;15:7186–7195. doi: 10.1158/1078-0432.CCR-09-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, Sellers WR. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 27.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy SG, Kandel ES, Cross TK, Hay N. Akt/Protein kinase B inhibits cell death by preventing the release of cytochrome c from mitochondria. Mol Cell Biol. 1999;19:5800–5810. doi: 10.1128/mcb.19.8.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, Li XM, Meinkoth J, Pittman RN. Akt regulates cell survival and apoptosis at a postmitochondrial level. J Cell Biol. 2000;151:483–494. doi: 10.1083/jcb.151.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitiello PF, Wu YC, Staversky RJ, O'Reilly MA. p21Cip1 protects against oxidative stress by suppressing ER-dependent activation of mitochondrial death pathways. Free Radic Biol Med. 2009;46:33–41. doi: 10.1016/j.freeradbiomed.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang MA, So EY, Simons AL, Spitz DR, Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3:e249. doi: 10.1038/cddis.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.