Abstract
Unlike many pathogens that are overtly toxic to their hosts, the primary virulence determinant of Mycobacterium tuberculosis appears to be its ability to persist for years or decades within humans in a clinically latent state. Since early in the 20th century latency has been linked to hypoxic conditions within the host, but the response of M. tuberculosis to a hypoxic signal remains poorly characterized. The M. tuberculosis α-crystallin (acr) gene is powerfully and rapidly induced at reduced oxygen tensions, providing us with a means to identify regulators of the hypoxic response. Using a whole genome microarray, we identified >100 genes whose expression is rapidly altered by defined hypoxic conditions. Numerous genes involved in biosynthesis and aerobic metabolism are repressed, whereas a high proportion of the induced genes have no known function. Among the induced genes is an apparent operon that includes the putative two-component response regulator pair Rv3133c/Rv3132c. When we interrupted expression of this operon by targeted disruption of the upstream gene Rv3134c, the hypoxic regulation of acr was eliminated. These results suggest a possible role for Rv3132c/3133c/3134c in mycobacterial latency.
Mycobacterium tuberculosis (MTB) is one of the world's most devastating human pathogens. Roughly 5% of all deaths worldwide are due to tuberculosis, more than from any other single infectious agent (1). Central to the pathogenic success of MTB is its ability to persist within humans for long periods in a clinically latent state without causing any overt disease symptoms. The magnitude of this disease reservoir is remarkable, with roughly one-third of all persons latently infected with MTB (2). Such people carry about a 10% lifetime chance of developing the active disease unless they become coinfected with HIV; then the risk increases to 8–10% per year (3). Largely because of the difficulties of studying an infection with no clinical symptoms, progress has been slow in understanding the mycobacterial genetic program and the host responses that are important in latent disease and reactivation (2). Reduced levels of inducible nitric oxide synthase and IFN-γ have been associated with reactivation of chronic/persistent tuberculosis in animal models (4–7). In addition, a few MTB genes have been implicated in long-term persistence (8–11). However, an overall picture of the adaptation to latency is lacking, and potential regulators of the process have remained elusive (12).
Several lines of evidence link latent tuberculosis and inhibition of MTB growth/metabolism with hypoxic conditions within the host. Tuberculosis infections are preferentially associated with the most oxygen-rich sites in the body (13), suggesting that reduced levels of O2 may limit MTB growth in vivo. This may explain why recrudescent tuberculosis occurs most often in the upper lobes of the lung (13), the single most-oxygenated region of the body. Conversely, inhibition of MTB growth in vivo is associated with the formation of fibrous granulomas, which are likely sites of low oxygen tension (14). One explanation proposed for the remarkable rates of reactivation TB among AIDS patients is that HIV infection impairs a person's ability to form and maintain hypoxic granulomas (15). Based on these observations, investigators have used hypoxic culture conditions to generate nonreplicating, persistent mycobacteria as an in vitro surrogate of the metabolic state of latent TB in vivo (9, 16–18).
One MTB gene induced under hypoxia and potentially involved in latency is acr (also known as hspX, Rv2031), which encodes α-crystallin (Acr). MTB Acr was originally described as a dominant antigen, frequently recognized by TB patient sera (19, 20). Further evidence of its role in TB pathogenesis comes from studies showing that Acr is needed for the growth of MTB in cultured macrophages (9). Acr localizes to the inner surface of the cell membrane (21) and is a member of the small heat shock protein family that forms high molecular weight aggregates and has chaperone activity in vitro (22). Under hypoxic conditions, Acr expression is dramatically increased (9, 22). In addition to its potential role in MTB latency, the powerful regulation of Acr under reduced O2 tension may provide insight into the nature and control of the whole genetic response/program by which MTB adapts to hypoxic microenvironments of the host. We describe here the subset of the MTB genome that responds rapidly to hypoxia. In addition, we report the identification and initial biological characterization of an MTB genetic system that regulates Acr expression in response to a hypoxic signal.
Materials and Methods
Manipulating Bacterial Strains.
H37Rv (ATCC 27294) and Mycobacterium bovis bacillus Calmette–Guérin (BCG) Montreal (ATCC 35735) were maintained on Middlebrook 7H9 medium (Difco) with 0.05% Tween 80 and ADC supplement or on Middlebrook 7H10 plates at 37°C as described (23). Bacilli were grown in roller bottles unless otherwise noted. When needed, kanamycin was used at 30 μg/ml and hygromycin at 50 μg/ml. Defined hypoxic culture was as previously described (9). Gas flow was maintained at ≈0.15 standard cubic feet (4.2 liters) per hour within each flask.
RNA Purification, Probe Synthesis, and Microarray Analysis.
Frozen cell pellets were suspended in 1 ml of Trizol reagent (GIBCO/BRL) and transferred to 2-ml screw-cap tubes containing 0.4-ml of 0.1-mm diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK). Cells were disrupted by three 30-sec pulses in a BioSpec Products bead beater. After 5 min at room temperature, samples were centrifuged at 13,000 × g for 45 sec, and the supernatants were extracted with 300 μl of chloroform/isoamyl alcohol (24:1) and precipitated with isopropyl alcohol. Samples were incubated 10 min to overnight at 4°C and centrifuged for 10 min at 4°C. The RNA pellets were washed with 1 ml of 75% ethanol, centrifuged at 13,000 × g for 5 min, and air-dried. After suspension of the RNA pellets in 90 μl of water, 10 μl of DNase I 10× buffer and 4 units of DNase I (Ambion, Austin, TX) were added and the samples were incubated for 30 min. Final purification of RNA was by RNeasy column (Qiagen, Chatsworth, CA). All steps in the MTB DNA microarray gene expression analysis were performed as described (24, 25). Controls for array quality were printed onto each slide and included a dilution series of MTB genomic DNA (positive control) and salmon sperm DNA (negative control). Additionally, DNA prelabeled with the cyanine dyes Cy3 or Cy5 were printed to ascertain whether spot-to-spot carryover contamination had occurred during the printing step. The quality of cDNA labeling was monitored by quantitatively measuring the intensities of Cy3- and Cy5-labeled samples hybridized to the MTB genomic positive control spots.
Targeted MTB Gene Disruptions.
Our targeted disruption strategy is based on the plasmid pEX18T (ampR with sacB for sucrose counterselection, kindly provided by H. Schweizer, Univ. of Colorado, Boulder; GenBank accession no. AF004910). A 1.2-kb fragment conferring kanamycin resistance was cloned from pMV306 into the XbaI site of the pEX18T polylinker to create pEX18TK. Then a 1.4-kb gene cassette containing the hygromycin phosphotransferase (hpt) gene from pRF498H (kindly provided by Khisimuzi Mdluli, Chiron) was cloned into the unique ScaI site of pEX18TK to create pKO. pKO no longer confers ampicillin resistance, but does confer resistance to hygromycin and kanamycin, has a functional sacB gene for sucrose counterselection, and cannot replicate autonomously in mycobacteria. Genomic regions (≈800 bp each) flanking the gene of interest were amplified by PCR and cloned into pKO so as to flank the kanR determinant. For targeted disruptions, constructs were electroporated into mycobacteria as described (26) and selected on 7H10 plates with hygromycin. In the first screening step, each colony was tested by PCR with two primer pairs, one specific for integration upstream of the gene of interest and the other specific for integration downstream. Underscoring the need for sucrose counterselection, we have not yet found any colonies in which both primer pairs were positive (a result that would indicate a single-step disruption). Colonies found to be positive at either end by PCR were grown to an OD600 = ≈1.0 and plated onto 7H10 plates containing 10% sucrose. Bacilli that grow on sucrose have generally either mutated copies of sacB or have lost the integrated plasmid. A portion of those in which the plasmid is lost will also lose the gene of interest. Colonies appearing on sucrose plates were picked into media and patched separately onto 7H10 plates with kanamycin and hygromycin. Sucrose-resistant, hygromycin-sensitive, kanamycin-resistant colonies (indicating loss of the integrated plasmid) were screened for loss of the gene of interest by PCR.
Other Procedures.
The acr-lux reporter construct and luciferase assays were described previously (9), except that a TD-20/20 luminometer (Turner Designs) was used in this work. Escherichia coli strain DH5α (GIBCO/BRL) was used for routine DNA manipulations.
Results
Expression of acr Is Rapidly Induced in Response to Reduced Oxygen Tension.
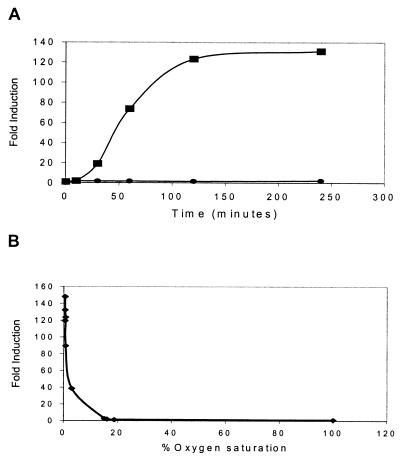
Acr is powerfully induced both during the stationary phase and when aerated logarithmic-phase cultures are allowed to settle (21, 22, 27). The same magnitude of induction is achieved when logarithmic-phase cultures are agitated under defined hypoxic conditions (9), and the signal in each case is believed to be reduced oxygen tension. However, the time span of those experiments (hours to days) could not distinguish between potentially direct or indirect mechanisms of induction. If the reduced O2 signal acts directly on acr expression without intervening protein synthesis, the effect would likely be evident very quickly. To investigate this possibility, we assessed acr expression in BCG, an attenuated vaccine strain of M. bovis. Because of their extensive homology at the genetic and molecular levels (28, 29), BCG can be selectively used as an avirulent surrogate of MTB (23), and previous results showed that the hypoxic induction of acr in MTB and BCG is indistinguishable (9, 27). Early logarithmic-phase cultures of BCG Montreal with an integrated copy of the acr promoter fused to the firefly luciferase (lux) gene (9) were placed in sealed tubes, flushed for 1 min with pressurized gas (0.2% O2 in N2), and then rotated at 37°C. Previous experiments showed that MTB growth is arrested in <1% O2, but even after 3 weeks of defined hypoxia, cell viability remains high and dormant cultures revive rapidly when ambient air (≈20% O2) is restored (9) (data not shown). At various times, individual tubes were removed and acr expression was monitored by luciferase assay. Enhanced acr expression was evident after only 10 min; within 2 h the induction was ≈125-fold (Fig. 1A). In contrast, lux values were unchanged in aerated controls over the same timeframe. This result is consistent with a direct effect of hypoxia on acr gene expression. To correlate acr induction with the amount of oxygen actually dissolved in the media, we placed early logarithmic-phase BCG acr-lux in defined hypoxic culture as described previously (9) and monitored conditions with a pO2 probe (Mettler). The acr response profile was notably sharp (Fig. 1B). The promoter did not respond to media O2 concentrations as low as 15% of full saturation (defined as the amount of dissolved O2 in a stirred culture maintained beneath ambient air). Partial induction (28-fold) was achieved at 3%, and complete induction (133-fold) occurred at 0.6% of full O2 saturation.
Figure 1.
Acr responds rapidly to reduced O2 tension. (A) Time course of acr induction, performed as described in the text. At the indicated times, individual sealed tubes were removed and assayed for lux activity in duplicate. Shown are representative data from one experiment of five. Squares represent BCG:acr-lux in sealed tubes flushed with 0.2% O2 in N2. Circles represent BCG:acr-lux treated identically, except the tubes were not sealed. (B) Induction of acr as a function of dissolved O2 concentration, performed as described in the text. Shown are representative data from one experiment of two.
Whole-Genome Expression Profiling Defines the Initial Transcriptional Response of MTB to Hypoxia.
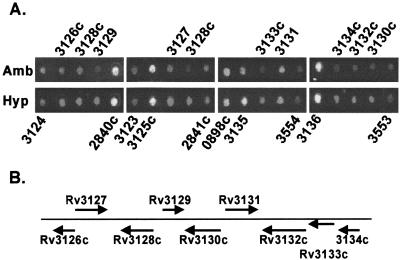
Having established that hypoxia can rapidly induce acr expression, we were interested in the extent of this effect on other MTB genes. Whole-genome microarray technology provides a robust tool to assess expression of many genes simultaneously in response to changing environmental conditions (30). Previously we generated a DNA-spotted glass slide microarray representing >97% of the 3,924 predicted ORFs of the MTB genome (29). The array provided a powerful way to monitor genome-wide changes in MTB gene expression in response to the drug isoniazid (24). As an initial characterization of the MTB response to reduced oxygen tension, MTB strain H37Rv was maintained for 2 h at 0.2% O2 in N2 as described previously (9). Medium pO2 under these conditions dropped to less than 1% of full saturation. RNA was isolated, labeled, and applied to the array surface. The resulting hybridized array was scanned and fluorescent intensities were used to identify regulated ORFs. Data for all ORFs on the microarray are available as supplementary material at http://schoolniklab.stanford.edu/projects/tb.html with highlights of the data described below. Expression of ≈100 genes was altered significantly; the 47 induced genes included acr, as expected (Table 1). Predicted functions for many of the 60 repressed genes indicate that low O2 is associated with broad adaptation to reduced metabolic activity (supplementary material, http://schoolniklab.stanford.edu/projects/tb.html). Repressed loci encode components of protein synthesis (6 genes), DNA synthesis/cell division (3 genes), lipid biosynthesis (5 genes), amino acid synthesis (3 genes), production of polyketides and other complex hydrophobic molecules (3 genes), and aerobic metabolism (7 genes). Fourteen (≈23%) of the repressed genes are of unknown function. In comparison, more than two-thirds (≈68%) of the 47 induced genes are of unknown function. Among the induced genes with motifs that predict a regulatory function is the putative two-component response regulator pair Rv3133c/Rv3132c (Table 1; Fig. 2). Because regulators induced by hypoxia are likely to have a role in the hypoxic response that controls acr expression, we chose to study this region in greater detail.
Table 1.
MTB genes induced by hypoxia
| Rv no. | Gene | ↑ Ratio | Gene product |
|---|---|---|---|
| Rv0079 | 11.6 ± 3.5 | HP | |
| Rv0080 | 7.6 ± 2.1 | HP | |
| Rv0081 | 3.5 ± 1.1 | Transcriptional regulator | |
| Rv0569 | 18.4 ± 4.0 | CHP | |
| Rv0572c | 13.9 ± 7.8 | HP | |
| Rv0574c | 3.8 ± 1.7 | CHP | |
| Rv1264 | 2.4 ± 0.2 | Similar to adenylate cyclases | |
| Rv1592c | 3.1 ± 0.7 | CHP | |
| Rv1733c | 12.6 ± 4.1 | Possible membrane protein | |
| Rv1734c | 9.8 ± 7.8 | HP | |
| Rv1736c | narX | 3.0 ± 0.8 | Fused nitrate reductase |
| Rv1737c | narK2 | 12.0 ± 3.0 | Nitrite extrusion protein |
| Rv1738 | 63.3 ± 37 | CHP | |
| Rv1739c | 3.9 ± 1.1 | Possible sulfate transporter | |
| Rv1813c | 14.7 ± 9.8 | CHP | |
| Rv1997 | ctpF | 8.8 ± 6.0 | Probable cation transport ATPase |
| Rv1998c | 4.7 ± 1.2 | CHP | |
| Rv2003c | 11.4 ± 7.2 | CHP | |
| Rv2005c | 8.5 ± 2.7 | CHP | |
| Rv2007c | fdxA | 22.0 ± 9.9 | Ferredoxin |
| Rv2028c | 3.3 ± 1.2 | CHP | |
| Rv2029c | pfkB | 12.2 ± 6.9 | Phosphofructokinase II |
| Rv2030c | 19.1 ± 14 | CHP | |
| Rv2031c | acr; hspX | 13.6 ± 3.1 | 14-kDa antigen, heat shock protein |
| Rv2032 | 43.9 ± 16 | CHP | |
| Rv2428 | ahpC | 3.8 ± 1.2 | Alkyl hydroperoxide reductase |
| Rv2623 | 6.8 ± 2.3 | CHP | |
| Rv2624c | 44.3 ± 34 | CHP | |
| Rv2625c | 6.3 ± 2.8 | CHP | |
| Rv2626c | 37.4 ± 7.4 | CHP | |
| Rv2627c | 17.0 ± 6.3 | CHP | |
| Rv2628 | 4.8 ± 1.1 | HP | |
| Rv2629 | 6.8 ± 1.3 | HP | |
| Rv2630 | 3.9 ± 1.1 | HP | |
| Rv2659c | 3.7 ± 1.5 | PhilRV2 integrase | |
| Rv3126c | 20.9 ± 7.3 | HP | |
| Rv3127 | 33.1 ± 14 | CHP | |
| Rv3128c | 11.7 ± 4.6 | CHP | |
| Rv3129 | 38.6 ± 15 | CHP | |
| Rv3130c | 26.6 ± 16 | CHP | |
| Rv3131 | 4.3 ± 1.1 | CHP | |
| Rv3132c | 9.1 ± 3.9 | Sensor histidine kinase | |
| Rv3133c | 13.8 ± 10 | Two-component response regulator | |
| Rv3134c | 10.6 ± 2.5 | CHP | |
| Rv3841 | bfrB | 8.1 ± 2.9 | Bacterioferritin |
| Rv3842c | glpQ1 | 6.9 ± 1.4 | Phosphodiesterase |
| Rv3908 | 3.7 ± 1.5 | CHP |
A DNA microarray was used to measure the change in mRNA levels in MTB cultures after a 2-h shift from ambient to 0.2% O2. Ratios were calculated by averaging the data from six microarray experiments of three biological sample sets. Genes were included if their level increased at least 2-fold in the hypoxic sample compared to the control sample after subtraction of the standard deviation. Genes are listed in genomic order and are grouped if two or more genes in the same region of the genome fulfill the above criteria. Genes are annotated as described by the Pasteur Institute on TubercuList (http://genolist.pasteur.fr/TubercuList). HP, hypothetical protein; CHP, conserved hypothetical protein.
Figure 2.
Hypoxia-induced expression of genes in the Rv3132/3133/3134 region of the MTB genome. (A) Composite of different portions of the MTB whole genome array, corresponding to genes in the region around Rv3133c. Shown are expression patterns after 2 h in ambient (Amb) or 0.2% O2 (Hyp) as described in the text. Text above and below the figure indicates the gene number as defined by the H37Rv sequencing project (3). Red spots indicate an increase in mRNA level, green a decrease, and yellow no change. (B) Schematic representation of the genomic region around Rv3133c. Arrows denote the relative size, position, and transcriptional orientation of each gene.
Characterization of the Rv3134c/3133c/3132c Gene Cluster.
Analysis of the predicted Rv3133c protein sequence indicates that it is a member of the LuxR family of two-component response regulators. Similarly, Rv3132c is predicted to encode a membrane-bound protein with histidine kinase activity (31). Upstream of Rv3133c is a conserved hypothetical gene, Rv3134c. The sequence in this region suggests that these three genes may together form a single transcriptional unit; only 27 bp separates the putative stop codon of Rv3134c from the putative initiator codon of Rv3133c, whereas the putative ORF of Rv3133c overlaps with that of Rv3132c by one nucleotide (Fig. 2B). Our array results (Fig. 2; Table 1) are consistent with the possibility that these genes are cotranscribed, as are recently published data from another group (31). To investigate their role in the control of Acr expression or in the MTB hypoxic response, we targeted each of these three genes for deletion from the mycobacterial chromosome. By using a two-step method and sacB/sucrose counterselection (32, 33), we generated strains of H37Rv and BCG Montreal in which Rv3134c and Rv3132c are replaced by a kanamycin-resistance determinant (data not shown). In the Δ3132 mycobacteria, the 1669-bp DNA fragment from sequence 5′-CCGCAATGCGTC-3′ to 5′-GCGGAACAGTGC-3′ was replaced by the kanamycin-resistance determinant. This deletion extends from 38 bp downstream of the putative translation start site (bp 3499226 of the H37Rv genome) to 26 bp upstream of the putative stop codon (bp 3497557 of the H37Rv genome). In the Δ3134 mycobacteria, the 814-bp DNA fragment from sequence 5′-GGATGTATCCGC-3′ to 5′-CGCCGTACTTA-3′ was replaced by the kanamycin-resistance determinant. This deletion extends from 31 bp upstream of the putative translation start site (bp 3500779 of the H37Rv genome) to 20 bp upstream of the putative stop codon (bp 3499965 of the H37Rv genome). Our inability to delete Rv3133c thus far in either strain may be the result of technical difficulties and should not be construed as evidence that this gene is essential.
Rv3133c Is Required for Induction of acr by Reduced Oxygen Tension.
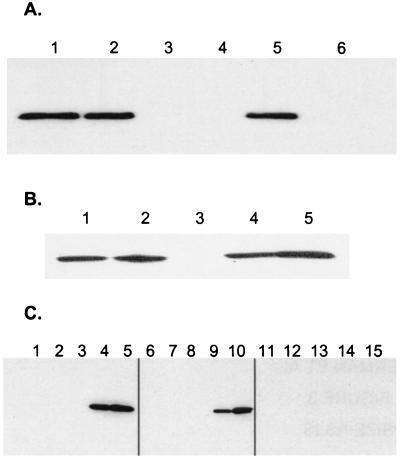
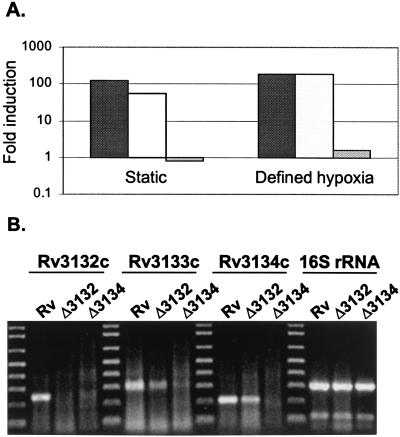
To test whether genes in the Rv3134c/3133c/3132c genomic region contribute to Acr induction, Western analysis with anti-Acr antibodies was performed. Logarithmic-phase cultures of H37Rv and isogenic deletion strains were left to settle overnight. Consistent with previous data (22), Acr expression was powerfully induced by settling in wild-type bacteria (Fig. 3A). Deletion of Rv3132c from H37Rv had little or no effect on Acr induction. However, in H37Rv:Δ3134 Acr was undetectable even after overnight settling, when it is one of the most abundant proteins in wild-type bacilli (22). Similar results were observed for BCG and its isogenic deletion mutants (Fig. 3B). Also, when BCG strains were monitored throughout the growth cycle, Acr could not be detected in wild-type or BCG:Δ3132 bacilli until the stationary phase, and the BCG:Δ3134 mutant produced no detectable Acr during any stage of growth, including the stationary phase (Fig. 3C). To quantitate the inducible acr expression in each strain, we introduced the acr-lux reporter construct into BCG:Δ3132 and BCG:Δ3134 and performed reporter gene assays. Under conditions of both settling and defined hypoxia, acr induction in BCG:Δ3132 was similar to that in wild-type BCG (≈100-fold, Fig. 4A). At the same time, induction in BCG:Δ3134 was less than 2-fold. Thus, deletion of Rv3134c completely eliminated the induction of Acr by hypoxia.
Figure 3.
Disruption of Acr expression in MTB and BCG. (A) Static MTB. Two-milliliter samples of H37Rv rolling cultures (OD600 ≈ 1.0) were left stationary in 15-ml tubes at 37°C overnight, and a portion was processed as below. Lanes: 1, H37Rv; 2, H37Rv:Δ3132; 3, H37Rv:Δ3134; 4, H37Rv:Δ3134∷3134; 5, H37Rv:Δ3134∷3133/3134; 6, H37Rv:Δ3134∷pCEB3 (vector control). Shown are representative data from one of three experiments. (B) Static BCG. Two-milliliter samples of BCG Montreal rolling cultures (OD600 ≈ 1.0) were left stationary in 15-ml tubes at 37°C overnight, and a portion was processed as below. Lanes: 1, BCG; 2, BCG:Δ3132; 3, BCG:Δ3134; 4, BCG:Δ3134∷3133P1 (expression controlled by 300 bp upstream of Rv3134c); 5, BCG:Δ3134∷3133P2 (expression controlled by 300 bp upstream of Rv3133c, form within the Rv3134c coding region). Shown are representative data from one of two experiments. (C) Growth curve in BCG Montreal (lanes 1–5), BCG:Δ3132 (lanes 6–10), and BCG: Δ3134 (lanes 11–15) were grown in 7H9 broth with aliquots removed at various times processed as below. Lanes 1, 6, and 11, early logarithmic phase (OD600 = 0.2–0.3); lanes 2, 7, and 12, mid-log phase (OD600 = 0.4–0.7); lanes 3, 8, and 13, late log/early stationary phase (OD600 = 0.9–1.9); lanes 4, 9, and 14, stationary phase (OD600 > 2.0); lanes 5, 10, and 15, stationary phase plus 1 week. Shown are representative data from one of three experiments. For each experiment, bacilli were lysed by bead beating (45 sec at maximum power) in a Fast Prep instrument (Bio 101). Five-microgram samples were separated by SDS/PAGE, transferred to Nitropure membrane (Osmonics, Minnetonka, MN), probed with anti-Acr monoclonal antibody IT-4 (supplied by John Belisle, Colorado State University, Fort Collins, as part of the National Institutes of Health Tuberculosis Materials Contract) and detected by SuperSignal West Pico (Pierce).
Figure 4.
Roles of Rv3132c, Rv3133c, and Rv3134c in hypoxic acr expression. (A) Quantitation of acr induction by lux assay. Cultures of rolling, log-phase BCG Montreal (black bars), BCGΔ3132 (stippled bars) and BCG:12Δ3134 (gray bars) were transformed with the acr-lux reporter construct and were either allowed to settle in 15- ml Falcon tubes overnight (static) or were placed for two hours in sealed tubes flushed with 0.2% O2 in N2 as described in Fig. 1. Plotted is the fold induction relative to the time 0 rolling culture. Shown are representative data from one of two experiments. (B) Reverse transcription-PCR analysis of RNA expression from Rv3132c, Rv3133c, and Rv3134c in wild-type and mutant H37Rv. RNA from each strain (indicated above each lane) was isolated. Expected size of each product: Rv3132, 278 bp; Rv3133, 299 bp; Rv3134, 206 bp; 16S RNA, 309 bp.
To confirm that acr disregulation in the Δ3134 strains was due to the mutation that we had introduced and to learn more about the roles of Rv3134c and Rv3133c in acr expression, we restored wild-type copies of the affected sequences into another region of the MTB chromosome. Because deletion of Rv3134c (which also removed a small portion of the upstream sequence) could have affected expression of the response regulator Rv3133c, we tested whether Rv3134c alone or Rv3134c/Rv3133c together could restore Acr regulation on H37Rv:Δ3134. Single-copy DNAs corresponding to Rv3134c alone and Rv3134c/Rv3133c together were integrated into the phage L5 attachment site (34), by using 300 bp of native Rv3134c upstream sequences to drive expression. Interestingly, reintroduction of Rv3134c alone failed to restore hypoxic expression of Acr, whereas the combination of Rv3134c and Rv3133c together restored Acr induction to wild-type levels (Fig. 3A). We then introduced Rv3133c alone and again regulated Acr expression was restored (Fig. 3B). These results suggest strongly that expression of Rv3133c is interrupted in the Δ3134 strains. To address this issue directly, we performed reverse transcription-PCR on RNA from H37Rv and on the two deletion mutants. Consistent with the complementation experiments above, Rv3133c RNA was detected in H37Rv and H37Rv:Δ3132, but not in H37Rv:Δ3134 under hypoxic conditions (Fig. 4B). Taken together, these experiments argue that the loss of Acr expression was because of targeted deletion of Rv3134c and that Rv3133c is sufficient to complement this phenotype. Further experiments are needed to determine whether Rv3134c plays an additional, subtler role in the hypoxic expression of Acr.
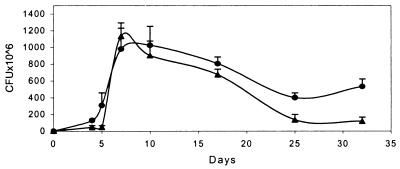
Disregulation of Acr would be expected to have deleterious consequences for MTB (9). In addition, because Rv3133c mediates the enhanced expression of one gene in response to a low oxygen signal, it seems reasonable that at least some of the other genes in the hypoxia regulon may also be under its control. Wild-type organisms respond to hypoxia with growth arrest. To begin testing whether deletion of Rv3134c affected this response, we compared the growth and survival rates of the parent strain BCG with those of BCG:Δ3134 (Fig. 5) in rolling culture. Growth rates were very similar until the stationary phase, when O2 tensions drop. After 3 weeks of the stationary phase, recoverable colony-forming units of BCG:Δ3134 were only ≈20% of the parent strain. Over a similar time course, growth and survival rates of an acr deletion mutant were indistinguishable from those of the wild-type (9), suggesting that acr is not the only disregulated gene in the Δ3134 mutants. Because colony-forming units can be influenced by multiple phenomena such as extent of clumping or susceptibility to damage by sonication, further experiments are required to determine whether BCG:Δ3134 is really less able to survive in stationary phase.
Figure 5.
Deletion of Rv3134c affects stationary phase survival of BCG. BCG Montreal (●) and BCG:Δ3134 (▴) were maintained in rolling culture with aliquots removed, sonicated, and plated at two concentrations in quadruplicate at the times indicated. Colonies were enumerated after 3 weeks at 37°C. Shown are representative data from one of two experiments.
Discussion
Nearly two billion people harbor latent TB infections (2), yet little is known about either the bacterial or host factors that alter the balance between latency and reactivation. The link between latency, altered MTB metabolism, and microaerophilic conditions within the host has been discussed in the literature for most of the last century (18, 22, 35–37). To update this long-standing concept, we hypothesize that hypoxia directly affects expression of a set of MTB genes, resulting in altered metabolism and contributing to latency in vivo. If so, regulators of the MTB hypoxic response are likely to be important components of the adaptation to latency. We reasoned that the potent induction of MTB acr by reduced oxygen tension could provide a means to dissect regulatory events that lead to this adaptation. The rapid acr induction time course (Fig. 1) showed that MTB can and does respond quickly to a hypoxic signal, encouraging us to search for other, similarly regulated genes by using whole-genome microarray.
The gene expression changes we identified indicate a broad adaptation to reduced metabolic activity. Among the repressed genes (supplementary material, http://schoolniklab.stanford.edu/projects/tb.html) at least 40% encode proteins with roles in biosynthesis, cell division, or aerobic metabolism. These are probably underestimates because ≈20% of the repressed genes are of unknown function. In comparison, the list of the genes rapidly induced by hypoxia (Table 1) tells a subtler story. More than two-thirds of the induced genes are of unknown function, underscoring that the MTB adaptation to hypoxia is not yet well described. Loci with postulated roles in MTB latency, including genes for glyoxylate shunt enzymes (8, 11) and alternative σ factors (10, 12), were not up-regulated in our experiments, emphasizing the early nature of the response that we have identified. Delineation of initial response genes suggests that adaptation to nonreplicating persistence may be a complex multistep process. Accordingly, genes involved with induction of the latent state may be different from those that maintain it over time. A potentially analogous process, sporulation in Bacillus subtilis, requires a cascade of regulatory events and gene expression changes (38).
Hypoxic induction of Rv3133c, identified in this study by microarray transcript profiling, has been confirmed recently by Boon and colleagues (39), who obtained protein expression profiles by two-dimensional electrophoresis of BCG Pasteur bacilli grown in the oxygen-limited in vitro dormancy model of Wayne (17). In addition to the up-regulation of Rv3133c, they identified increased expression of two other proteins, Rv2623 and Rv2626c; both of these, annotated as conserved hypothetical proteins, were also identified at the transcriptional level by microarray analysis (Table 1).
Our observation that the Rv3134c/3133c/3132c genomic region is rapidly induced suggested that this two-component response regulator system could play a role in the adaptation to hypoxia. Because these genes are proposed to form a single operon, it is not surprising that deletion of the sequence just upstream of and including the Rv3134c coding sequence disrupted expression of the downstream genes. The protein products of an operon often function together as well. However, whereas complementation experiments clearly indicate a role for the Rv3133c protein in acr expression, we have yet to demonstrate a role for the Rv3134c or Rv3132c proteins in this process. The lack of a phenotype in the Rv3132c disruptions could be due to the presence of a homologous sensor kinase gene elsewhere in the genome. The similarity between the predicted proteins Rv3132c and Rv2027 was noted elsewhere recently (31). Alternatively, Rv3132c may activate Rv3133c in response to a different signal. Rv3134c is a conserved hypothetical gene, with six related sequences in the MTB genome. Two-component response systems frequently have additional proteins that participate in the phosphorelay (40), and Rv3134c may play such a role in this case.
The work described here is an initial report of the MTB response to hypoxia. To better define the MTB hypoxic response and to see whether a cascade of transcriptional responses is required for the adaptation to nonreplicating persistence, we need to analyze global gene expression in cultures maintained under defined hypoxic conditions for different, longer periods of time. In addition, we want to determine whether other hypoxic response genes are also under control of this regulatory switch. In B. subtilis, regulators of sporulation are among the first genes induced, and mutations in these genes block the entire cascade (38). Array experiments with H37Rv:Δ3134 should help determine how much additional phenotypic characterization of this mutant is warranted.
Undoubtedly, factors in addition to reduced oxygen tension are important in the processes of MTB latency and reactivation. For example, reactive nitrogen intermediates are clearly involved in preventing MTB reactivation in mice (5, 6), and possibly in humans (41). We have focused here on hypoxia as but one aspect of a complex and important biological phenomenon. A molecular description of the response to hypoxia, along with mutant(s) in which portions of the hypoxic response are effectively masked, should provide insight into MTB latency and could help reveal the nature and extent of other latency mechanisms.
Acknowledgments
We thank Kristi Guinn for complementing plasmids and Anu Karnataki for assessing the growth of the mutant strains. This work is funded in part by the Tuberculosis Vaccine Collaboration of the Sequella Global Tuberculosis Foundation, of which D.R.S. is a Core Scientist, and by grants to G.K.S. from the Action TB Program of Glaxo SmithKline and the National Institutes of Health (Grant AI 44826). M.V. was supported by a fellowship from the American Lung Association; D.S. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
Abbreviations
- MTB
Mycobacterium tuberculosis
- Acr
α-crystallin protein
- BCG
bacillus Calmette–Guérin
References
- 1.Bloom B R, Small P M. N Engl J Med. 1998;338:677–678. doi: 10.1056/NEJM199803053381008. [DOI] [PubMed] [Google Scholar]
- 2.Parrish N M, Dick J D, Bishai W R. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 3.Zumla A, Mwaba P, Squire S B, Grange J M. J Infect. 1999;38:74–79. doi: 10.1016/s0163-4453(99)90072-5. [DOI] [PubMed] [Google Scholar]
- 4.Chan J, Tanaka K, Carroll D, Flynn J, Bloom B R. Infect Immun. 1995;63:736–740. doi: 10.1128/iai.63.2.736-740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacMicking J D, North R J, LaCourse R, Mudgett J S, Shah S K, Nathan C F. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn J L, Scanga C A, Tanaka K E, Chan J. J Immunol. 1998;160:1796–1803. [PubMed] [Google Scholar]
- 7.Scanga C A, Mohan V P, Yu K, Joseph H, Tanaka K, Chan J, Flynn J L. J Exp Med. 2000;192:347–358. doi: 10.1084/jem.192.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wayne L G, Lin K Y. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Y, C, C D, Simpson R M, Zhu Y, Hickey M J, Sherman D R, Barry C E., III Proc Natl Acad Sci USA. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMaio J, Zhang Y, Ko C, Young D B, Bishai W R. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney J D, Honer zu Bentrup K, Munoz-Elias E J, Miczak A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Nature (London) 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 12.Manabe Y C, Bishai W R. Nat Med. 2000;6:1327–1329. doi: 10.1038/82139. [DOI] [PubMed] [Google Scholar]
- 13.Adler J J, Rose D N. In: in Tuberculosis. Rom W N, Garay S M, editors. Brown, Boston: Little; 1996. pp. 129–140. [Google Scholar]
- 14.Dannenberg A M., Jr Hosp Pract. 1993;28:51–58. doi: 10.1080/21548331.1993.11442738. [DOI] [PubMed] [Google Scholar]
- 15.Doenhoff M J. Immunol Today. 1998;19:462–467. doi: 10.1016/s0167-5699(98)01310-3. [DOI] [PubMed] [Google Scholar]
- 16.Wayne L G, Diaz G A. J Bacteriol. 1967;93:1374–1381. doi: 10.1128/jb.93.4.1374-1381.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne L G, Hayes L G. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imboden P, Schoolnik G K. Gene. 1998;213:107–117. doi: 10.1016/s0378-1119(98)00192-9. [DOI] [PubMed] [Google Scholar]
- 19.Verbon A, Hartskeerl R A, Schuitema A, Kolk A H, Young D B, Lathigra R. J Bacteriol. 1992;174:1352–1359. doi: 10.1128/jb.174.4.1352-1359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B Y, Hefta S A, Brennan P J. Infect Immun. 1992;60:2066–2074. doi: 10.1128/iai.60.5.2066-2074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham A F, Spreadbury C L. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y, Crane D D, Barry C E., III J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman D R, Sabo P J, Hickey M J, Arain T M, Mahairas G G, Yuan Y, Barry C E, III, Stover C K. Proc Natl Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoolnik, G. K., Voskuil, M. I., Schnappinger, D., Yildiz, F. H., Meibom, K., Dolganov, N. A., Wilson, M. A. & Chong, K. H. (2001) Methods Enzymol. 336, in press. [DOI] [PubMed]
- 26.Wards B J, Collins D M. FEMS Microbiol Lett. 1996;145:101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x. [DOI] [PubMed] [Google Scholar]
- 27.Lim A, Eleuterio M, Hutter B, Murugasu-Oei B, Dick T. J Bacteriol. 1999;181:2252–2256. doi: 10.1128/jb.181.7.2252-2256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosch R, Philipp W J, Stavropoulos E, Colston M J, Cole S T, Gordon S V. Infect Immun. 1999;67:5768–5774. doi: 10.1128/iai.67.11.5768-5774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 30.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 31.Dasgupta N, Kapur V, Singh K K, Das T K, Sachdeva S, Jyothisri K, Tyagi J S. Tuber Lung Dis. 2000;80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 32.Pelicic V, Reyrat J M, Gicquel B. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 33.Parish T, Stoker N G. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 34.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, et al. Nature (London) 1991;351:456–60. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 35.Kempner W. Am Rev Tuberc. 1939;40:157–168. [Google Scholar]
- 36.Canetti G. The Tubercle Bacillus in the Pulmonary Lesion of Man. New York: Springer; 1955. pp. 111–126. [Google Scholar]
- 37.Wayne L G. Eur J Clin Microbiol Infect Dis. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 38.Errington J. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boon C, Li R, Qi R, Dick T. J Bacteriol. 2001;183:2672–2676. doi: 10.1128/JB.183.8.2672-2676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoch J A. In: Two Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 129–144. [Google Scholar]
- 41.Nathan C, Shiloh M U. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]