Abstract
In the title complex, {[Zn(NCS)2(C15H15N9)2]·H2O}n, the ZnII ion is located on an inversion centre and is six-coordinated in a distorted octahedral geometry, coordinated by N atoms from four bridging 1,3,5-tris(1,2,4-triazol-1-ylmethyl)benzene (ttmb) ligands and two terminal SCN− counter-anions. Two of the three triazol groups in each ttmb ligand link the ZnII atoms, forming a looped-chain structure along [0-11]. The lattice water molecule shows half-occupancy due to disorder around an inversion centre.
Related literature
For background to the use of flexible tripodal compounds in the design and construction of compounds with metal-organic framework structures, see: Moon et al. (2006 ▶); Xu et al. (2009 ▶). For similar structures, see: Yin et al. (2009 ▶); Shi et al. (2011 ▶).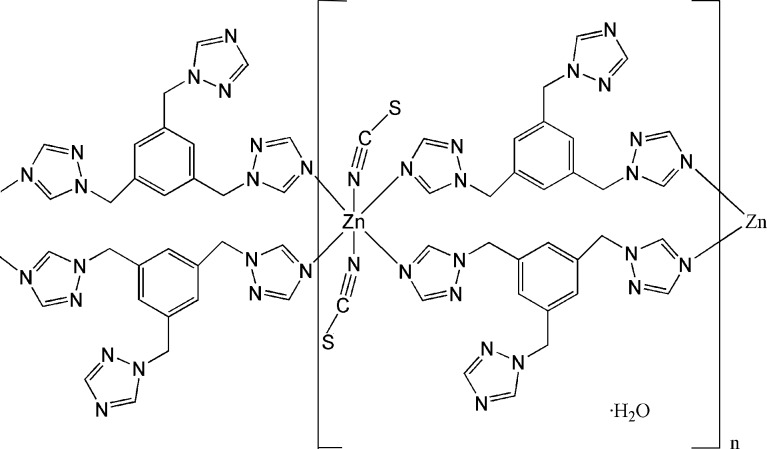
Experimental
Crystal data
[Zn(NCS)2(C15H15N9)2]·H2O
M r = 842.27
Triclinic,

a = 8.5766 (17) Å
b = 9.5036 (19) Å
c = 11.723 (2) Å
α = 80.01 (3)°
β = 85.40 (3)°
γ = 89.55 (3)°
V = 938.0 (3) Å3
Z = 1
Mo Kα radiation
μ = 0.83 mm−1
T = 293 K
0.20 × 0.18 × 0.16 mm
Data collection
Rigaku Saturn724 diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2006 ▶) T min = 0.852, T max = 0.879
11566 measured reflections
4444 independent reflections
3972 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.108
S = 1.05
4444 reflections
265 parameters
9 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.61 e Å−3
Δρmin = −0.39 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2006 ▶); cell refinement: CrystalClear (Rigaku/MSC, 2006 ▶); data reduction: CrystalClear (Rigaku/MSC, 2006 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: CrystalStructure (Rigaku/MSC, 2006 ▶); software used to prepare material for publication: CrystalStructure (Rigaku/MSC, 2006 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812039256/ds2211sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039256/ds2211Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported financially by the National Natural Science Foundation (No. 20971110) and the program for the construction of Puyang Key Laboratory.
supplementary crystallographic information
Comment
Flexible tripodal compounds are known to be the versatile structural constructors in the rational design and construction of novel metal-organic frameworks (MOFs), in respect that its three potential coordination groups can bend and rotate freely to satisfy various coordination preferences and facilitate the formation of various complexes with diverse structures and properties (Moon et al., 2006; Xu et al., 2009). Therefore, the prospect of exploring the influential principles of tripodal compounds on the resulting framework structures provides an impetus for further researches on tripodal compounds. We were thus engaged in the synthesis of a flexible tripodal N-heterocyclic compound 1,3,5-tris(1,2,4-triazol-1-ylmethyl)-benzene (ttmb) (Yin et al., 2009; Shi et al., 2011), and employed it as a ligand to construct a new complex {[Zn(SCN)2(ttmb)2].H2O}n. In the title complex, ZnII ion is six-coordinated in a distorted octahedral geometry, coordinated by N2, N2A, N8 and N8A from four ttmb ligands, and N1, N1A from two terminal counter-anion SCN- (Fig. 1). In ttmb, the center of one triazol ring lies inside the benzene plane with the dihedral angle of 89.4 °, and the other two triazol rings lie in the opposite orientation outside the plane to form an infrequent trans conformation. Two of the three triazol groups in each ttmb link the ZnII centers together to form an one-dimensional looped-chain structure (Fig. 2), in which the ZnII ions are collinear with the adjacent Zn···Zn distance of 13.8 Å.
Experimental
A reaction mixture of ZnSO4.7H2O (29 mg, 0.1 mmol), 1,3,5-tris(1,2,4-triazol-1-ylmethyl)-benzene (ttmb) (32.1 mg, 0.1 mmol), KSCN (19.4 mg, 0.2 mmol), and 10 ml water was sealed in a Teflon-lined stainless steel vessel, which was heated at 130 °C for 72 h, and then cooled to room temperature, obtaining colorless crystals of the title complex. Yield (based on Zn): 31%.
Refinement
H atoms were generated geometrically and refined as riding atoms with C-H = 0.93 Å, 0.97 (CH2) Å and Uiso(H) = 1.2 times Ueq(C).
Figures
Fig. 1.
A fragment of the title complex, showing the coordination environment of ZnII center with atom labelling of the non-H atoms and 30% probability ellipsoids. H atoms have been omitted.
Fig. 2.
View of the one-dimensional looped-chain structure of the title complex.
Crystal data
| [Zn(NCS)2(C15H15N9)2]·H2O | Z = 1 |
| Mr = 842.27 | F(000) = 434 |
| Triclinic, P1 | Dx = 1.491 Mg m−3 |
| a = 8.5766 (17) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.5036 (19) Å | Cell parameters from 2773 reflections |
| c = 11.723 (2) Å | θ = 2.2–27.9° |
| α = 80.01 (3)° | µ = 0.83 mm−1 |
| β = 85.40 (3)° | T = 293 K |
| γ = 89.55 (3)° | Prism, colorless |
| V = 938.0 (3) Å3 | 0.20 × 0.18 × 0.16 mm |
Data collection
| Rigaku Saturn724 diffractometer | 4444 independent reflections |
| Radiation source: fine-focus sealed tube | 3972 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.024 |
| Detector resolution: 28.5714 pixels mm-1 | θmax = 27.9°, θmin = 2.2° |
| ω and φ scans | h = −11→11 |
| Absorption correction: multi-scan ? | k = −12→12 |
| Tmin = 0.852, Tmax = 0.879 | l = −15→15 |
| 11566 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.108 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0517P)2 + 0.3475P] where P = (Fo2 + 2Fc2)/3 |
| 4444 reflections | (Δ/σ)max < 0.001 |
| 265 parameters | Δρmax = 0.61 e Å−3 |
| 9 restraints | Δρmin = −0.39 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Zn1 | 0.0000 | 1.0000 | 0.0000 | 0.03753 (14) | |
| N1 | −0.1335 (3) | 1.0823 (3) | 0.1368 (2) | 0.0516 (6) | |
| N2 | 0.1925 (2) | 0.9400 (2) | 0.11113 (18) | 0.0398 (5) | |
| N3 | 0.3170 (2) | 0.8611 (2) | 0.26349 (17) | 0.0374 (4) | |
| N4 | 0.4133 (3) | 0.8369 (3) | 0.1727 (2) | 0.0561 (7) | |
| N5 | 0.2223 (3) | 0.2854 (2) | 0.3054 (2) | 0.0431 (5) | |
| N8 | 0.0899 (2) | 1.2105 (2) | −0.06854 (18) | 0.0397 (5) | |
| N9 | 0.0847 (3) | 1.4448 (3) | −0.1391 (2) | 0.0579 (7) | |
| N10 | 0.2207 (3) | 1.3860 (2) | −0.17586 (18) | 0.0395 (5) | |
| O1 | 0.5990 (11) | 0.4298 (9) | 0.9817 (9) | 0.135 (3) | 0.50 |
| C2 | 0.1872 (3) | 0.9224 (3) | 0.2261 (2) | 0.0404 (5) | |
| H2 | 0.1043 | 0.9494 | 0.2736 | 0.049* | |
| C3 | 0.3328 (3) | 0.8857 (4) | 0.0833 (2) | 0.0527 (7) | |
| H3 | 0.3701 | 0.8829 | 0.0070 | 0.063* | |
| C4 | 0.3613 (4) | 0.8227 (3) | 0.3821 (2) | 0.0444 (6) | |
| H4B | 0.3159 | 0.8913 | 0.4273 | 0.053* | |
| H4A | 0.4742 | 0.8302 | 0.3812 | 0.053* | |
| C5 | 0.3110 (3) | 0.6738 (2) | 0.4421 (2) | 0.0343 (5) | |
| C6 | 0.3413 (3) | 0.6377 (2) | 0.5581 (2) | 0.0350 (5) | |
| H6 | 0.3910 | 0.7036 | 0.5933 | 0.042* | |
| C7 | 0.2990 (3) | 0.5056 (3) | 0.6222 (2) | 0.0357 (5) | |
| C8 | 0.2262 (3) | 0.4067 (3) | 0.5681 (2) | 0.0396 (5) | |
| H8 | 0.1982 | 0.3171 | 0.6103 | 0.048* | |
| C9 | 0.1954 (3) | 0.4408 (3) | 0.4526 (2) | 0.0385 (5) | |
| C10 | 0.2379 (3) | 0.5756 (3) | 0.3897 (2) | 0.0381 (5) | |
| H10 | 0.2168 | 0.5993 | 0.3121 | 0.046* | |
| C11 | 0.1173 (3) | 0.3323 (3) | 0.3945 (3) | 0.0466 (6) | |
| H11B | 0.0839 | 0.2503 | 0.4527 | 0.056* | |
| H11A | 0.0251 | 0.3744 | 0.3599 | 0.056* | |
| C14 | 0.3404 (3) | 0.4707 (3) | 0.7471 (2) | 0.0458 (6) | |
| H14A | 0.4382 | 0.4187 | 0.7498 | 0.055* | |
| H14B | 0.3564 | 0.5592 | 0.7753 | 0.055* | |
| C15 | 0.0108 (4) | 1.3348 (3) | −0.0744 (3) | 0.0528 (7) | |
| H15 | −0.0874 | 1.3420 | −0.0360 | 0.063* | |
| C16 | 0.2207 (3) | 1.2478 (3) | −0.1330 (2) | 0.0412 (6) | |
| H16 | 0.3017 | 1.1857 | −0.1465 | 0.049* | |
| C1 | −0.1657 (3) | 1.0544 (3) | 0.2354 (2) | 0.0422 (6) | |
| N6 | 0.3491 (3) | 0.2042 (3) | 0.3333 (2) | 0.0560 (6) | |
| N7 | 0.3519 (4) | 0.2694 (3) | 0.1395 (3) | 0.0728 (8) | |
| C12 | 0.4207 (4) | 0.1989 (4) | 0.2304 (3) | 0.0599 (8) | |
| H12 | 0.5134 | 0.1490 | 0.2217 | 0.072* | |
| C13 | 0.2266 (5) | 0.3210 (4) | 0.1909 (3) | 0.0661 (9) | |
| H13 | 0.1508 | 0.3756 | 0.1514 | 0.079* | |
| H1A | 0.517 (7) | 0.391 (8) | 1.021 (7) | 0.099* | 0.50 |
| H1B | 0.588 (10) | 0.5203 (18) | 0.969 (8) | 0.099* | 0.50 |
| S1 | −0.21123 (13) | 1.00586 (11) | 0.37384 (7) | 0.0710 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Zn1 | 0.0382 (2) | 0.0368 (2) | 0.0329 (2) | 0.00232 (16) | −0.00183 (16) | 0.00624 (15) |
| N1 | 0.0540 (14) | 0.0583 (15) | 0.0387 (12) | 0.0090 (11) | 0.0033 (11) | −0.0010 (11) |
| N2 | 0.0371 (11) | 0.0417 (11) | 0.0377 (11) | 0.0018 (9) | −0.0051 (9) | 0.0024 (9) |
| N3 | 0.0422 (11) | 0.0363 (10) | 0.0313 (10) | 0.0001 (8) | −0.0048 (8) | 0.0018 (8) |
| N4 | 0.0436 (13) | 0.0825 (19) | 0.0403 (12) | 0.0183 (12) | −0.0039 (10) | −0.0054 (12) |
| N5 | 0.0470 (12) | 0.0392 (11) | 0.0464 (12) | 0.0006 (9) | −0.0147 (10) | −0.0112 (9) |
| N8 | 0.0418 (11) | 0.0370 (11) | 0.0364 (10) | 0.0007 (9) | −0.0020 (9) | 0.0040 (8) |
| N9 | 0.0670 (16) | 0.0378 (12) | 0.0624 (16) | 0.0086 (11) | 0.0138 (13) | 0.0002 (11) |
| N10 | 0.0450 (12) | 0.0347 (10) | 0.0358 (10) | −0.0016 (9) | −0.0034 (9) | 0.0023 (8) |
| O1 | 0.147 (7) | 0.097 (5) | 0.142 (7) | −0.010 (5) | 0.029 (6) | 0.012 (5) |
| C2 | 0.0390 (13) | 0.0412 (13) | 0.0391 (13) | 0.0037 (10) | −0.0015 (10) | −0.0019 (10) |
| C3 | 0.0417 (15) | 0.079 (2) | 0.0343 (13) | 0.0104 (14) | −0.0010 (11) | −0.0011 (13) |
| C4 | 0.0587 (16) | 0.0396 (13) | 0.0333 (12) | −0.0075 (12) | −0.0109 (11) | 0.0017 (10) |
| C5 | 0.0352 (12) | 0.0323 (11) | 0.0340 (11) | 0.0014 (9) | −0.0028 (9) | −0.0019 (9) |
| C6 | 0.0365 (12) | 0.0337 (11) | 0.0345 (12) | −0.0018 (9) | −0.0060 (9) | −0.0034 (9) |
| C7 | 0.0360 (12) | 0.0356 (12) | 0.0336 (11) | −0.0013 (9) | −0.0033 (9) | −0.0004 (9) |
| C8 | 0.0412 (13) | 0.0328 (12) | 0.0425 (13) | −0.0024 (10) | −0.0027 (11) | −0.0003 (10) |
| C9 | 0.0358 (12) | 0.0368 (12) | 0.0439 (13) | −0.0017 (10) | −0.0028 (10) | −0.0098 (10) |
| C10 | 0.0435 (13) | 0.0380 (12) | 0.0330 (12) | 0.0014 (10) | −0.0068 (10) | −0.0055 (10) |
| C11 | 0.0446 (15) | 0.0441 (14) | 0.0535 (16) | −0.0052 (11) | −0.0081 (12) | −0.0131 (12) |
| C14 | 0.0505 (15) | 0.0461 (14) | 0.0368 (13) | −0.0133 (12) | −0.0084 (11) | 0.0072 (11) |
| C15 | 0.0564 (17) | 0.0432 (15) | 0.0531 (16) | 0.0054 (12) | 0.0138 (13) | −0.0011 (12) |
| C16 | 0.0390 (13) | 0.0381 (13) | 0.0426 (13) | 0.0035 (10) | −0.0054 (11) | 0.0050 (10) |
| C1 | 0.0423 (14) | 0.0388 (13) | 0.0457 (14) | 0.0095 (10) | −0.0019 (11) | −0.0090 (11) |
| N6 | 0.0552 (15) | 0.0525 (14) | 0.0631 (16) | 0.0080 (11) | −0.0175 (13) | −0.0127 (12) |
| N7 | 0.089 (2) | 0.074 (2) | 0.0569 (17) | 0.0002 (17) | 0.0022 (16) | −0.0172 (15) |
| C12 | 0.0532 (18) | 0.0542 (18) | 0.078 (2) | −0.0016 (14) | −0.0063 (16) | −0.0266 (16) |
| C13 | 0.086 (2) | 0.063 (2) | 0.0518 (18) | 0.0154 (18) | −0.0215 (17) | −0.0102 (15) |
| S1 | 0.0963 (6) | 0.0730 (5) | 0.0395 (4) | 0.0230 (5) | 0.0105 (4) | −0.0065 (4) |
Geometric parameters (Å, º)
| Zn1—N8 | 2.146 (2) | C4—H4B | 0.9700 |
| Zn1—N8i | 2.146 (2) | C4—H4A | 0.9700 |
| Zn1—N1 | 2.149 (2) | C5—C10 | 1.383 (3) |
| Zn1—N1i | 2.149 (2) | C5—C6 | 1.390 (3) |
| Zn1—N2i | 2.196 (2) | C6—C7 | 1.382 (3) |
| Zn1—N2 | 2.196 (2) | C6—H6 | 0.9300 |
| N1—C1 | 1.152 (3) | C7—C8 | 1.398 (3) |
| N2—C2 | 1.327 (3) | C7—C14 | 1.515 (3) |
| N2—C3 | 1.344 (3) | C8—C9 | 1.383 (4) |
| N3—C2 | 1.322 (3) | C8—H8 | 0.9300 |
| N3—N4 | 1.346 (3) | C9—C10 | 1.398 (3) |
| N3—C4 | 1.455 (3) | C9—C11 | 1.518 (3) |
| N4—C3 | 1.316 (4) | C10—H10 | 0.9300 |
| N5—C13 | 1.324 (4) | C11—H11B | 0.9700 |
| N5—N6 | 1.357 (3) | C11—H11A | 0.9700 |
| N5—C11 | 1.452 (4) | C14—N10iii | 1.459 (3) |
| N8—C16 | 1.316 (3) | C14—H14A | 0.9700 |
| N8—C15 | 1.353 (4) | C14—H14B | 0.9700 |
| N9—C15 | 1.314 (4) | C15—H15 | 0.9300 |
| N9—N10 | 1.361 (3) | C16—H16 | 0.9300 |
| N10—C16 | 1.322 (3) | C1—S1 | 1.625 (3) |
| N10—C14ii | 1.459 (3) | N6—C12 | 1.317 (4) |
| O1—H1A | 0.853 (13) | N7—C13 | 1.322 (5) |
| O1—H1B | 0.853 (13) | N7—C12 | 1.334 (5) |
| C2—H2 | 0.9300 | C12—H12 | 0.9300 |
| C3—H3 | 0.9300 | C13—H13 | 0.9300 |
| C4—C5 | 1.516 (3) | ||
| N8—Zn1—N8i | 180.0 | C10—C5—C6 | 119.5 (2) |
| N8—Zn1—N1 | 89.99 (9) | C10—C5—C4 | 124.7 (2) |
| N8i—Zn1—N1 | 90.01 (9) | C6—C5—C4 | 115.9 (2) |
| N8—Zn1—N1i | 90.01 (9) | C7—C6—C5 | 121.1 (2) |
| N8i—Zn1—N1i | 89.99 (9) | C7—C6—H6 | 119.5 |
| N1—Zn1—N1i | 180.0 | C5—C6—H6 | 119.5 |
| N8—Zn1—N2i | 85.29 (8) | C6—C7—C8 | 119.0 (2) |
| N8i—Zn1—N2i | 94.71 (8) | C6—C7—C14 | 118.5 (2) |
| N1—Zn1—N2i | 88.53 (9) | C8—C7—C14 | 122.4 (2) |
| N1i—Zn1—N2i | 91.47 (9) | C9—C8—C7 | 120.6 (2) |
| N8—Zn1—N2 | 94.71 (8) | C9—C8—H8 | 119.7 |
| N8i—Zn1—N2 | 85.29 (8) | C7—C8—H8 | 119.7 |
| N1—Zn1—N2 | 91.47 (9) | C8—C9—C10 | 119.4 (2) |
| N1i—Zn1—N2 | 88.53 (9) | C8—C9—C11 | 120.2 (2) |
| N2i—Zn1—N2 | 180.00 (11) | C10—C9—C11 | 120.4 (2) |
| C1—N1—Zn1 | 140.3 (2) | C5—C10—C9 | 120.4 (2) |
| C2—N2—C3 | 102.6 (2) | C5—C10—H10 | 119.8 |
| C2—N2—Zn1 | 127.61 (18) | C9—C10—H10 | 119.8 |
| C3—N2—Zn1 | 128.64 (18) | N5—C11—C9 | 111.5 (2) |
| C2—N3—N4 | 109.9 (2) | N5—C11—H11B | 109.3 |
| C2—N3—C4 | 128.9 (2) | C9—C11—H11B | 109.3 |
| N4—N3—C4 | 121.2 (2) | N5—C11—H11A | 109.3 |
| C3—N4—N3 | 102.6 (2) | C9—C11—H11A | 109.3 |
| C13—N5—N6 | 108.7 (3) | H11B—C11—H11A | 108.0 |
| C13—N5—C11 | 129.8 (3) | N10iii—C14—C7 | 113.3 (2) |
| N6—N5—C11 | 121.1 (2) | N10iii—C14—H14A | 108.9 |
| C16—N8—C15 | 103.1 (2) | C7—C14—H14A | 108.9 |
| C16—N8—Zn1 | 128.82 (18) | N10iii—C14—H14B | 108.9 |
| C15—N8—Zn1 | 126.93 (19) | C7—C14—H14B | 108.9 |
| C15—N9—N10 | 102.7 (2) | H14A—C14—H14B | 107.7 |
| C16—N10—N9 | 109.5 (2) | N9—C15—N8 | 114.2 (3) |
| C16—N10—C14ii | 128.6 (2) | N9—C15—H15 | 122.9 |
| N9—N10—C14ii | 121.8 (2) | N8—C15—H15 | 122.9 |
| H1A—O1—H1B | 109 (3) | N8—C16—N10 | 110.5 (2) |
| N2—C2—N3 | 110.2 (2) | N8—C16—H16 | 124.8 |
| N2—C2—H2 | 124.9 | N10—C16—H16 | 124.8 |
| N3—C2—H2 | 124.9 | N1—C1—S1 | 176.9 (3) |
| N4—C3—N2 | 114.6 (2) | C12—N6—N5 | 102.2 (3) |
| N4—C3—H3 | 122.7 | C13—N7—C12 | 101.6 (3) |
| N2—C3—H3 | 122.7 | N6—C12—N7 | 115.9 (3) |
| N3—C4—C5 | 114.5 (2) | N6—C12—H12 | 122.1 |
| N3—C4—H4B | 108.6 | N7—C12—H12 | 122.1 |
| C5—C4—H4B | 108.6 | N5—C13—N7 | 111.6 (3) |
| N3—C4—H4A | 108.6 | N5—C13—H13 | 124.2 |
| C5—C4—H4A | 108.6 | N7—C13—H13 | 124.2 |
| H4B—C4—H4A | 107.6 |
Symmetry codes: (i) −x, −y+2, −z; (ii) x, y+1, z−1; (iii) x, y−1, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DS2211).
References
- Moon, D., Kang, S., Park, J., Lee, K., John, R. P., Won, H., Seong, G. H., Kim, Y. S., Kim, G. H., Rhee, H. & Lah, M. S. (2006). J. Am. Chem. Soc. 128, 3530–3531. [DOI] [PubMed]
- Rigaku/MSC (2006). CrystalClear and CrystalStructure Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, X.-J., Zhang, X.-H., Li, X.-X., Hou, H.-W. & Fan, Y.-T. (2011). J. Mol. Struct. 996, 110–114.
- Xu, G.-C., Ding, Y.-J., Okamura, T., Huang, Y.-Q., Bai, Z.-S., Hua, Q., Liu, G.-X. & Sun, W.-Y. (2009). Cryst. Growth Des. 9, 395–403.
- Yin, X.-J., Zhou, X.-H., Gu, Z.-G., Zuo, J.-L. & You, X.-Z. (2009). Inorg. Chem. Commun. 12, 548–551.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812039256/ds2211sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039256/ds2211Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




