Abstract
In the title compound, C9H8O3, the molecular skeleton is almost planar [r.m.s. deviation = 0.016 (2) Å]. Weak intermolecular C—H⋯O and C—H⋯π interactions consolidate the crystal packing, with the molecules stacking in the [101] direction.
Related literature
For the biological activity of isobenzofuran-1(3H)-one, see: Brady et al. (2000 ▶); Huang et al. (2012) ▶; Cardozo et al. (2005 ▶); Yoganathan et al. (2003 ▶); Demuner et al. (2006 ▶). For related structures, see: Sun et al. (2009 ▶); Mendenhall et al. (2003 ▶).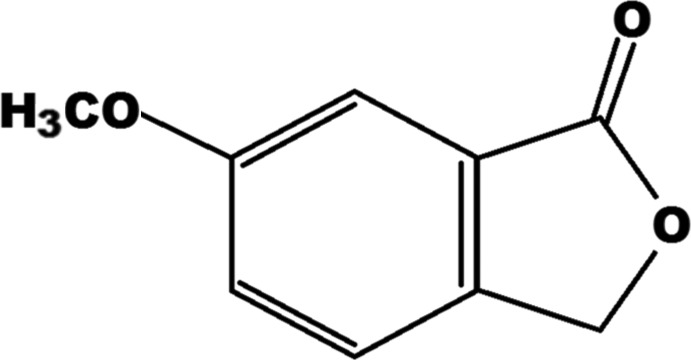
Experimental
Crystal data
C9H8O3
M r = 164.15
Monoclinic,
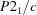
a = 9.2922 (19) Å
b = 8.4982 (12) Å
c = 9.786 (2) Å
β = 90.471 (15)°
V = 772.8 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 293 K
0.28 × 0.17 × 0.12 mm
Data collection
Nonius KappaCCD diffractometer
3304 measured reflections
1741 independent reflections
1285 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.050
wR(F 2) = 0.166
S = 1.12
1741 reflections
109 parameters
H-atom parameters constrained
Δρmax = 0.20 e Å−3
Δρmin = −0.18 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: DENZO-SMN (Otwinowski & Minor, 1997 ▶); data reduction: DENZO-SMN; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812039074/cv5338sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039074/cv5338Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812039074/cv5338Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of C2–C7 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C9—H9B⋯O2i | 0.96 | 2.55 | 3.490 (2) | 165 |
| C8—H8B⋯Cg1ii | 0.97 | 2.84 | 3.637 (2) | 140 |
| C9—H9C⋯Cg1iii | 0.96 | 2.91 | 3.744 (2) | 146 |
Symmetry codes: (i) 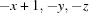 ; (ii)
; (ii) 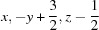 ; (iii)
; (iii) 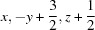 .
.
Acknowledgments
The authors thank Professor Dr Javier Ellena of the IFSC, USP, Brazil, for the X-ray data collection. This work was supported financially by CAPES, CNPq, FUNARBE and FAPEMIG. This work is also a collaborative research project with members of the Rede Mineira de Química (RQ—MG), also supported by FAPEMIG.
supplementary crystallographic information
Comment
Isobenzofuran-1(3H)-one fragment, γ-lactone fused to aromatic ring, occurs in several compounds that exhibit biological activities ranging from antibacterial (Brady et al., 2000), antioxidant (Zhu et al., 2012), anticonvulsant (Cardozo et al., 2005), and anti-HIV (Yoganathan et al., 2003) to inhibition of the photosynthetic electron transport (Demuner et al., 2006). In a research program devoted to finding the phytotoxic compounds containing the isobenzofuran-1(3H)-one core, the title compound was synthesized.
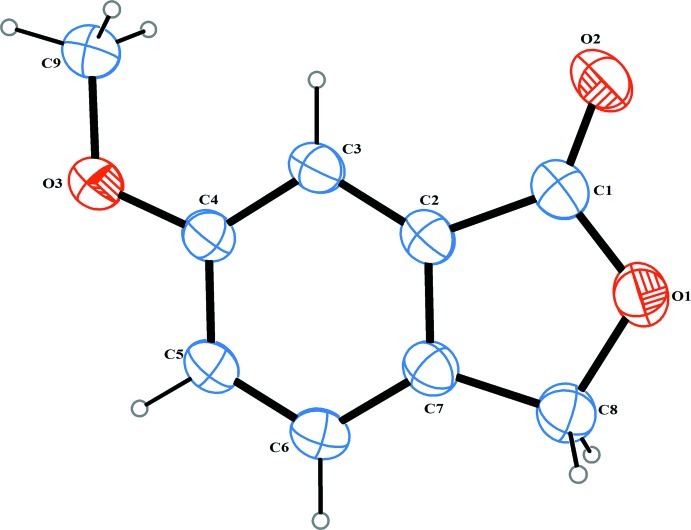
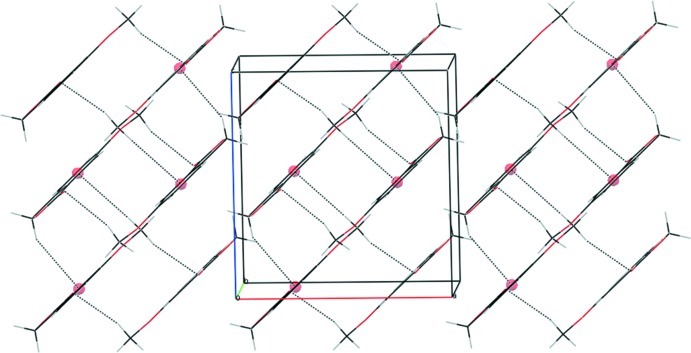
The title molecule (Fig. 1) is essentially planar with a mean deviation of 0.016 (2) Å from the best plane formed by 12 non-H atoms. All bond lengths and angles are normal and correspond to those observed in the related compounds (Sun et al., 2009; Mendenhall et al., 2003). The crystal packing is stabilized by weak intermolecular C—H···O and C—H···π interactions (Table 1, Fig. 2), with a stacking direction of the molecules in [101].
Experimental
Starting materials were commercially available and were used without further purification. The synthesis of the title compound was carried out as follows. A tube of 40 ml equipped with a magnetic stir bar was charged with Palladium (II) acetate (156.8 mg, 0.70 mmol), potassium dihydrogen phosphate (3654 mg, 21.0 mmol), 3-methoxy benzoic acid (1064 mg, 7.00 mmol) and dibromomethane (28 ml). The tube was sealed with a teflon cap and the reaction mixture was stirred at 140 °C for 36 h. After this time, the mixture was filtered through a pad of celite. The filtrate was concentrated under reduced pressure and the residue was purified by silica gel column chromatography (hexane-ethyl acetate 2:1 v/v) to afford the title compound in 59% yield (681 mg, 4.15 mmol) as a white solid. The crystals suitable for X-ray crystallographic analysis were obtained by slow evaporation from acetone solution at room temperature. M.p. 116.9–118.4 °C. IR (selected bands, cm-1): 3003, 2925, 2837, 1733, 1600, 1585, 1488, 1454, 1431, 1267, 1206, 1028, 973, 869, 749, 690, 545. 1H NMR (300 MHz, MeOH-d4) δ 3.87 (s, 3H, H-9), 5.26 (s, 2H, H-8), 7.22–7.38 (m, 3H, H-3, H-5 and H-6); 13C NMR (75 MHz, MeOH-d4) δ 56.0 (C-9), 69.7 (C-8), 107.7 (C-3), 123.1; 123.3 (C-5 and C-6), 127.3 (C-2), 139.1 (C-7), 160.8 (C-4), 171.4 (C-1). HREIMS m/z (M+H+): Calcd for C9H8O3, 165.0552; found: 165.0608.
Figures
Fig. 1.
The molecular structure of the title compound, showing the atom labeling and displacement ellipsoids at the 30% probability level.
Fig. 2.
A portion of the crystal packing viewed approximately down the c axis. Dotted lines denote intermolecular C—H···O and C—H···π interactions. Centroids of six-membered rings are drawn as red balls.
Crystal data
| C9H8O3 | F(000) = 344 |
| Mr = 164.15 | Dx = 1.411 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.2922 (19) Å | Cell parameters from 3082 reflections |
| b = 8.4982 (12) Å | θ = 2.1–27.5° |
| c = 9.786 (2) Å | µ = 0.11 mm−1 |
| β = 90.471 (15)° | T = 293 K |
| V = 772.8 (2) Å3 | Prism, colourless |
| Z = 4 | 0.28 × 0.17 × 0.12 mm |
Data collection
| Nonius KappaCCD diffractometer | 1285 reflections with I > 2σ(I) |
| Radiation source: Enraf Nonius FR590 | Rint = 0.036 |
| Graphite monochromator | θmax = 27.5°, θmin = 2.2° |
| Detector resolution: 9 pixels mm-1 | h = −12→12 |
| CCD rotation images, thick slices scans | k = −10→11 |
| 3304 measured reflections | l = −12→12 |
| 1741 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.166 | H-atom parameters constrained |
| S = 1.12 | w = 1/[σ2(Fo2) + (0.0943P)2 + 0.032P] where P = (Fo2 + 2Fc2)/3 |
| 1741 reflections | (Δ/σ)max < 0.001 |
| 109 parameters | Δρmax = 0.20 e Å−3 |
| 0 restraints | Δρmin = −0.18 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.93834 (12) | 0.04009 (14) | 0.16871 (11) | 0.0695 (4) | |
| O2 | 0.79489 (14) | −0.11746 (14) | 0.04458 (12) | 0.0752 (4) | |
| O3 | 0.54600 (12) | 0.40633 (13) | −0.19372 (11) | 0.0699 (4) | |
| C1 | 0.83141 (17) | 0.0134 (2) | 0.07489 (15) | 0.0612 (4) | |
| C2 | 0.77893 (16) | 0.16711 (17) | 0.02593 (13) | 0.0552 (4) | |
| C3 | 0.67205 (15) | 0.19885 (18) | −0.07089 (14) | 0.0569 (4) | |
| H3 | 0.6212 | 0.1185 | −0.114 | 0.068* | |
| C4 | 0.64497 (16) | 0.35569 (18) | −0.09995 (14) | 0.0568 (4) | |
| C5 | 0.72174 (17) | 0.47477 (18) | −0.03217 (16) | 0.0632 (4) | |
| H5 | 0.7013 | 0.5794 | −0.0524 | 0.076* | |
| C6 | 0.82660 (17) | 0.44069 (19) | 0.06357 (16) | 0.0639 (4) | |
| H6 | 0.8766 | 0.5208 | 0.108 | 0.077* | |
| C7 | 0.85594 (16) | 0.28364 (18) | 0.09213 (14) | 0.0572 (4) | |
| C8 | 0.96195 (18) | 0.2080 (2) | 0.18656 (16) | 0.0663 (5) | |
| H8A | 1.0596 | 0.2365 | 0.1624 | 0.08* | |
| H8B | 0.9448 | 0.2392 | 0.2804 | 0.08* | |
| C9 | 0.45913 (19) | 0.2893 (2) | −0.25875 (18) | 0.0730 (5) | |
| H9A | 0.3942 | 0.3389 | −0.3222 | 0.11* | |
| H9B | 0.4051 | 0.2336 | −0.191 | 0.11* | |
| H9C | 0.5197 | 0.2169 | −0.3067 | 0.11* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0805 (7) | 0.0576 (7) | 0.0702 (7) | 0.0088 (5) | −0.0046 (5) | 0.0056 (5) |
| O2 | 0.0934 (9) | 0.0475 (7) | 0.0847 (8) | 0.0000 (5) | 0.0030 (6) | 0.0033 (5) |
| O3 | 0.0797 (7) | 0.0492 (7) | 0.0804 (7) | −0.0024 (5) | −0.0172 (6) | 0.0022 (5) |
| C1 | 0.0700 (9) | 0.0536 (9) | 0.0602 (8) | 0.0018 (7) | 0.0085 (7) | 0.0018 (6) |
| C2 | 0.0635 (8) | 0.0479 (8) | 0.0541 (7) | 0.0004 (6) | 0.0070 (6) | 0.0000 (6) |
| C3 | 0.0651 (8) | 0.0457 (8) | 0.0599 (8) | −0.0047 (6) | 0.0027 (6) | −0.0034 (6) |
| C4 | 0.0635 (8) | 0.0480 (8) | 0.0588 (8) | −0.0014 (6) | −0.0003 (6) | −0.0001 (6) |
| C5 | 0.0757 (9) | 0.0428 (8) | 0.0708 (9) | 0.0005 (6) | −0.0035 (7) | −0.0013 (6) |
| C6 | 0.0734 (9) | 0.0496 (8) | 0.0686 (9) | −0.0049 (7) | −0.0032 (7) | −0.0081 (7) |
| C7 | 0.0633 (8) | 0.0524 (9) | 0.0561 (7) | −0.0001 (6) | 0.0046 (6) | −0.0021 (6) |
| C8 | 0.0728 (9) | 0.0610 (10) | 0.0649 (9) | 0.0037 (7) | −0.0022 (7) | −0.0042 (7) |
| C9 | 0.0790 (10) | 0.0593 (10) | 0.0805 (10) | −0.0097 (8) | −0.0147 (8) | 0.0019 (8) |
Geometric parameters (Å, º)
| O1—C1 | 1.366 (2) | C5—C6 | 1.377 (2) |
| O1—C8 | 1.454 (2) | C5—H5 | 0.93 |
| O2—C1 | 1.199 (2) | C6—C7 | 1.390 (2) |
| O3—C4 | 1.3635 (19) | C6—H6 | 0.93 |
| O3—C9 | 1.427 (2) | C7—C8 | 1.491 (2) |
| C1—C2 | 1.473 (2) | C8—H8A | 0.97 |
| C2—C7 | 1.380 (2) | C8—H8B | 0.97 |
| C2—C3 | 1.393 (2) | C9—H9A | 0.96 |
| C3—C4 | 1.385 (2) | C9—H9B | 0.96 |
| C3—H3 | 0.93 | C9—H9C | 0.96 |
| C4—C5 | 1.402 (2) | ||
| C1—O1—C8 | 110.59 (12) | C5—C6—H6 | 120.8 |
| C4—O3—C9 | 117.14 (13) | C7—C6—H6 | 120.8 |
| O2—C1—O1 | 121.50 (16) | C2—C7—C6 | 119.63 (15) |
| O2—C1—C2 | 130.52 (17) | C2—C7—C8 | 108.60 (14) |
| O1—C1—C2 | 107.98 (14) | C6—C7—C8 | 131.77 (14) |
| C7—C2—C3 | 122.97 (15) | O1—C8—C7 | 104.50 (12) |
| C7—C2—C1 | 108.34 (15) | O1—C8—H8A | 110.9 |
| C3—C2—C1 | 128.69 (14) | C7—C8—H8A | 110.9 |
| C4—C3—C2 | 116.94 (14) | O1—C8—H8B | 110.9 |
| C4—C3—H3 | 121.5 | C7—C8—H8B | 110.9 |
| C2—C3—H3 | 121.5 | H8A—C8—H8B | 108.9 |
| O3—C4—C3 | 124.20 (14) | O3—C9—H9A | 109.5 |
| O3—C4—C5 | 115.37 (14) | O3—C9—H9B | 109.5 |
| C3—C4—C5 | 120.43 (15) | H9A—C9—H9B | 109.5 |
| C6—C5—C4 | 121.64 (15) | O3—C9—H9C | 109.5 |
| C6—C5—H5 | 119.2 | H9A—C9—H9C | 109.5 |
| C4—C5—H5 | 119.2 | H9B—C9—H9C | 109.5 |
| C5—C6—C7 | 118.38 (15) | ||
| C8—O1—C1—O2 | 179.83 (13) | O3—C4—C5—C6 | −178.97 (12) |
| C8—O1—C1—C2 | −0.01 (16) | C3—C4—C5—C6 | 0.7 (2) |
| O2—C1—C2—C7 | −179.93 (15) | C4—C5—C6—C7 | 0.2 (2) |
| O1—C1—C2—C7 | −0.11 (16) | C3—C2—C7—C6 | 0.6 (2) |
| O2—C1—C2—C3 | −0.6 (3) | C1—C2—C7—C6 | 179.99 (12) |
| O1—C1—C2—C3 | 179.23 (12) | C3—C2—C7—C8 | −179.21 (12) |
| C7—C2—C3—C4 | 0.2 (2) | C1—C2—C7—C8 | 0.18 (16) |
| C1—C2—C3—C4 | −179.01 (12) | C5—C6—C7—C2 | −0.8 (2) |
| C9—O3—C4—C3 | 4.2 (2) | C5—C6—C7—C8 | 178.95 (14) |
| C9—O3—C4—C5 | −176.13 (13) | C1—O1—C8—C7 | 0.11 (15) |
| C2—C3—C4—O3 | 178.74 (12) | C2—C7—C8—O1 | −0.18 (15) |
| C2—C3—C4—C5 | −0.9 (2) | C6—C7—C8—O1 | −179.96 (14) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of C2–C7 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C9—H9B···O2i | 0.96 | 2.55 | 3.490 (2) | 165 |
| C8—H8B···Cg1ii | 0.97 | 2.84 | 3.637 (2) | 140 |
| C9—H9C···Cg1iii | 0.96 | 2.91 | 3.744 (2) | 146 |
Symmetry codes: (i) −x+1, −y, −z; (ii) x, −y+3/2, z−1/2; (iii) x, −y+3/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5338).
References
- Brady, S. F., Wagenaar, M. M., Sing, M. P., Janson, J. E. & Clardy, J. (2000). Org. Lett. 2, 4043–4046. [DOI] [PubMed]
- Cardozo, J. A., Filho, R. B., Velandia, J. R. & Pabón, M. F. G. (2005). Rev. Col. Cienc. Quim. Farm. 34, 69–79.
- Demuner, A. J., Barbosa, L. C. A., Veiga, T. A. M., Barreto, R. W., King-Diaz, B. & Lotina-Hennsen, B. (2006). Biochem. Syst. Ecol. 34, 790–795.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Huang, X.-Z., Zhu, Y., Guan, X.-L., Tian, K., Guo, J.-M., Wang, H.-B. & Fu, G.-M. (2012). Molecules, 17, 4219–4224. [DOI] [PMC free article] [PubMed]
- Mendenhall, G. D., Luck, R. L., Bohn, R. K. & Castejon, H. J. (2003). J. Mol. Struct. 645, 249–258.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sun, M.-X., Li, X., Liu, W.-Y. & Xiao, K. (2009). Acta Cryst. E65, o2146. [DOI] [PMC free article] [PubMed]
- Yoganathan, K., Rossant, C., Ng, S., Huang, Y., Butler, M. S. & Buss, A. D. (2003). J. Nat. Prod. 66, 1116–1117. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812039074/cv5338sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039074/cv5338Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812039074/cv5338Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report