Abstract
In the title compound, C26H24N2O2, the planar 1H-imidazole ring makes dihedral angles of 35.78 (4), 26.35 (5) and 69.75 (5)°, respectively, with the dimethoxyphenyl ring and the phenyl rings in the 4- and 5-positions. In the crystal, C—H⋯O hydrogen bonds connect neighbouring molecules, forming infinite chains running along the b axis. Furthermore, the crystal structure exhibits a C—H-⋯π interaction between a methyl H atom and a phenyl ring from an adjacent molecule.
Related literature
For the synthesis of imidazole compounds, see: Shalini et al. (2010 ▶). For the medicinal properties of imidazole derivatives, see: Adams et al. (2001 ▶); Nakamura et al. (2004 ▶); Venkatesan et al. (2008 ▶); Nanterment et al. (2004 ▶); Roman et al. (2007 ▶); Congiu et al. (2008 ▶). For standard bond distances, see: Allen et al. (1987 ▶).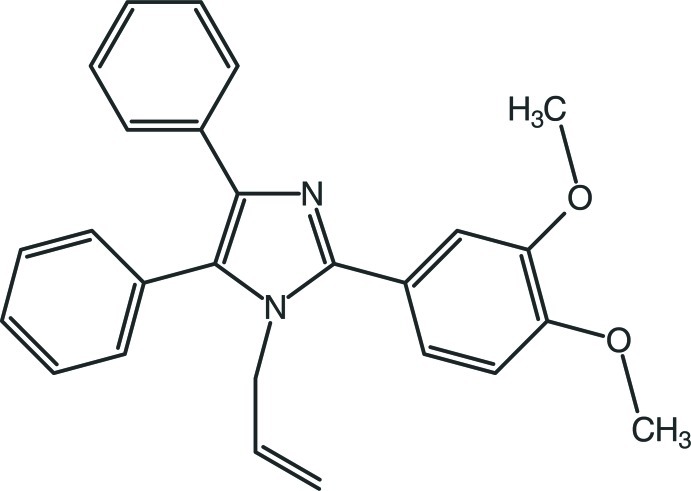
Experimental
Crystal data
C26H24N2O2
M r = 396.47
Triclinic,
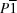
a = 8.9683 (4) Å
b = 10.7916 (5) Å
c = 11.7219 (5) Å
α = 110.174 (2)°
β = 106.267 (2)°
γ = 91.991 (3)°
V = 1011.32 (8) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 90 K
0.36 × 0.12 × 0.06 mm
Data collection
Bruker Kappa APEXII DUO diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.971, T max = 0.995
17346 measured reflections
6149 independent reflections
4408 reflections with I > 2σ(I)
R int = 0.039
Standard reflections: 0
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.127
S = 1.02
6149 reflections
273 parameters
H-atom parameters constrained
Δρmax = 0.46 e Å−3
Δρmin = −0.25 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PARST (Nardelli, 1983 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812039566/bt6835sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039566/bt6835Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812039566/bt6835Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg3 is the centroid of the C15–C20 phenyl ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C7—H9B⋯O1i | 0.96 | 2.57 | 3.515 (2) | 170 |
| C8—H8B⋯Cg3ii | 0.96 | 2.98 | 3.8316 (17) | 149 |
Symmetry codes: (i) 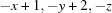 ; (ii)
; (ii) 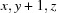 .
.
Acknowledgments
Manchester Metropolitan University, Erciyes University and Louisiana State University are gratefully acknowledged for supporting this study.
supplementary crystallographic information
Comment
The high therapeutic properties of the imidazole related drugs have encouraged the medicinal chemists to synthesize a large number of novel chemotherapeutic agents incorporating the imidazole nucleus (Shalini et al., 2010). The broad medicinal properties of imidazole drugs include anticancer, β-lactamase inhibitors, 20-HETE (20-hydroxy-5,8,11,14-eicosatetraenoic acid) synthase inhibitors, carboxypeptidase inhibitors, hemeoxygenase inhibitors, anti-aging agents, anticoagulants, anti-inflammatory, antibacterial, antifungal, antiviral, anti-tubercular, anti-diabetic and antimalarial (Congiu et al., 2008; Venkatesan et al., 2008; Nakamura et al., 2004; Roman et al., 2007; Nanterment et al., 2004 and Adams et al., 2001). In this respect and in continuation of our on-going study for synthesis of bioactive molecules, we herein report synthesis and crystal structure of the title compound (I) among series of other imidazole derivatives.
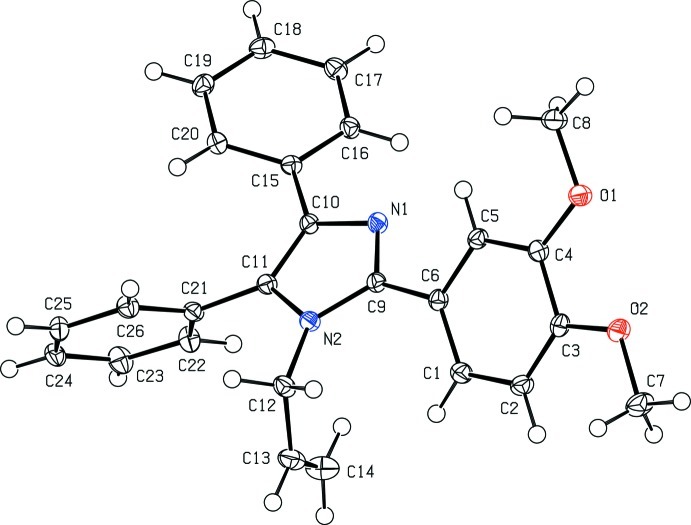
The molecular structure of (I), (Fig. 1), has not a planar conformation. The (N1/N2/C9—C11) 1H-imidazole ring which is planar [maximum deviation = 0.005 (1) Å for C11] forms dihedral angles of 35.78 (4), 26.35 (5) and 69.75 (5)°, respectively, with the C1–C6 benzene ring and the C15–20 and C21–C26 phenyl rings. The plane of the allyl group makes a dihedral angle of 82.09 (12)° with the plane of the 1H-imidazole ring.
In (I), all bond lengths and bond angles are within normal range (Allen et al., 1987). The C3–C4–O1–C8 and C4–C3–O2–C7 torsion angles are 171.98 (13) and -176.30 (13)°, respectively.
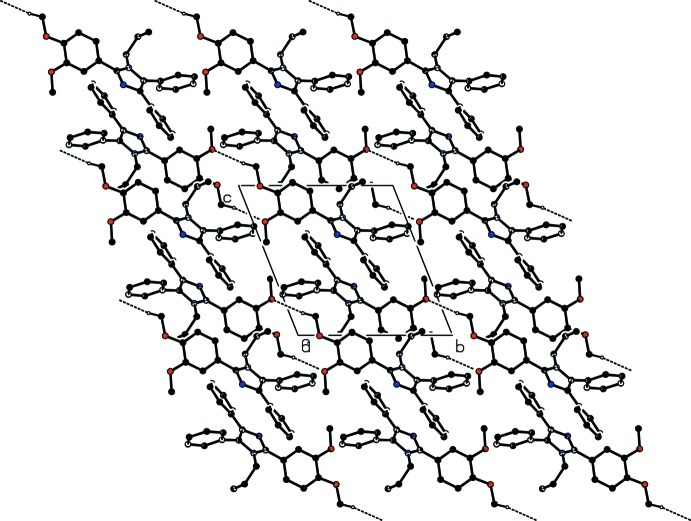
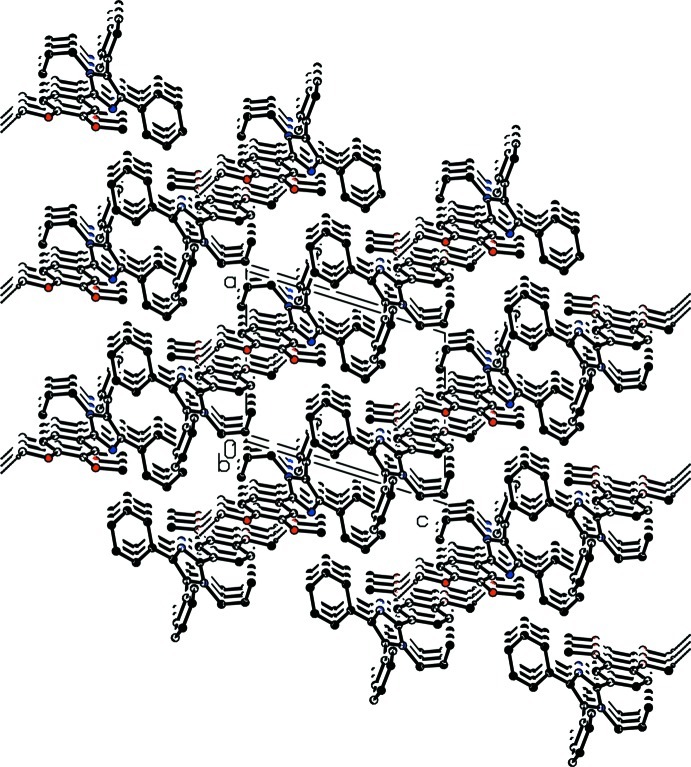
In the crystal packing, molecules are linked by C—H···O hydrogen bonds, forming infinite chains running along the b axis (Table 1, Figs. 2 & 3). In addition, a C–H···π interaction is observed between the (C8)H8B methyl H atom and the C15–C20 phenyl ring of the adjacent molecule (Table 1).
Experimental
A mixture of 25 ml. of dimethyl sulfoxide and 2.4 g. (40 mmol) of potassium hydroxide was added in a 50-ml. volumetric flask equipped with a magnetic stirring bar. The mixture was stirred at room temperature for 5 minutes before adding of 3.26 g. (10 mmole) of 2-(3,4-dimethoxyphenyl)-4,5-diphenyl-1H-imidazole. Stirring was continued for 45 minutes, then 4.80 g. (20 mmol) of allylbromide was added. After being stirred for an additional 45 minutes the mixture was diluted with 20 ml. of water. The organic product was extracted with three 20-ml. portions of diethyl ether, and each ether layer was washed with three 10-ml. portions of water. The combined ether layers were dried over calcium chloride, and the solvent was removed at slightly reduced pressure. The excess allyl bromide was removed by distillation at approximately 15 mm. The residue was solidified upon cooling and scratching to furnish the title compound (3.22 g; 88%). Mono crystals suitable for X-ray analyses were obtained by slow evaporation method from ethanol at room temperature. M.p. 486 – 488 K.
Refinement
Hydrogen atoms were located geometrically and refined using a riding model with C—H = 0.93–0.97 Å, and with Uiso =1.2Ueq(C) or 1.5Ueq(Cmethyl). The methyl groups were allowed to rotate but not to tip.
Figures
Fig. 1.
The molecular structure of (I) with the atom numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 50% probability level.
Fig. 2.
View of the crystal packing and hydrogen bonding of (I) down the a axis. H atoms not involved in hydrogen bonds have been omitted for clarity.
Fig. 3.
View of the crystal packing and hydrogen bonding of (I) down the b axis. H atoms not involved in hydrogen bonds have been omitted for clarity.
Crystal data
| C26H24N2O2 | Z = 2 |
| Mr = 396.47 | F(000) = 420 |
| Triclinic, P1 | Dx = 1.302 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.9683 (4) Å | Cell parameters from 3877 reflections |
| b = 10.7916 (5) Å | θ = 2.2–30.5° |
| c = 11.7219 (5) Å | µ = 0.08 mm−1 |
| α = 110.174 (2)° | T = 90 K |
| β = 106.267 (2)° | Needle, colourless |
| γ = 91.991 (3)° | 0.36 × 0.12 × 0.06 mm |
| V = 1011.32 (8) Å3 |
Data collection
| Bruker Kappa APEXII DUO diffractometer | 6149 independent reflections |
| Radiation source: sealed tube | 4408 reflections with I > 2σ(I) |
| TRIUMPH curved graphite monochromator | Rint = 0.039 |
| φ and ω scans | θmax = 30.5°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −12→12 |
| Tmin = 0.971, Tmax = 0.995 | k = −15→15 |
| 17346 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.127 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0575P)2 + 0.309P] where P = (Fo2 + 2Fc2)/3 |
| 6149 reflections | (Δ/σ)max < 0.001 |
| 273 parameters | Δρmax = 0.46 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.60501 (12) | 0.92104 (10) | 0.24135 (9) | 0.0174 (3) | |
| O2 | 0.57242 (12) | 0.86263 (10) | 0.00369 (9) | 0.0180 (3) | |
| N1 | 0.72481 (13) | 0.48753 (11) | 0.32705 (10) | 0.0136 (3) | |
| N2 | 0.88572 (13) | 0.40893 (11) | 0.21328 (10) | 0.0135 (3) | |
| C1 | 0.70264 (16) | 0.55194 (14) | 0.03650 (12) | 0.0160 (4) | |
| C2 | 0.65044 (16) | 0.64264 (14) | −0.02155 (12) | 0.0163 (4) | |
| C3 | 0.62078 (15) | 0.76560 (13) | 0.04910 (12) | 0.0144 (3) | |
| C4 | 0.63977 (16) | 0.79788 (13) | 0.18038 (12) | 0.0141 (3) | |
| C5 | 0.69306 (15) | 0.70881 (13) | 0.23773 (12) | 0.0138 (3) | |
| C6 | 0.72605 (15) | 0.58461 (13) | 0.16638 (12) | 0.0134 (3) | |
| C7 | 0.5608 (2) | 0.83768 (15) | −0.12690 (13) | 0.0229 (4) | |
| C8 | 0.64201 (19) | 0.96524 (14) | 0.37715 (13) | 0.0205 (4) | |
| C9 | 0.77680 (15) | 0.49333 (13) | 0.23350 (12) | 0.0129 (3) | |
| C10 | 0.80286 (15) | 0.39533 (13) | 0.36895 (12) | 0.0125 (3) | |
| C11 | 0.90182 (15) | 0.34498 (13) | 0.29931 (12) | 0.0129 (3) | |
| C12 | 0.97719 (16) | 0.39151 (14) | 0.12516 (13) | 0.0162 (4) | |
| C13 | 0.91694 (18) | 0.26993 (15) | 0.00627 (13) | 0.0207 (4) | |
| C14 | 0.7876 (2) | 0.18741 (17) | −0.02524 (15) | 0.0277 (5) | |
| C15 | 0.77327 (15) | 0.36358 (12) | 0.47378 (12) | 0.0127 (3) | |
| C16 | 0.62980 (16) | 0.38237 (13) | 0.49728 (12) | 0.0146 (3) | |
| C17 | 0.59989 (16) | 0.36083 (14) | 0.59963 (13) | 0.0167 (4) | |
| C18 | 0.71297 (17) | 0.31669 (14) | 0.67889 (13) | 0.0172 (4) | |
| C19 | 0.85580 (17) | 0.29638 (14) | 0.65568 (13) | 0.0167 (4) | |
| C20 | 0.88689 (16) | 0.32016 (13) | 0.55478 (12) | 0.0150 (3) | |
| C21 | 0.99892 (16) | 0.23750 (13) | 0.30191 (12) | 0.0139 (3) | |
| C22 | 0.92335 (17) | 0.10790 (14) | 0.26220 (14) | 0.0180 (4) | |
| C23 | 1.00989 (18) | 0.00593 (14) | 0.27258 (14) | 0.0207 (4) | |
| C24 | 1.17247 (17) | 0.03201 (14) | 0.32038 (13) | 0.0181 (4) | |
| C25 | 1.24860 (16) | 0.16004 (14) | 0.35776 (13) | 0.0175 (4) | |
| C26 | 1.16261 (16) | 0.26308 (14) | 0.34948 (13) | 0.0162 (3) | |
| H1 | 0.72180 | 0.46930 | −0.01200 | 0.0190* | |
| H2 | 0.63550 | 0.62000 | −0.10840 | 0.0200* | |
| H5 | 0.70730 | 0.73110 | 0.32450 | 0.0170* | |
| H8A | 0.57790 | 0.90890 | 0.39830 | 0.0310* | |
| H8B | 0.62230 | 1.05560 | 0.41010 | 0.0310* | |
| H8C | 0.75070 | 0.96090 | 0.41430 | 0.0310* | |
| H9A | 0.66190 | 0.82570 | −0.13840 | 0.0340* | |
| H9B | 0.52540 | 0.91210 | −0.14830 | 0.0340* | |
| H9C | 0.48740 | 0.75850 | −0.18160 | 0.0340* | |
| H12A | 1.08480 | 0.38680 | 0.16900 | 0.0190* | |
| H12B | 0.97800 | 0.46930 | 0.10170 | 0.0190* | |
| H13 | 0.97540 | 0.25080 | −0.05030 | 0.0250* | |
| H14A | 0.72580 | 0.20300 | 0.02870 | 0.0330* | |
| H14B | 0.75810 | 0.11360 | −0.10140 | 0.0330* | |
| H16 | 0.55270 | 0.40990 | 0.44340 | 0.0170* | |
| H17 | 0.50460 | 0.37590 | 0.61490 | 0.0200* | |
| H18 | 0.69340 | 0.30080 | 0.74690 | 0.0210* | |
| H19 | 0.93150 | 0.26650 | 0.70840 | 0.0200* | |
| H20 | 0.98340 | 0.30720 | 0.54110 | 0.0180* | |
| H22 | 0.81450 | 0.08960 | 0.22860 | 0.0220* | |
| H23 | 0.95870 | −0.08010 | 0.24740 | 0.0250* | |
| H24 | 1.23020 | −0.03640 | 0.32730 | 0.0220* | |
| H25 | 1.35770 | 0.17720 | 0.38850 | 0.0210* | |
| H26 | 1.21420 | 0.34920 | 0.37570 | 0.0190* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0249 (5) | 0.0137 (5) | 0.0147 (4) | 0.0065 (4) | 0.0063 (4) | 0.0060 (4) |
| O2 | 0.0244 (5) | 0.0182 (5) | 0.0155 (4) | 0.0069 (4) | 0.0064 (4) | 0.0108 (4) |
| N1 | 0.0149 (5) | 0.0132 (5) | 0.0146 (5) | 0.0035 (4) | 0.0050 (4) | 0.0068 (4) |
| N2 | 0.0159 (5) | 0.0133 (5) | 0.0135 (5) | 0.0036 (4) | 0.0061 (4) | 0.0064 (4) |
| C1 | 0.0183 (7) | 0.0148 (6) | 0.0152 (6) | 0.0045 (5) | 0.0061 (5) | 0.0052 (5) |
| C2 | 0.0192 (7) | 0.0179 (7) | 0.0128 (6) | 0.0030 (5) | 0.0054 (5) | 0.0064 (5) |
| C3 | 0.0138 (6) | 0.0155 (6) | 0.0155 (6) | 0.0023 (5) | 0.0034 (5) | 0.0085 (5) |
| C4 | 0.0143 (6) | 0.0132 (6) | 0.0154 (6) | 0.0022 (5) | 0.0048 (5) | 0.0057 (5) |
| C5 | 0.0146 (6) | 0.0154 (6) | 0.0126 (5) | 0.0024 (5) | 0.0039 (5) | 0.0067 (5) |
| C6 | 0.0132 (6) | 0.0140 (6) | 0.0144 (6) | 0.0024 (5) | 0.0042 (5) | 0.0069 (5) |
| C7 | 0.0328 (8) | 0.0236 (8) | 0.0159 (6) | 0.0058 (6) | 0.0071 (6) | 0.0117 (6) |
| C8 | 0.0295 (8) | 0.0180 (7) | 0.0150 (6) | 0.0078 (6) | 0.0081 (6) | 0.0058 (5) |
| C9 | 0.0130 (6) | 0.0123 (6) | 0.0130 (5) | 0.0026 (5) | 0.0035 (5) | 0.0047 (5) |
| C10 | 0.0134 (6) | 0.0113 (6) | 0.0132 (5) | 0.0024 (5) | 0.0039 (5) | 0.0050 (5) |
| C11 | 0.0144 (6) | 0.0112 (6) | 0.0133 (5) | 0.0022 (5) | 0.0043 (5) | 0.0046 (5) |
| C12 | 0.0178 (6) | 0.0171 (7) | 0.0176 (6) | 0.0054 (5) | 0.0094 (5) | 0.0078 (5) |
| C13 | 0.0272 (8) | 0.0202 (7) | 0.0177 (6) | 0.0065 (6) | 0.0123 (6) | 0.0063 (6) |
| C14 | 0.0337 (9) | 0.0261 (8) | 0.0203 (7) | 0.0011 (7) | 0.0109 (7) | 0.0032 (6) |
| C15 | 0.0153 (6) | 0.0100 (6) | 0.0128 (5) | 0.0015 (5) | 0.0047 (5) | 0.0041 (5) |
| C16 | 0.0157 (6) | 0.0138 (6) | 0.0158 (6) | 0.0043 (5) | 0.0056 (5) | 0.0067 (5) |
| C17 | 0.0171 (6) | 0.0170 (7) | 0.0184 (6) | 0.0033 (5) | 0.0086 (5) | 0.0071 (5) |
| C18 | 0.0230 (7) | 0.0157 (6) | 0.0146 (6) | 0.0016 (5) | 0.0070 (5) | 0.0069 (5) |
| C19 | 0.0197 (7) | 0.0150 (6) | 0.0150 (6) | 0.0033 (5) | 0.0030 (5) | 0.0070 (5) |
| C20 | 0.0148 (6) | 0.0145 (6) | 0.0164 (6) | 0.0038 (5) | 0.0052 (5) | 0.0060 (5) |
| C21 | 0.0168 (6) | 0.0133 (6) | 0.0134 (5) | 0.0048 (5) | 0.0064 (5) | 0.0056 (5) |
| C22 | 0.0151 (6) | 0.0157 (7) | 0.0229 (7) | 0.0029 (5) | 0.0051 (5) | 0.0072 (6) |
| C23 | 0.0237 (7) | 0.0124 (6) | 0.0264 (7) | 0.0040 (6) | 0.0082 (6) | 0.0073 (6) |
| C24 | 0.0219 (7) | 0.0167 (7) | 0.0187 (6) | 0.0097 (6) | 0.0084 (5) | 0.0079 (5) |
| C25 | 0.0143 (6) | 0.0200 (7) | 0.0188 (6) | 0.0053 (5) | 0.0045 (5) | 0.0083 (6) |
| C26 | 0.0170 (6) | 0.0134 (6) | 0.0175 (6) | 0.0022 (5) | 0.0053 (5) | 0.0051 (5) |
Geometric parameters (Å, º)
| O1—C4 | 1.3685 (18) | C22—C23 | 1.388 (2) |
| O1—C8 | 1.4286 (17) | C23—C24 | 1.387 (2) |
| O2—C3 | 1.3618 (18) | C24—C25 | 1.384 (2) |
| O2—C7 | 1.4321 (17) | C25—C26 | 1.390 (2) |
| N1—C9 | 1.3250 (18) | C1—H1 | 0.9300 |
| N1—C10 | 1.3835 (19) | C2—H2 | 0.9300 |
| N2—C9 | 1.3721 (19) | C5—H5 | 0.9300 |
| N2—C11 | 1.3854 (18) | C7—H9A | 0.9600 |
| N2—C12 | 1.4583 (18) | C7—H9B | 0.9600 |
| C1—C2 | 1.398 (2) | C7—H9C | 0.9600 |
| C1—C6 | 1.3899 (18) | C8—H8A | 0.9600 |
| C2—C3 | 1.381 (2) | C8—H8B | 0.9600 |
| C3—C4 | 1.4134 (18) | C8—H8C | 0.9600 |
| C4—C5 | 1.379 (2) | C12—H12A | 0.9700 |
| C5—C6 | 1.406 (2) | C12—H12B | 0.9700 |
| C6—C9 | 1.470 (2) | C13—H13 | 0.9300 |
| C10—C11 | 1.3721 (19) | C14—H14A | 0.9300 |
| C10—C15 | 1.4712 (19) | C14—H14B | 0.9300 |
| C11—C21 | 1.478 (2) | C16—H16 | 0.9300 |
| C12—C13 | 1.491 (2) | C17—H17 | 0.9300 |
| C13—C14 | 1.318 (3) | C18—H18 | 0.9300 |
| C15—C16 | 1.397 (2) | C19—H19 | 0.9300 |
| C15—C20 | 1.3991 (19) | C20—H20 | 0.9300 |
| C16—C17 | 1.391 (2) | C22—H22 | 0.9300 |
| C17—C18 | 1.389 (2) | C23—H23 | 0.9300 |
| C18—C19 | 1.392 (2) | C24—H24 | 0.9300 |
| C19—C20 | 1.390 (2) | C25—H25 | 0.9300 |
| C21—C22 | 1.393 (2) | C26—H26 | 0.9300 |
| C21—C26 | 1.396 (2) | ||
| C4—O1—C8 | 116.86 (11) | C1—C2—H2 | 120.00 |
| C3—O2—C7 | 116.81 (12) | C3—C2—H2 | 120.00 |
| C9—N1—C10 | 105.70 (12) | C4—C5—H5 | 120.00 |
| C9—N2—C11 | 106.90 (11) | C6—C5—H5 | 120.00 |
| C9—N2—C12 | 128.55 (12) | O2—C7—H9A | 109.00 |
| C11—N2—C12 | 124.46 (12) | O2—C7—H9B | 109.00 |
| C2—C1—C6 | 120.37 (13) | O2—C7—H9C | 110.00 |
| C1—C2—C3 | 120.52 (12) | H9A—C7—H9B | 109.00 |
| O2—C3—C2 | 125.76 (12) | H9A—C7—H9C | 109.00 |
| O2—C3—C4 | 114.84 (12) | H9B—C7—H9C | 109.00 |
| C2—C3—C4 | 119.40 (13) | O1—C8—H8A | 109.00 |
| O1—C4—C3 | 114.91 (12) | O1—C8—H8B | 109.00 |
| O1—C4—C5 | 125.16 (12) | O1—C8—H8C | 109.00 |
| C3—C4—C5 | 119.92 (13) | H8A—C8—H8B | 110.00 |
| C4—C5—C6 | 120.68 (12) | H8A—C8—H8C | 109.00 |
| C1—C6—C5 | 119.06 (13) | H8B—C8—H8C | 109.00 |
| C1—C6—C9 | 123.53 (13) | N2—C12—H12A | 109.00 |
| C5—C6—C9 | 117.34 (11) | N2—C12—H12B | 109.00 |
| N1—C9—N2 | 111.38 (12) | C13—C12—H12A | 109.00 |
| N1—C9—C6 | 122.61 (13) | C13—C12—H12B | 109.00 |
| N2—C9—C6 | 125.96 (12) | H12A—C12—H12B | 108.00 |
| N1—C10—C11 | 110.18 (12) | C12—C13—H13 | 117.00 |
| N1—C10—C15 | 120.50 (12) | C14—C13—H13 | 117.00 |
| C11—C10—C15 | 129.32 (13) | C13—C14—H14A | 120.00 |
| N2—C11—C10 | 105.85 (12) | C13—C14—H14B | 120.00 |
| N2—C11—C21 | 124.07 (12) | H14A—C14—H14B | 120.00 |
| C10—C11—C21 | 129.91 (13) | C15—C16—H16 | 119.00 |
| N2—C12—C13 | 113.96 (13) | C17—C16—H16 | 119.00 |
| C12—C13—C14 | 125.34 (15) | C16—C17—H17 | 120.00 |
| C10—C15—C16 | 119.34 (12) | C18—C17—H17 | 120.00 |
| C10—C15—C20 | 122.15 (13) | C17—C18—H18 | 120.00 |
| C16—C15—C20 | 118.45 (12) | C19—C18—H18 | 120.00 |
| C15—C16—C17 | 121.38 (13) | C18—C19—H19 | 120.00 |
| C16—C17—C18 | 119.72 (14) | C20—C19—H19 | 120.00 |
| C17—C18—C19 | 119.41 (13) | C15—C20—H20 | 120.00 |
| C18—C19—C20 | 120.92 (14) | C19—C20—H20 | 120.00 |
| C15—C20—C19 | 120.11 (14) | C21—C22—H22 | 120.00 |
| C11—C21—C22 | 118.45 (13) | C23—C22—H22 | 120.00 |
| C11—C21—C26 | 122.14 (13) | C22—C23—H23 | 120.00 |
| C22—C21—C26 | 119.31 (14) | C24—C23—H23 | 120.00 |
| C21—C22—C23 | 120.26 (14) | C23—C24—H24 | 120.00 |
| C22—C23—C24 | 120.17 (15) | C25—C24—H24 | 120.00 |
| C23—C24—C25 | 119.91 (15) | C24—C25—H25 | 120.00 |
| C24—C25—C26 | 120.24 (14) | C26—C25—H25 | 120.00 |
| C21—C26—C25 | 120.08 (14) | C21—C26—H26 | 120.00 |
| C2—C1—H1 | 120.00 | C25—C26—H26 | 120.00 |
| C6—C1—H1 | 120.00 | ||
| C8—O1—C4—C3 | −171.98 (13) | C5—C6—C9—N2 | −143.78 (14) |
| C8—O1—C4—C5 | 6.6 (2) | C1—C6—C9—N1 | −143.64 (15) |
| C7—O2—C3—C4 | 176.30 (13) | C5—C6—C9—N1 | 33.3 (2) |
| C7—O2—C3—C2 | −4.1 (2) | N1—C10—C11—N2 | 0.70 (15) |
| C10—N1—C9—N2 | −0.18 (15) | C11—C10—C15—C20 | 28.2 (2) |
| C9—N1—C10—C11 | −0.34 (15) | N1—C10—C15—C20 | −152.15 (13) |
| C10—N1—C9—C6 | −177.67 (12) | C11—C10—C15—C16 | −154.50 (15) |
| C9—N1—C10—C15 | 179.97 (11) | C15—C10—C11—C21 | 5.1 (2) |
| C12—N2—C11—C21 | −8.3 (2) | N1—C10—C15—C16 | 25.13 (19) |
| C11—N2—C9—C6 | 178.00 (13) | N1—C10—C11—C21 | −174.60 (13) |
| C9—N2—C11—C21 | 174.87 (13) | C15—C10—C11—N2 | −179.65 (13) |
| C12—N2—C11—C10 | 176.06 (12) | N2—C11—C21—C22 | −109.09 (16) |
| C11—N2—C9—N1 | 0.61 (15) | C10—C11—C21—C26 | −110.82 (18) |
| C12—N2—C9—N1 | −176.05 (13) | N2—C11—C21—C26 | 74.64 (18) |
| C11—N2—C12—C13 | 81.73 (17) | C10—C11—C21—C22 | 65.5 (2) |
| C9—N2—C12—C13 | −102.15 (17) | N2—C12—C13—C14 | 4.5 (2) |
| C9—N2—C11—C10 | −0.78 (15) | C10—C15—C16—C17 | −176.38 (13) |
| C12—N2—C9—C6 | 1.3 (2) | C20—C15—C16—C17 | 1.0 (2) |
| C2—C1—C6—C9 | 178.37 (14) | C10—C15—C20—C19 | 177.46 (13) |
| C2—C1—C6—C5 | 1.4 (2) | C16—C15—C20—C19 | 0.2 (2) |
| C6—C1—C2—C3 | −0.3 (2) | C15—C16—C17—C18 | −1.5 (2) |
| C1—C2—C3—C4 | −1.6 (2) | C16—C17—C18—C19 | 0.9 (2) |
| C1—C2—C3—O2 | 178.77 (14) | C17—C18—C19—C20 | 0.3 (2) |
| C2—C3—C4—C5 | 2.4 (2) | C18—C19—C20—C15 | −0.8 (2) |
| O2—C3—C4—O1 | 0.68 (19) | C11—C21—C22—C23 | −174.93 (13) |
| O2—C3—C4—C5 | −177.97 (13) | C26—C21—C22—C23 | 1.5 (2) |
| C2—C3—C4—O1 | −178.96 (13) | C11—C21—C26—C25 | 175.78 (13) |
| C3—C4—C5—C6 | −1.2 (2) | C22—C21—C26—C25 | −0.5 (2) |
| O1—C4—C5—C6 | −179.74 (13) | C21—C22—C23—C24 | −1.2 (2) |
| C4—C5—C6—C1 | −0.7 (2) | C22—C23—C24—C25 | 0.0 (2) |
| C4—C5—C6—C9 | −177.80 (13) | C23—C24—C25—C26 | 1.0 (2) |
| C1—C6—C9—N2 | 39.2 (2) | C24—C25—C26—C21 | −0.8 (2) |
Hydrogen-bond geometry (Å, º)
Cg3 is a centroid of the C15–C20 phenyl ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H9B···O1i | 0.96 | 2.57 | 3.515 (2) | 170 |
| C14—H14A···N2 | 0.93 | 2.53 | 2.859 (2) | 101 |
| C8—H8B···Cg3ii | 0.96 | 2.98 | 3.8316 (17) | 149 |
Symmetry codes: (i) −x+1, −y+2, −z; (ii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT6835).
References
- Adams, J. L., Boehm, J. C., Gallagher, T. F., Kassis, S., Webb, E. F., Hall, R., Sorenson, M., Garigipati, R., Don, E., Griswold, D. E. & Lee, J. C. (2001). Bioorg. Med. Chem. Lett. 11, 2867–2870. [DOI] [PubMed]
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst. 32, 115–119.
- Bruker (2005). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Congiu, C., Cocco, M. T. & Onnis, V. (2008). Bioorg. Med. Chem. Lett. 18, 989–993. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Nakamura, T., Kakinuma, H., Umemiya, H., Amada, H., Miyata, N., Taniguchi, K., Bando, K. & Sato, M. (2004). Bioorg. Med. Chem. Lett. 14, 333–336. [DOI] [PubMed]
- Nanterment, P. G., Barrow, J. C., Lindsley, S. R., Young, M., Mao, S., Carroll, S., Bailey, C., Bosserman, M., Colussi, D., McMasters, D. R., Vacca, J. P. & Selnick, H. G. (2004). Bioorg. Med. Chem. Lett. 14, 2141–2145. [DOI] [PubMed]
- Nardelli, M. (1983). Comput. Chem. 7, 95–98.
- Roman, G., Riley, J. G., Vlahakis, J. Z., Kinobe, R. T., Brien, J. F., Nakatsu, K. & Szarek, W. A. (2007). Bioorg. Med. Chem. 15, 3225–3234. [DOI] [PubMed]
- Shalini, K., Sharma, P. K. & Kumar, N. (2010). Chem. Sin. 1, 36-47.
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Venkatesan, A. M., Agarwal, A., Abe, T., Ushirogochi, H. O., Santos, D., Li, Z., Francisco, G., Lin, Y. I., Peterson, P. J., Yang, Y., Weiss, W. J., Shales, D. M. & Mansour, T. S. (2008). Bioorg. Med. Chem. 16, 1890–1902. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812039566/bt6835sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039566/bt6835Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812039566/bt6835Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report