Abstract
Background: Heme oxygenase-1 (HO-1) is a cytoprotective and antiapoptotic enzyme, which has been involved in maintaining cellular homeostasis, and plays an important protective role by modulating oxidative injury. Up-regulation of (HO-1) has contributed to tumorogenicity of some cancers. In this study we investigated the expression pattern of the HO-1, in five different human-derived cancer cell lines with high incidence in Iran.
Methods: Total cell RNA were extracted from HepG2 (hepato carcinoma), A549 (lung adenocarcinoma), MCF-7 (breast cancer), K562 (myeloid leukemia) and LS174T (colon cancer) cell lines. Human embryonic kidney (HEK293) cell line was used as a control. cDNAs were synthesized and expression of HO-1 was examined using RT-PCR.
Results: The expression of HO-1 was not detected in the control cell line (HEK293), but it was observed to express following ultraviolet (UV) exposure indicating that HO-1 is not constantly expressed. The examined cancer cell lines constitutively expressed different variety of HO-1 on mRNA level. Strong expression of HO-1 was observed in HepG2, MCF-7 and A549 cells. A moderate expression of HO-1 was observed in K562 cells, and LS174T cells showed no expression of HO-1.
Conclusion: Heme oxygenase-1 could be considered as a new marker in the diagnosis of some cancers, especially hepatomacarcinoma. Our results also suggest that up-regulation of HO-1 may contribute to tumorogenicity of some cancers. Therefore, the combination of gene-silencing effect of HO-1 and chemotherapy might be considered as a new modality for the treatment of cancers in which the expression HO-1 is up-regulated.
Key Words: Heme oxygenase-1, gene expression, HepG2, A549, MCF7, K562, cancer cells
Introduction
A complex variation in gene expression patterns was found occur in the development and progression of cancer, and the experimental reversal of tumorigenicity.1 Thus, it seems that there is a relation between malignancies and alterations in the expression pattern of some genes. One of the genes, which have been discovered to be involved in the ‘rescue response’ of the tumor, is heme oxygenase-1 (HO-1).2 Heme oxygenase-1 acts as a cytoprotective agent against oxidative injury and cellular stress both in vitro and in vivo.3 This stress protein, which catalyzes the degradation of heme to biliverdin, carbon monoxide (CO) and free iron, is the inducible isoform of the three heme oxygenases (HO-1, HO-2 and HO-3). Heme oxygenase-1 and its derivatives also possess anti-inflammatory properties.2,4
Expression of HO-1 is low under normal physiologic conditions, and a variety of stimuli and activated signalling molecules such as HO-1 substrate heme, reactive oxygen species (ROS), nitric oxide species, prostaglandins, cytokines, growth factors such as insulin, and lipopolysaccharide can up-regulate its expression.5 Important roles for the HO-1 and its products in tumor progression and formation of metastases as well as resistance to anticancer therapy have been hypothesized.2,6 Thus, the high levels of HO-1 in tumor cells may, at least partly, be responsible for their resistance to anticancer treatment.2 Moreover, HO-1 accelerates vascularization of tumors and increases the metastatic potential of cancer cells, because of its proangiogenic properties. Therefore, the expression of HO-1 is usually increased in tumors, compared with surrounding healthy tissues,7 This was shown in lymphosarcoma, adenocarcinoma, hepatoma, glioblastoma, melanoma, prostate cancers, Kaposi sarcoma, squamous carcinoma, pancreatic cancer, and brain tumors.8-13 Generally, it seems that tumor growth and metastasis is accelerated by HO-1, though it may vary according to the type of cancer.
In order to extend the knowledge on the expression pattern of HO-1 in the human cancers, we investigated the expression of HO-1 in different cancerous and normal cells so far by measuring its mRNA by RT-PCR in five cancer cell lines that are commonly used in Iran. We examined cell lines of hepatocarcinoma (HEP G2), lung adenocarcinoma (A549), breast cancer (MCF-7), myeloid leukemia-derived cell line (K562) and colon cancer (LS174T).
Until now, only limited data are available on the expression of HO-1 in the cell lines investigated herein. Our findings might suggest HO-1 as a promising marker for the diagnosis of cancers.
Materials and Methods
Cell Culture
All the cell lines used were obtained from national cell bank of (table 1). Briefly, all cells were cultured in RPMI-1640 medium (Gibco-BRL, Germany) with 10% fetal bovine serum (Gibco-BRL, Germany) at 37°C in the presence of 5% CO2.
Table1.
Characteristics of cell lines used
| NCBI Code | Designation | Species | Tissue | Morphology |
|---|---|---|---|---|
| C497 | HEK 293 | Human | Kidney | Epithelial-like |
| C158 | HEP G2 | Human | Liver | Epithelial-like |
| C137 | A549 | Human | Lung | Epithelial-like |
| C135 | MCF-7 | Human | Breast | Epithelial-like |
| C122 | K562 | Human | Pleural effusion | Lymphoblast-like |
| C568 | LS174T | Human | Epithelial-like |
NCBI; national cell bank of Iran
RNA Extraction and cDNA Synthesis
Total RNA was extracted from 106 cells using Trizol reagent (Invitrogen, ) according to the manufacturer's instruction. Total cellular RNA was eluted in 60 µl RNase free water and stored at -20°C. One mg of Total RNA was treated with SuperScript III reverse transcriptase (Invitrogen) followed by DNase I (Invitrogen, Carlsbad, CA, USA) treatment and heat inactivation. The Synthesized cDNAs were stored at 20°C for further expression analysis.
Semiquantitative RT-PCR
Expression analysis of HO-1 was performed under optimized reaction conditions using gene specific primers designed by Primer 3 (http://primer3.sourceforge.net/). The Primer pair for amplification of the 864 bp HO-1 fragment was: forward 5' ATG ACA CCA AGG ACC AGA GC□3΄and reverse 5΄□GTG TAA GGA CCC ATC GGA GA□3΄. For normalization, expression of β-actin was examined with the primer pair of: forward 5’-TTC TAC AAT GAG CTG CGT GTG G -3’ and reverse 5’-GTG TTG AAG GTC TCA AAC ATG AT-3’.
The PCR condition included an initial denaturation at 94°C for 5 min followed by 30 amplification cycles consisting of denaturation for 30 sec at 94°C, annealing 30 sec at 59°C and extension of 30 sec at 72°C. The annealing temperature was 59°C for beta-actin. All reactions were performed in triplicates. Then the PCR products were separated on agarose gel and visualized using ethidium-bromide (Roth, ). Then, the expression pattern of HO-1 gene was analyzed by UVIdoc Gel Documentation System (Avebury House 36a Union Lane Cambridge CB4 1QB-uk).
Real-Time PCR
Real-time PCR analysis was performed in a Rotor-Gene RG 3000 (Corbett Research, ) thermocycler. Amplification was conducted using ABsolute SYBR Green mix (ABgene, ) according to the manufacturer’s instructions. Briefly, 25 µl of total PCR reaction was prepared containing 12.5 µl of the 2× SYBR Green mix, 10 pmole of each forward and reverse primers, and 1 µl of cDNA template. The Primer pair for amplification of the 153 bp HO-1 fragments was: forward 5'□ ATGACACCAAGGACCAGAGC □3΄and reverse 5΄□GTGTAAGGACCCATCGGAGA□3΄. Threshold cycle values were normalized with respect to the β-actin expression. The reaction condition included an initial denaturation at 94°C for 15 minutes followed by 40 amplification cycles of denaturation at 94°C for 30 seconds, annealing at suitable temperature for 30 seconds and extension at 72°C for 30 seconds. Triplicate reactions were set up for each sample and their mean values were used for calculations. The values related to the HO-1 expression were normalized against those of β-actin and the relative expressions were calculated by comparative Ct (threshold cycle) method.
Results
Expression of HO-1 mRNA in Different Cancer Cell Lines
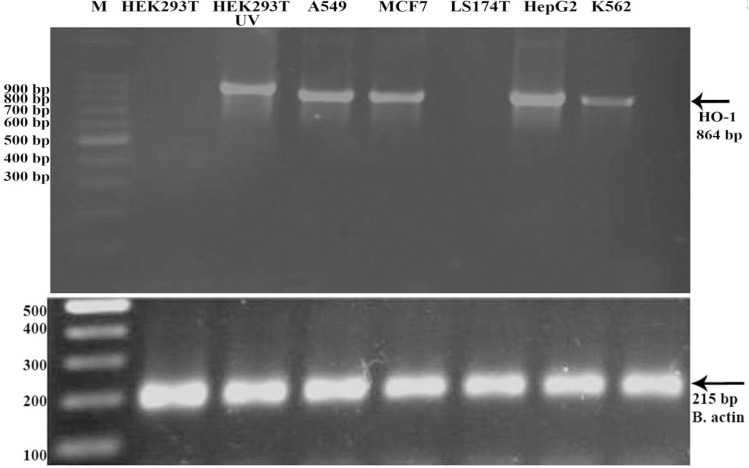
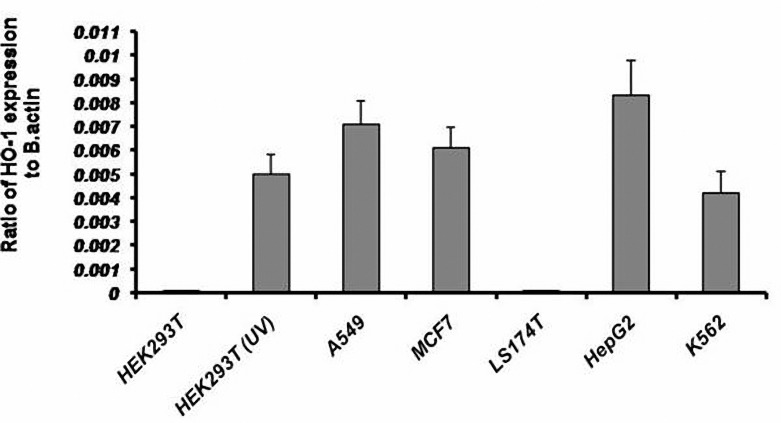
Total mRNA from the following cancer cell lines was extracted and used as template for RT-PCR analysis. Different expression patterns of HO-1 could be observed on mRNA level, depend on the cell line investigated (figure 1). A very strong expression of the HO-1 mRNA was detected in the HEPG2 cell line. A strong expression of HO-1 was found in the MCF7 and A549 cell lines and a moderate expression of HO-1 mRNA was detected in the k562 cell line. The LS174T cell line was the only one amongst the investigated cancer cell lines which showed no expression for HO-1. Next, we quantified the expression of HO-1 by Real time PCR analysis. The highest expression level of HO-1 was detected in HEPG2 cell line followed by MCF-7, A549 and k562 cells, respectively (figure 2). Furthermore, the results revealed no HO-1 expression in LS174T cell line.
Figure 1.
The expression pattern of HO-1 in different cancer cell lines in vitro by RT-PCR. The examined cell lines are shown at the top of figure. For each sample, the amount of RNA was normalized according to the amount of β-actin mRNA as a housekeeping gene. UV; Ultraviolent irradiation, M; 100-bp ladder marker
Figure 2.
Quantification of HO-1 expression by Real-time PCR. The values (Mean±SD) of HO-1 were normalized against β-actin values and their relative expressions were calculated by comparative Ct (threshold cycle) method. UV; Ultraviolent irradiation
Induction of HO-1 in HEK293T Cell Line by Ultraviolet
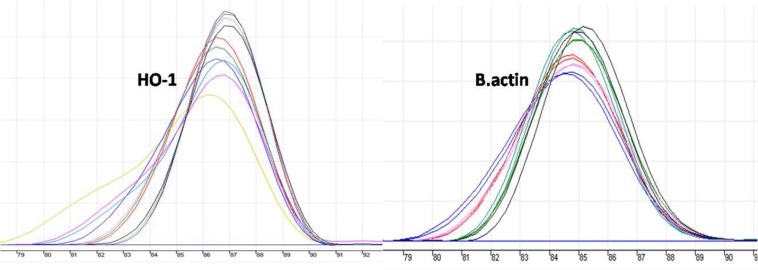
Ultraviolet (UV) irradiation was used to induce oxidative stress to HEK293T cells, in order to compare the HO-1 gene expression in normal physiological and oxidative stress conditions. For this purpose, HEK293T cells were exposed to UV for one hour followed by RNA extraction and cDNA synthesis. Finally, HO-1 expression was analyzed by RT-PCR. In contrast to the normal cells, which revealed no HO-1 mRNA expression, a strong expression of HO-1 was seen in the UV-irradiated HEK293T cells (HEK293T-UV) showing that HO-1 expression could be induced by oxidative injuries (figure 1). This finding was also confirmed by real time PCR analysis (figure 2). Melt curve analysis has been shown in figure 3. The overlap of beta-actin and HO-1 curves indicates that they are due to a single band. Considering this result and the expression of HO-1 in the cancer cells, it seems that continued expression of HO-1 in the cancer cells is a strategy for survival and proliferation.
Figure 3.
Melt curve analysis for HO-1 (left) and beta-actin (right). The overlap of beta-actin and HO-1 curves indicates that they are due to single band.
Discussion
It has been shown that HO-1 is an inducible enzyme, whose expression is often increased by those oxidative stresses, which produce reactive oxygen species.3,14 In spite of the cytoprotective affects of HO-1 on healthy tissues following their exposure to harmful stimuli, it can also protect tumor cells. Such a protection can result in the progression of the disease. Different studies have shown that higher levels of HO-1 expression are associated with faster growth of tumors, as indicated by bigger volumes of nodules or by more numerous cancer cells.11,15 Although some studies have reported a selective decrease in the expression of HO-1 in a few malignant cells such as adenocarcinoma or tongue squamous carcinoma, most studies have shown that the expression of HO-1 is strongly up-regulated in various tumors. Therefore, it seems that HO-1 represent a molecular target in some cancers.
The current study was designed to determine the expression pattern of HO-1 gene in five cancer cell lines that are highly prevalent in . Until now, only limited data are available on the expression of HO-1 in the cell lines investigated here. Among the cell line studied, HEPG2 cell line showed the highest expression of HO-1 based on the level of mRNA measured. The increased expression of HO-1 mRNA in HEPG2 cell line in the present study is in agreement with the high levels of expression of HO-1 in tissues of hepatocellular carcinoma described by Doi and colleagues.16 Thus, based on these results, we suggest HEPG2 cell line is the best model for future analysis of biology and regulation of HO-1 in hepatocellular carcinoma cell lines. The gene of HO-1 was found by RT-PCR to be expressed in MCF7 cell line as well as in A549 cell line. Human lung adenocarcinoma A549 cells constitutively express HO-1, which help them to resist against toxic compounds and antitumor drugs.17 Concerning MCF7, our results are in line with the results obtained by Hill et al. who reported overexpression of HO-1 protein in MCF-7 cells.18
A moderate level of HO-1 expression could be observed in myeloid leukemia-derived cell line K562. Given the up-regulation of HO-1 expression in myeloid leukemia cell line and based on the function of HO-1 as a survival factor in chronic myeloid leukemia,19 it can be suggested that there is an authentic pattern of HO-1 expression related to chronic myeloid leukemia. Based on our results the LS174T cell line was the only one amongst the investigated cancer cell lines to reveal no HO-1 expression. In contrast to our results, Fang and co-workers indicated that up-regulation of HO-1 expression in colon cancer was a main factor for resistance against anticancer therapy, since the HO-1 inhibitors or targeted knocking down of the HO-1 expression made the cultured tumor cell lines much more sensitive to anticancer therapy.20 This discrepancy could be due to difference in the colon cell lines. Taking these results together, it seems that an actual pattern of HO-1 expression may be related to cancer cells.
We detected an expression pattern of HO-1 in different cancer cell lines. HepG2 cells showed the strongest HO-1 mRNA expression among the cancer cell lines examined. Therefore, it might be possible to suggest that increased expression of HO-1 in hepatocellular carcinoma is a new cancer marker. HepG2 may also be the best choice for all future investigations on the effects of HO-1 up-regulation, and its likely role in different types of cancer. Gene-silencing of HO-1 would be an area of future investigations. Previous studies showed the HO-1 overexpression in some cancers, however, no data was available about the HO-1 expression in four cell lines that were investigated herein. The finding of the present study might be taken as evidence that the expression of HO-1 in different cancer cell lines might serve as a useful cancer marker. However, further studies on the role and expression pattern of HO-1 in more cancer cell lines are necessary. Moreover, the results presented here might establish a basis for the analysis of the regulation of HO-1 expression in some tumors.
Conclusion
The present study showed that HO-1 could be considered as a new marker in diagnosis of some cancers specially hepatocellular carcinoma. The results also suggest that up-regulation of HO-1 may contribute to tumorogenicity of some cancers.
Acknowledgment
This research was supported financially by Rafsanjan University of Medical Sciences.
Conflict of Interest: None declared
References
- 1.DeRisi J, Penland L, Brown PO, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–60. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 2.Nuhn P, Künzli BM, Hennig R, et al. Heme oxygenase-1 and its metabolites affect pancreatic tumor growth in vivo. Mol Cancer. 2009;8 doi: 10.1186/1476-4598-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank J, Lornejad-Schäfer MR, Schöffl H, et al. Inhibition of heme oxygenase-1 increases responsiveness of melanoma cells to ALA-based photodynamic therapy. Int J Oncol. 2007;31:1539–45. [PubMed] [Google Scholar]
- 4.Majewska M, Zajac K, Dulak J, Szczepanik M. Heme oxygenase (HO-1) is involved in the negative regulation of contact sensitivity reaction. Pharmacol Rep. 2008;60:933–40. [PubMed] [Google Scholar]
- 5.Tauber S, Jais A, Jeitler M, et al. Transcriptome analysis of human cancer reveals a functional role of heme oxygenase-1 in tumor cell adhesion. Mol Cancer. 2010;9 doi: 10.1186/1476-4598-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dulak J. Changing faces of heme oxygenases. Antioxid Redox Signal. 2007;9:2043–7. doi: 10.1089/ars.2007.1833. [DOI] [PubMed] [Google Scholar]
- 7.Rushworth SA, Bowles KM, Raninga P, MacEwan DJ. NF-kappaB-inhibited acute myeloid leukemia cells are rescued from apoptosis by heme oxygenase-1 induction. Tumor and Stem Cell Biology. 2010;70:2973–83. doi: 10.1158/0008-5472.CAN-09-3407. [DOI] [PubMed] [Google Scholar]
- 8.Jozkowicz A, Was H, Dulak J. Heme oxygenase-1 in tumors: is it a false friend? Antioxid Redox Signal. 2007;9:2099–117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruel CR, Pinto LF, Blanco TC, et al. Evaluation of the heme oxygenase-1 expression in esophagitis and esophageal cancer induced by different reflux experimental models and diethylnitrosamine. Acta Cir Bras. 2010;25:304–10. doi: 10.1590/s0102-86502010000300015. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Su J, DingZhang X, et al. PTEN deletion and heme oxygenase-1 overexpression cooperate in prostate cancer progression and are associated with adverse clinical outcome. J Pathol. 2011;224:90–100. doi: 10.1002/path.2855. [DOI] [PubMed] [Google Scholar]
- 11.Miyazaki T, Kirino Y, Takeno M, et al. Expression of heme oxygenase-1 in human leukemic cells and its regulation by transcriptional repressor Bach1. Cancer Science. 2010;101:1409–16. doi: 10.1111/j.1349-7006.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirley S, Micheau O. The heme oxygenase-1 and c-FLIP in acute myeloid leukemias: two non-redundant but mutually exclusive cellular safeguards protecting cells against TNF-induced cell death? Oncotarget. 2010;1:317–9. doi: 10.18632/oncotarget.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Was H, Dulak J, Jozkowicz A. Heme oxygenase-1 in tumor biology and therapy. Curr Drug Targets. 2010;11:1551–70. doi: 10.2174/1389450111009011551. [DOI] [PubMed] [Google Scholar]
- 14.Ewing P, Wilke A, Eissner G, et al. Expression of heme oxygenase-1 protects endothelial cells from irradiation-induced apoptosis. Endothelium. 2005;12:113–9. doi: 10.1080/10623320500189814. [DOI] [PubMed] [Google Scholar]
- 15.Deshane J, Chen S, Caballero S, et al. Stromal cell-derived factor 1 promotes angiogenesis via a heme oxygenase 1-dependent mechanism. J Exp Med. 2007;204:605–18. doi: 10.1084/jem.20061609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi K, Akaike T, Fujii S, et al. Induction of haem oxygenase-1 by nitric oxide and ischaemia in experimental solid tumours and implications for tumour growth. Br J Cancer. 1999;80:1945–54. doi: 10.1038/sj.bjc.6690624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kweon MH, Adhami VM, Lee JS, Mukhtar H. Constitutive overexpression of Nrf2-dependent heme oxygenase-1 in A549 cells contributes to resistance to apoptosis induced by epigallocatechin 3-gallate. J Biol Chem. 2006;281:33761–72. doi: 10.1074/jbc.M604748200. [DOI] [PubMed] [Google Scholar]
- 18.Hill M, Pereira V, Chauveau C, et al. Heme oxygenase-1 inhibits rat and human breast cancer cell proliferation: mutual cross inhibition with indoleamine 2, 3-dioxygenase. FASEB J. 2005;19:1957–68. doi: 10.1096/fj.05-3875com. [DOI] [PubMed] [Google Scholar]
- 19.Mayerhofer M, Florian S, Krauth MT, et al. Identification of heme oxygenase-1 as a novel BCR/ABL-dependent survival factor in chronic myeloid leukemia. Cancer Res. 2004;64:3148–54. doi: 10.1158/0008-5472.can-03-1200. [DOI] [PubMed] [Google Scholar]
- 20.Fang J, Sawa T, Akaike T, et al. Enhancement of chemotherapeutic response of tumor cells by a heme oxygenase inhibitor, pegylated zinc protoporphyrin. Int J Cancer. 2004;109:1–8. doi: 10.1002/ijc.11644. [DOI] [PubMed] [Google Scholar]