Abstract
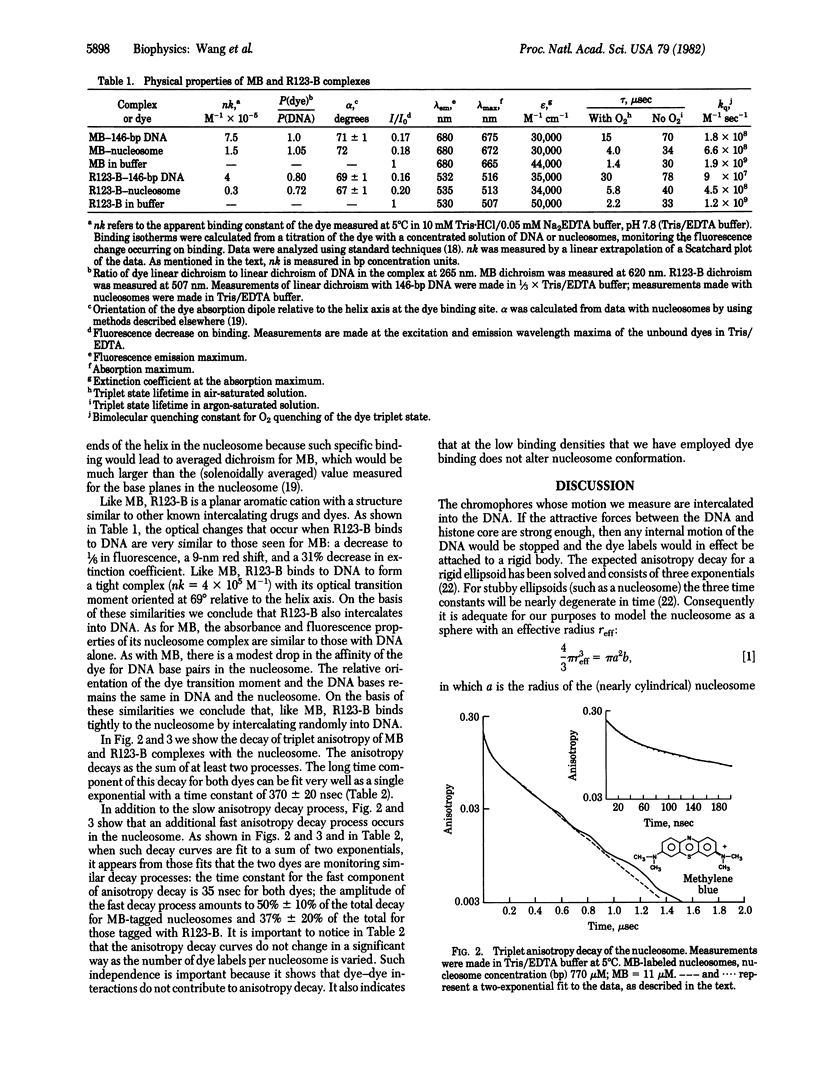
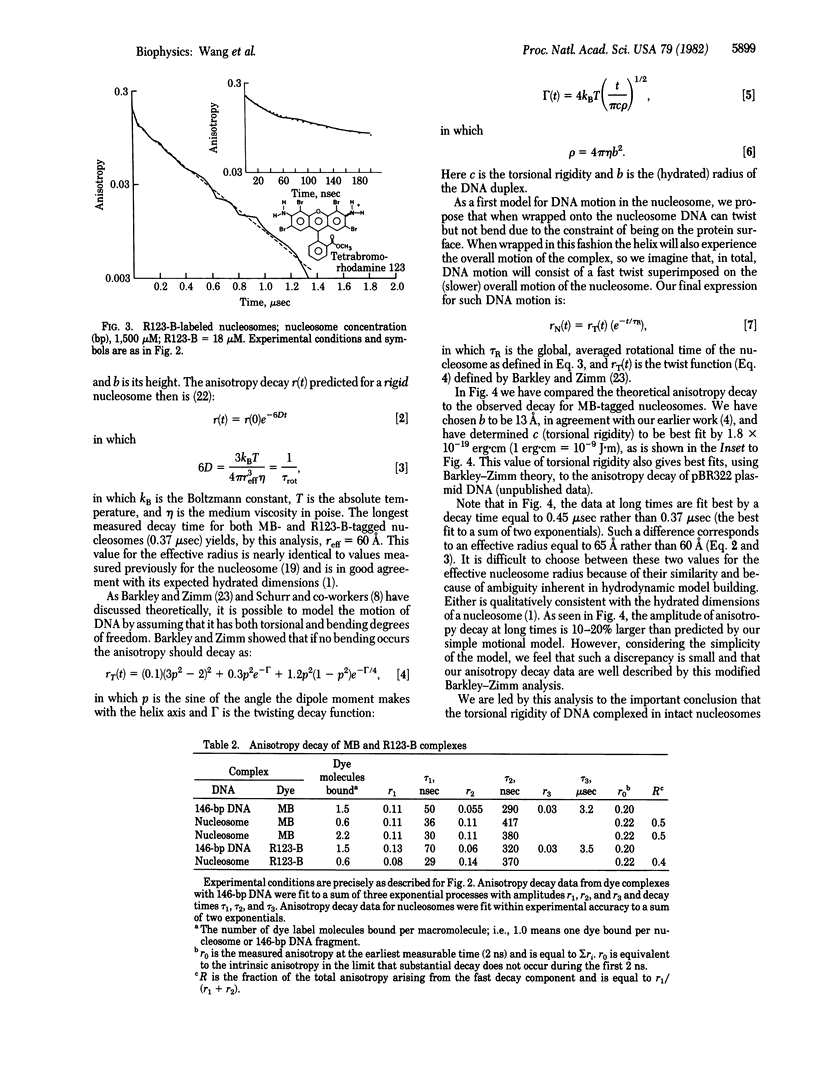
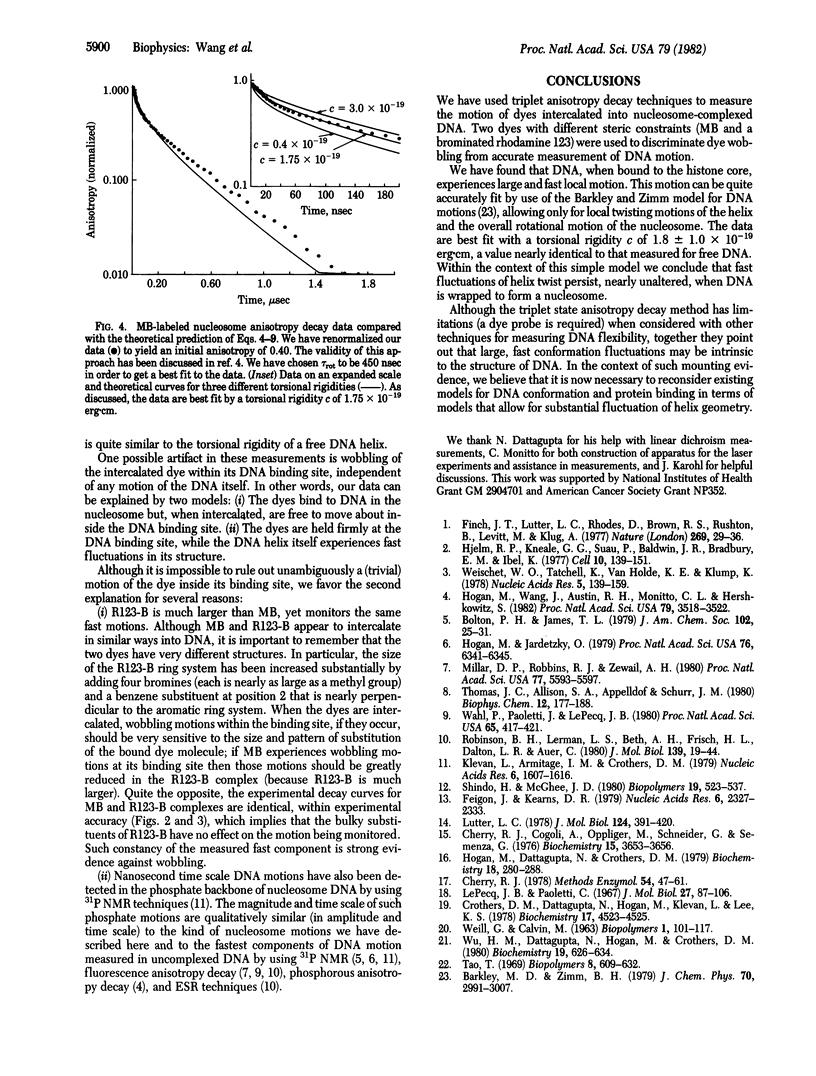
We have used time-resolved triplet state anisotropy decay techniques to measure the conformational flexibility of DNA in the nucleosome. From these measurements we conclude that, in a nucleosome, the DNA helix experiences substantial internal flexibility, which occurs with a time constant near 30 nsec. We find that our data can be fit well by a modified version of the Barkley-Zimm model for DNA motion, allowing only DNA twisting motions and the overall tumbling of the nucleosome. That fit yields a calculated torsional rigidity equal to 1.8 X 10(-19) erg X cm, a value equal to that measured for uncomplexed DNA. We conclude from such similarity that large, fast twisting motions of the DNA helix persist, nearly unaltered, when DNA is wrapped to form a nucleosome.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cherry R. J., Cogoli A., Oppliger M., Schneider G., Semenza G. A spectroscopic technique for measuring slow rotational diffusion of macromolecules. 1: Preparation and properties of a triplet probe. Biochemistry. 1976 Aug 24;15(17):3653–3656. doi: 10.1021/bi00662a001. [DOI] [PubMed] [Google Scholar]
- Cherry R. J. Measurement of protein rotational diffusion in membranes by flash photolysis. Methods Enzymol. 1978;54:47–61. doi: 10.1016/s0076-6879(78)54007-x. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Dattagupta N., Hogan M., Klevan L., Lee K. S. Transient electric dichroism studies of nucleosomal particles. Biochemistry. 1978 Oct 17;17(21):4525–4533. doi: 10.1021/bi00614a026. [DOI] [PubMed] [Google Scholar]
- Feigon J., Kearns D. R. 1H NMR investigation of the conformational states of DNA in nucleosome core particles. Nucleic Acids Res. 1979;6(6):2327–2337. doi: 10.1093/nar/6.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Hjelm R. P., Kneale G. G., Sauau P., Baldwin J. P., Bradbury E. M., Ibel K. Small angle neutron scattering studies of chromatin subunits in solution. Cell. 1977 Jan;10(1):139–151. doi: 10.1016/0092-8674(77)90148-9. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Jardetzky O. Internal motions in DNA. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6341–6345. doi: 10.1073/pnas.76.12.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transient electric dichroism studies of the structure of the DNA complex with intercalated drugs. Biochemistry. 1979 Jan 23;18(2):280–288. doi: 10.1021/bi00569a007. [DOI] [PubMed] [Google Scholar]
- Hogan M., Wang J., Austin R. H., Monitto C. L., Hershkowitz S. Molecular motion of DNA as measured by triplet anisotropy decay. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3518–3522. doi: 10.1073/pnas.79.11.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevan L., Armitage I. M., Crothers D. M. 31P NMR studies of the solution structure and dynamics of nucleosomes and DNA. Nucleic Acids Res. 1979 Apr;6(4):1607–1616. doi: 10.1093/nar/6.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Millar D. P., Robbins R. J., Zewail A. H. Direct observation of the torsional dynamics of DNA and RNA by picosecond spectroscopy. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5593–5597. doi: 10.1073/pnas.77.10.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. H., Lerman L. S., Beth A. H., Frisch H. L., Dalton L. R., Auer C. Analysis of double-helix motions with spin-labeled probes: binding geometry and the limit of torsional elasticity. J Mol Biol. 1980 May 5;139(1):19–44. doi: 10.1016/0022-2836(80)90113-8. [DOI] [PubMed] [Google Scholar]
- Shindo H., McGhee J. D., Cohen J. S. 31P-NMR studies of DNA in nucleosome core particles. Biopolymers. 1980 Mar;19(3):523–537. doi: 10.1002/bip.1980.360190307. [DOI] [PubMed] [Google Scholar]
- Thomas J. C., Allison S. A., Appellof C. J., Schurr J. M. Torison dynamics and depolarization of fluorescence of linear macromolecules. II. Fluorescence polarization anisotropy measurements on a clean viral phi 29 DNA. Biophys Chem. 1980 Oct;12(2):177–188. doi: 10.1016/0301-4622(80)80050-0. [DOI] [PubMed] [Google Scholar]
- Wahl P., Paoletti J., Le Pecq J. B. Decay of fluorescence emission anisotropy of the ethidium bromide-DNA complex. Evidence for an internal motion in DNA. Proc Natl Acad Sci U S A. 1970 Feb;65(2):417–421. doi: 10.1073/pnas.65.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Dattagupta N., Hogan M., Crothers D. M. Unfolding of nucleosomes by ethidium binding. Biochemistry. 1980 Feb 19;19(4):626–634. doi: 10.1021/bi00545a004. [DOI] [PubMed] [Google Scholar]