Abstract
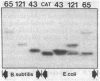
The Gram-negative product-encoding Tn9-derived chloramphenicol-resistance (Cmr) gene can be cloned but not phenotypically expressed in Bacillus subtilis. We show that, even when transcribed from B. subtilis promoters, the ribosomal binding site for the Cmr gene does not function well in B. subtilis. The Cmr gene product, chloramphenicol acetyltransferase (CmAcTase; acetyl-CoA:chloramphenicol 3-O-acetyltransferase, EC 2.3.1.28), is detected in B. subtilis when the promoters, ribosomal binding sites, and initiation codons of B. subtilis genes are fused to the Cmr gene. These gene fusions lead to the in vivo production of mRNAs containing B. subtilis translation start signals followed in an open reading frame by the translation start site normally used by Escherichia coli to initiate translation of Cmr mRNA. Both fusion and native CmAcTase proteins are produced in E. coli, but only fusion CmAcTase is produced in B. subtilis. We conclude that the absence of native CmAcTase in B. subtilis is due to inability of the E. coli ribosomal binding site to function well in B. subtilis. Since fusion CmAcTase polypeptides are produced in E. coli, we conclude that these particular B. subtilis regulatory elements function heterologously in E. coli. The absence of a suitable binding site on the Cmr gene for B. subtilis ribosomes is consistent with reports that many E. coli genes are not expressed in B. subtilis and that E. coli mRNA functions poorly in B. subtilis in vitro translation systems. The functioning of B. subtilis regulatory sequences in E. coli is consistent with in vivo and in vitro data showing the expression of B. subtilis genes in E. coli. To confirm the hypothesis that the large CmAcTase proteins are NH2-terminal fusions of native CmAcTase we partially determined the sequence of one CmAcTase fusion protein.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Genome construction between bacterial species in vitro: replication and expression of Staphylococcus plasmid genes in Escherichia coli. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1030–1034. doi: 10.1073/pnas.71.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N. Y., Ehrlich S. D., Lederberg J. Functional expression of two Bacillus subtilis chromosomal genes in Escherichia coli. J Bacteriol. 1978 Feb;133(2):816–821. doi: 10.1128/jb.133.2.816-821.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann J. L., Dahmus M. E. Monoclonal antibody specific for calf thymus RNA polymerases IIO and IIA. J Biol Chem. 1981 Nov 25;256(22):11798–11803. [PubMed] [Google Scholar]
- Davison B. L., Murray C. L., Rabinowitz J. C. Specificity of promoter site utilization in vitro by bacterial RNA polymerases on Bacillus phage phi 29 DNA. Transcription mapping with exonuclease III. J Biol Chem. 1980 Sep 25;255(18):8819–8830. [PubMed] [Google Scholar]
- Doi R. H. Multiple RNA polymerase holoenzymes exert transcriptional specificity in Bacillus subtilis. Arch Biochem Biophys. 1982 Apr 1;214(2):772–781. doi: 10.1016/0003-9861(82)90084-4. [DOI] [PubMed] [Google Scholar]
- Ehrlich S. D., Bursztyn-Pettegrew H., Stroynowski I., Lederberg J. Expression of the thymidylate synthetase gene of the Bacillus subtilis bacteriophage Phi-3-T in Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4145–4149. doi: 10.1073/pnas.73.11.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraker P. J., Speck J. C., Jr Protein and cell membrane iodinations with a sparingly soluble chloroamide, 1,3,4,6-tetrachloro-3a,6a-diphrenylglycoluril. Biochem Biophys Res Commun. 1978 Feb 28;80(4):849–857. doi: 10.1016/0006-291x(78)91322-0. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Goldfarb D. S., Doi R. H., Rodriguez R. L. Expression of Tn9-derived chloramphenicol resistance in Bacillus subtilis. Nature. 1981 Sep 24;293(5830):309–311. doi: 10.1038/293309a0. [DOI] [PubMed] [Google Scholar]
- Gray O., Chang S. Molecular cloning and expression of Bacillus licheniformis beta-lactamase gene in Escherichia coli and Bacillus subtilis. J Bacteriol. 1981 Jan;145(1):422–428. doi: 10.1128/jb.145.1.422-428.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura K., Hirose T., Crea R., Riggs A. D., Heyneker H. L., Bolivar F., Boyer H. W. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977 Dec 9;198(4321):1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- Kreft J., Bernhard K., Goebel W. Recombinant plasmids capable to replication in B. subtilis and E. coli. Mol Gen Genet. 1978 Jun 1;162(1):59–67. doi: 10.1007/BF00333851. [DOI] [PubMed] [Google Scholar]
- Kulbe K. D. Micropolyamide thin-layer chromatography of phenylthiohydantoin amino acids (PTH) at subnanomolar level. A rapid microtechnique for simultaneous multisample identification after automated Edman degradations. Anal Biochem. 1974 Jun;59(2):564–573. doi: 10.1016/0003-2697(74)90310-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Legualt-Démare L., Chambliss G. H. Selective messenger translation by Bacillus subtilis ribosomes. Mol Gen Genet. 1976 Dec 31;142(4):277–287. doi: 10.1007/BF00271252. [DOI] [PubMed] [Google Scholar]
- Leventhal J. M., Chambliss G. H. DNA-directed cell-free protein-synthesizing system of Bacillus subtilis. Biochim Biophys Acta. 1979 Aug 29;564(1):162–171. doi: 10.1016/0005-2787(79)90197-7. [DOI] [PubMed] [Google Scholar]
- Mahler I., Halvorson H. O. Transformation of Escherichia coli and Bacillus subtilis with a hybrid plasmid molecule. J Bacteriol. 1977 Jul;131(1):374–377. doi: 10.1128/jb.131.1.374-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Initiation factor-independent translation of mRNAs from Gram-positive bacteria. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4912–4916. doi: 10.1073/pnas.78.8.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Nagahari K., Sakaguchi K. Cloning of Bacillus subtilis leucina A, B and C genes with Escherichia coli plasmids and expression of the leuC gene in E. coli. Mol Gen Genet. 1978 Jan 17;158(3):263–270. doi: 10.1007/BF00267197. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Munoz L., Doi R. H. A procedure to remove protease activities from Bacillus subtilis sporulating cells and their crude extracts. Anal Biochem. 1977 Mar;78(1):165–170. doi: 10.1016/0003-2697(77)90020-3. [DOI] [PubMed] [Google Scholar]
- Neve R. L., West R. W., Rodriguez R. L. Eukaryotic DNA fragments which act as promoters for a plasmid gene. Nature. 1979 Jan 25;277(5694):324–325. doi: 10.1038/277324a0. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Shaw W. V. Identification of "buried" lysine residues in two variants of chloramphenicol acetyltransferase specified by R-factors. Biochem J. 1981 Feb 1;193(2):525–539. doi: 10.1042/bj1930525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano J. J., Bronzert T. J. Analysis of amino acid phenylthiohydantoins by gas chromatography. J Biol Chem. 1969 Oct 25;244(20):5597–5607. [PubMed] [Google Scholar]
- Rubin E. M., Wilson G. A., Young F. E. Expression of thymidylate synthetase activity in Bacillus subtilis upon integration of a cloned gene from Escherichia coli. Gene. 1980 Aug;10(3):227–235. doi: 10.1016/0378-1119(80)90052-9. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Packman L. C., Burleigh B. D., Dell A., Morris H. R., Hartley B. S. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979 Dec 20;282(5741):870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- Stallcup M. R., Sharrock W. J., Rabinowitz J. C. Specificity of bacterial ribosomes and messenger ribonucleic acids in protein synthesis reactions in vitro. J Biol Chem. 1976 Apr 25;251(8):2499–2510. [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wiggs J. L., Bush J. W., Chamberlin M. J. Utilization of promoter and terminator sites on bacteriophage T7 DNA by RNA polymerases from a variety of bacterial orders. Cell. 1979 Jan;16(1):97–109. doi: 10.1016/0092-8674(79)90191-0. [DOI] [PubMed] [Google Scholar]
- Wiggs J. L., Gilman M. Z., Chamberlin M. J. Heterogeneity of RNA polymerase in Bacillus subtilis: evidence for an additional sigma factor in vegetative cells. Proc Natl Acad Sci U S A. 1981 May;78(5):2762–2766. doi: 10.1073/pnas.78.5.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Schoner R. G., Duvall E. J., Preis L. H., Lovett P. S. Expression of Escherichia coli trp genes and the mouse dihydrofolate reductase gene cloned in Bacillus subtilis. Gene. 1981 Dec;16(1-3):199–206. doi: 10.1016/0378-1119(81)90076-7. [DOI] [PubMed] [Google Scholar]
- Zaidenzaig Y., Shaw W. V. Affinity and hydrophobic chromatography of three variants of chloramphenicol acetyltransferases specified by R factors in Escherichia coli. FEBS Lett. 1976 Mar 1;62(3):266–271. doi: 10.1016/0014-5793(76)80072-5. [DOI] [PubMed] [Google Scholar]