Abstract
In the title molecule, C24H20N2, the pyrazine ring is significantly distorted from planarity, presumably due to steric crowding, and its conformation is well described as a flattened twist-boat. The benzene ring adjacent to the ethyl group forms dihedral angles of 53.79 (13) and 85.47 (12)° with the other benzene rings; the dihedral angle between adjacent benzene rings is 57.90 (12)°. The ethyl group is disordered over two positions; the site-occupancy factor of the major component is 0.546 (4). No hydrogen bonds are found in the crystal structure.
Related literature
For the biological properties of pyrazines and for a closely related crystal structure, see: Anuradha et al. (2009 ▶).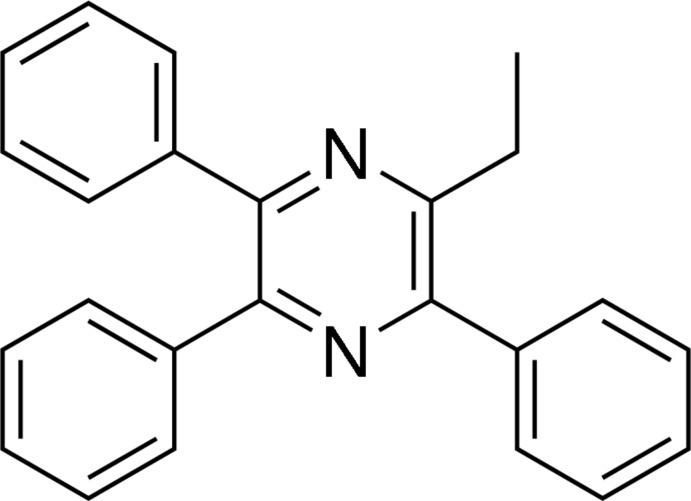
Experimental
Crystal data
C24H20N2
M r = 336.42
Triclinic,
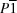
a = 9.2327 (9) Å
b = 9.8708 (11) Å
c = 10.6787 (14) Å
α = 79.604 (10)°
β = 70.351 (11)°
γ = 87.848 (8)°
V = 901.20 (19) Å3
Z = 2
Cu Kα radiation
μ = 0.56 mm−1
T = 123 K
0.44 × 0.37 × 0.24 mm
Data collection
Agilent Xcalibur Ruby Gemini diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2012 ▶) T min = 0.845, T max = 1.000
5769 measured reflections
3576 independent reflections
2622 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.063
wR(F 2) = 0.198
S = 1.05
3576 reflections
244 parameters
H-atom parameters constrained
Δρmax = 0.29 e Å−3
Δρmin = −0.21 e Å−3
Data collection: CrysAlis PRO (Agilent, 2012 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812039827/tk5151sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039827/tk5151Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812039827/tk5151Isup3.cdx
Supplementary material file. DOI: 10.1107/S1600536812039827/tk5151Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
RJB acknowledges the NSF MRI program (grant No. CHE-0619278) for funds to purchase an X-ray diffractometer.
supplementary crystallographic information
Comment
As part of our investigations of pyrazine derivatives (Anuradha et al., 2009) to compare their chemical and biological activities, we have undertaken the X-ray crystal structure analysis of the title compound.
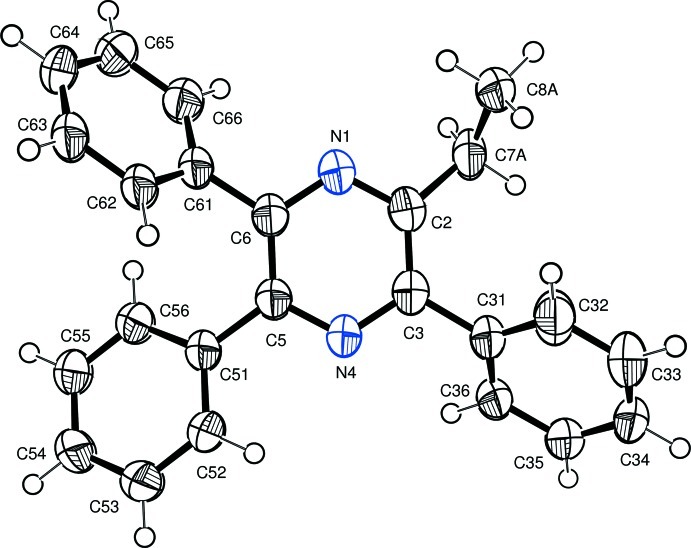
In the title molecule, Fig.1, the pyrazine ring adopts a flattened twist-boat conformation. The phenyl ring at position 5 makes a dihedral angle of 53.79 (13)° and 57.90 (12)° with the phenyl rings at position 3 and 6 respectively. The dihedral angle between the phenyl rings at positions 3 and 6 is 85.47 (12)°. The ethyl group is found to be disordered over two positions; the site occupancy factors refined to 0.546 (4) and 0.454 (4). No classical hydrogen bonds are found in the crystal structure.
Experimental
To a homogeneous solution of benzil (1.05 g, 0.005 mol) and 1-ethyl-2-phenyl-1,2-ethanediaminedihydrochloride (1.45 g, 0.005 mol) in ethanol (20 ml), sodium acetate trihydrate (2.04 g, 0.015 mol) was added. The precipitated sodium chloride was filtered off and the filtrate was refluxed for 2 h. On completion of the reaction, as indicated by TLC, the reaction mixture was poured into crushed ice and the resulting solid was filtered and purified by column chromatography on silica gel. Elution with benzene–petroleum ether (3:2 v/v) at 333–353 K gave the pure product. Yield 1.54 g (70%). The pure product was recrystallized in ethyl acetate, to obtain crystals suitable for X-ray diffraction studies.
Refinement
The H atoms were positioned geometrically and allowed to ride on their parent atoms, with C—H = 0.95–0.99 Å , and with Uiso(H) = 1.2–1.5Ueq(C). The ethyl group is found to be disordered over two positions. The anisotropic displacement parameters of equivalent atoms were constrained to be equal; the site occupancy factors refined to 0.546 (4) and 0.454 (4).
Figures
Fig. 1.
The molecular structure of the title compound, with displacement ellipsoids drawn at the 50% probability level. H atoms are shown as small spheres of arbitrary radius. Only the major disorder component of ethyl group is shown.
Crystal data
| C24H20N2 | Z = 2 |
| Mr = 336.42 | F(000) = 356 |
| Triclinic, P1 | Dx = 1.240 Mg m−3 |
| Hall symbol: -P 1 | Melting point: 423 K |
| a = 9.2327 (9) Å | Cu Kα radiation, λ = 1.54184 Å |
| b = 9.8708 (11) Å | Cell parameters from 1596 reflections |
| c = 10.6787 (14) Å | θ = 4.6–76.1° |
| α = 79.604 (10)° | µ = 0.56 mm−1 |
| β = 70.351 (11)° | T = 123 K |
| γ = 87.848 (8)° | Prism, colourless |
| V = 901.20 (19) Å3 | 0.44 × 0.37 × 0.24 mm |
Data collection
| Agilent Xcalibur Ruby Gemini diffractometer | 3576 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 2622 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.027 |
| Detector resolution: 10.5081 pixels mm-1 | θmax = 76.3°, θmin = 4.6° |
| ω scans | h = −11→11 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2012) | k = −6→12 |
| Tmin = 0.845, Tmax = 1.000 | l = −12→13 |
| 5769 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.063 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.198 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.1016P)2 + 0.1391P] where P = (Fo2 + 2Fc2)/3 |
| 3576 reflections | (Δ/σ)max = 0.001 |
| 244 parameters | Δρmax = 0.29 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.3945 (2) | 0.3594 (2) | 0.4200 (2) | 0.0631 (6) | |
| N4 | 0.6170 (2) | 0.19621 (19) | 0.48492 (19) | 0.0528 (5) | |
| C2 | 0.5434 (3) | 0.3820 (3) | 0.3449 (3) | 0.0645 (8) | |
| C3 | 0.6537 (3) | 0.2913 (3) | 0.3721 (2) | 0.0580 (7) | |
| C5 | 0.4699 (2) | 0.1805 (2) | 0.5654 (2) | 0.0498 (6) | |
| C6 | 0.3549 (3) | 0.2558 (2) | 0.5259 (2) | 0.0534 (6) | |
| C7A | 0.5713 (15) | 0.5231 (13) | 0.2548 (9) | 0.063 (3) | 0.546 (4) |
| C8A | 0.4983 (5) | 0.5205 (5) | 0.1381 (5) | 0.0663 (11) | 0.546 (4) |
| C31 | 0.8188 (3) | 0.2978 (2) | 0.2824 (2) | 0.0563 (7) | |
| C32 | 0.8599 (3) | 0.3073 (3) | 0.1431 (3) | 0.0720 (9) | |
| C33 | 1.0135 (3) | 0.3172 (3) | 0.0618 (3) | 0.0736 (9) | |
| C34 | 1.1255 (3) | 0.3181 (3) | 0.1184 (3) | 0.0656 (8) | |
| C35 | 1.0868 (3) | 0.3074 (3) | 0.2567 (3) | 0.0603 (7) | |
| C36 | 0.9331 (3) | 0.2953 (2) | 0.3381 (2) | 0.0549 (7) | |
| C51 | 0.4413 (2) | 0.0844 (2) | 0.6962 (2) | 0.0496 (6) | |
| C52 | 0.5414 (2) | −0.0232 (2) | 0.7054 (2) | 0.0542 (6) | |
| C53 | 0.5192 (3) | −0.1138 (3) | 0.8258 (3) | 0.0634 (8) | |
| C54 | 0.3977 (3) | −0.0970 (3) | 0.9402 (2) | 0.0661 (8) | |
| C55 | 0.2987 (3) | 0.0105 (3) | 0.9334 (2) | 0.0614 (8) | |
| C56 | 0.3201 (3) | 0.1008 (2) | 0.8133 (2) | 0.0555 (7) | |
| C61 | 0.1863 (2) | 0.2299 (2) | 0.5919 (2) | 0.0518 (6) | |
| C62 | 0.1196 (3) | 0.0985 (2) | 0.6206 (2) | 0.0534 (6) | |
| C63 | −0.0386 (3) | 0.0797 (3) | 0.6767 (2) | 0.0573 (7) | |
| C64 | −0.1311 (3) | 0.1920 (3) | 0.7057 (2) | 0.0609 (7) | |
| C65 | −0.0657 (3) | 0.3224 (3) | 0.6778 (3) | 0.0634 (8) | |
| C66 | 0.0919 (3) | 0.3419 (3) | 0.6198 (3) | 0.0600 (7) | |
| C8B | 0.4650 (6) | 0.5857 (6) | 0.2183 (6) | 0.0663 (11) | 0.454 (4) |
| C7B | 0.5880 (19) | 0.4955 (17) | 0.2180 (12) | 0.063 (3) | 0.454 (4) |
| H2A | 0.68302 | 0.54456 | 0.21418 | 0.0759* | 0.546 (4) |
| H1A | 0.52231 | 0.59471 | 0.30839 | 0.0759* | 0.546 (4) |
| H5A | 0.55242 | 0.45393 | 0.08165 | 0.0998* | 0.546 (4) |
| H3A | 0.38908 | 0.49358 | 0.17936 | 0.0998* | 0.546 (4) |
| H4A | 0.50889 | 0.61234 | 0.08217 | 0.0998* | 0.546 (4) |
| H34 | 1.23057 | 0.32608 | 0.06258 | 0.0787* | |
| H35 | 1.16506 | 0.30835 | 0.29591 | 0.0724* | |
| H36 | 0.90676 | 0.28515 | 0.43334 | 0.0659* | |
| H52 | 0.62619 | −0.03460 | 0.62782 | 0.0650* | |
| H53 | 0.58741 | −0.18763 | 0.82996 | 0.0760* | |
| H54 | 0.38243 | −0.15891 | 1.02299 | 0.0793* | |
| H55 | 0.21525 | 0.02227 | 1.01182 | 0.0737* | |
| H56 | 0.25193 | 0.17485 | 0.81016 | 0.0665* | |
| H62 | 0.18296 | 0.02151 | 0.60148 | 0.0640* | |
| H63 | −0.08372 | −0.00978 | 0.69533 | 0.0688* | |
| H64 | −0.23956 | 0.17912 | 0.74471 | 0.0730* | |
| H65 | −0.12926 | 0.39879 | 0.69861 | 0.0761* | |
| H66 | 0.13612 | 0.43203 | 0.59889 | 0.0720* | |
| H32 | 0.78209 | 0.30691 | 0.10332 | 0.0864* | |
| H33 | 1.04047 | 0.32336 | −0.03311 | 0.0883* | |
| H6B | 0.61707 | 0.45254 | 0.13633 | 0.0759* | 0.454 (4) |
| H7B | 0.67843 | 0.54886 | 0.21448 | 0.0759* | 0.454 (4) |
| H8B | 0.42780 | 0.61962 | 0.30408 | 0.0998* | 0.454 (4) |
| H9B | 0.50093 | 0.66372 | 0.14346 | 0.0998* | 0.454 (4) |
| H10B | 0.38122 | 0.53611 | 0.20745 | 0.0998* | 0.454 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0495 (10) | 0.0628 (12) | 0.0572 (11) | 0.0002 (8) | −0.0045 (9) | 0.0135 (9) |
| N4 | 0.0437 (9) | 0.0559 (10) | 0.0440 (9) | 0.0028 (7) | −0.0019 (7) | 0.0032 (7) |
| C2 | 0.0504 (12) | 0.0650 (14) | 0.0596 (14) | −0.0040 (10) | −0.0071 (10) | 0.0148 (11) |
| C3 | 0.0484 (12) | 0.0617 (13) | 0.0491 (12) | −0.0004 (9) | −0.0039 (9) | 0.0037 (10) |
| C5 | 0.0461 (11) | 0.0481 (11) | 0.0434 (10) | 0.0039 (8) | −0.0041 (8) | 0.0000 (8) |
| C6 | 0.0469 (11) | 0.0496 (11) | 0.0490 (11) | 0.0025 (8) | −0.0030 (9) | 0.0027 (9) |
| C7A | 0.056 (3) | 0.067 (5) | 0.046 (5) | −0.006 (3) | 0.003 (4) | 0.004 (3) |
| C8A | 0.0526 (18) | 0.061 (2) | 0.062 (2) | 0.0075 (15) | −0.0023 (16) | 0.0137 (14) |
| C31 | 0.0470 (11) | 0.0547 (12) | 0.0492 (12) | −0.0005 (9) | −0.0011 (9) | 0.0073 (9) |
| C32 | 0.0575 (14) | 0.0932 (19) | 0.0529 (14) | −0.0104 (13) | −0.0094 (11) | 0.0032 (13) |
| C33 | 0.0674 (16) | 0.0890 (19) | 0.0448 (12) | 0.0002 (13) | 0.0012 (11) | −0.0007 (12) |
| C34 | 0.0462 (12) | 0.0663 (14) | 0.0578 (14) | 0.0037 (10) | 0.0065 (10) | 0.0085 (11) |
| C35 | 0.0470 (12) | 0.0590 (13) | 0.0598 (13) | 0.0027 (9) | −0.0077 (10) | 0.0075 (10) |
| C36 | 0.0523 (12) | 0.0479 (11) | 0.0483 (11) | 0.0042 (9) | −0.0031 (9) | 0.0049 (9) |
| C51 | 0.0447 (10) | 0.0514 (11) | 0.0409 (10) | 0.0042 (8) | −0.0038 (8) | 0.0006 (8) |
| C52 | 0.0448 (11) | 0.0610 (12) | 0.0435 (10) | 0.0097 (9) | −0.0023 (8) | −0.0018 (9) |
| C53 | 0.0590 (13) | 0.0684 (14) | 0.0510 (12) | 0.0170 (11) | −0.0109 (10) | 0.0013 (10) |
| C54 | 0.0655 (14) | 0.0749 (16) | 0.0421 (11) | 0.0110 (12) | −0.0083 (10) | 0.0089 (10) |
| C55 | 0.0559 (13) | 0.0727 (15) | 0.0400 (11) | 0.0087 (11) | −0.0007 (9) | −0.0024 (10) |
| C56 | 0.0508 (11) | 0.0591 (12) | 0.0440 (11) | 0.0109 (9) | −0.0035 (9) | −0.0038 (9) |
| C61 | 0.0449 (11) | 0.0534 (11) | 0.0410 (10) | 0.0047 (8) | −0.0008 (8) | 0.0047 (8) |
| C62 | 0.0487 (11) | 0.0532 (12) | 0.0409 (10) | 0.0065 (9) | 0.0008 (8) | 0.0035 (8) |
| C63 | 0.0511 (12) | 0.0597 (12) | 0.0437 (11) | −0.0021 (9) | 0.0005 (9) | 0.0036 (9) |
| C64 | 0.0425 (11) | 0.0780 (15) | 0.0441 (11) | 0.0050 (10) | 0.0029 (9) | 0.0001 (10) |
| C65 | 0.0527 (13) | 0.0662 (14) | 0.0562 (13) | 0.0146 (10) | −0.0022 (10) | −0.0068 (11) |
| C66 | 0.0538 (12) | 0.0553 (12) | 0.0579 (13) | 0.0056 (9) | −0.0062 (10) | −0.0022 (10) |
| C8B | 0.0526 (18) | 0.061 (2) | 0.062 (2) | 0.0075 (15) | −0.0023 (16) | 0.0137 (14) |
| C7B | 0.056 (3) | 0.067 (5) | 0.046 (5) | −0.006 (3) | 0.003 (4) | 0.004 (3) |
Geometric parameters (Å, º)
| N1—C2 | 1.342 (4) | C63—C64 | 1.389 (4) |
| N1—C6 | 1.338 (3) | C64—C65 | 1.382 (4) |
| N4—C3 | 1.337 (3) | C65—C66 | 1.382 (4) |
| N4—C5 | 1.338 (3) | C7A—H1A | 0.9900 |
| C2—C3 | 1.400 (4) | C7A—H2A | 0.9900 |
| C2—C7A | 1.519 (12) | C7B—H6B | 0.9900 |
| C2—C7B | 1.540 (14) | C7B—H7B | 0.9900 |
| C3—C31 | 1.500 (4) | C8A—H5A | 0.9800 |
| C5—C6 | 1.409 (3) | C8A—H3A | 0.9800 |
| C5—C51 | 1.488 (3) | C8A—H4A | 0.9800 |
| C6—C61 | 1.487 (3) | C8B—H8B | 0.9800 |
| C7A—C8A | 1.608 (13) | C8B—H9B | 0.9800 |
| C7B—C8B | 1.416 (19) | C8B—H10B | 0.9800 |
| C31—C36 | 1.372 (4) | C32—H32 | 0.9500 |
| C31—C32 | 1.392 (4) | C33—H33 | 0.9500 |
| C32—C33 | 1.388 (4) | C34—H34 | 0.9500 |
| C33—C34 | 1.363 (4) | C35—H35 | 0.9500 |
| C34—C35 | 1.383 (4) | C36—H36 | 0.9500 |
| C35—C36 | 1.389 (4) | C52—H52 | 0.9500 |
| C51—C52 | 1.393 (3) | C53—H53 | 0.9500 |
| C51—C56 | 1.400 (3) | C54—H54 | 0.9500 |
| C52—C53 | 1.384 (4) | C55—H55 | 0.9500 |
| C53—C54 | 1.384 (4) | C56—H56 | 0.9500 |
| C54—C55 | 1.382 (4) | C62—H62 | 0.9500 |
| C55—C56 | 1.382 (3) | C63—H63 | 0.9500 |
| C61—C66 | 1.394 (4) | C64—H64 | 0.9500 |
| C61—C62 | 1.395 (3) | C65—H65 | 0.9500 |
| C62—C63 | 1.385 (4) | C66—H66 | 0.9500 |
| C2—N1—C6 | 119.3 (2) | C8B—C7B—H6B | 109.00 |
| C3—N4—C5 | 118.9 (2) | C8B—C7B—H7B | 109.00 |
| N1—C2—C3 | 119.7 (3) | H6B—C7B—H7B | 108.00 |
| N1—C2—C7A | 111.8 (6) | C2—C7B—H7B | 109.00 |
| N1—C2—C7B | 119.3 (7) | C2—C7B—H6B | 109.00 |
| C3—C2—C7A | 127.5 (6) | H3A—C8A—H5A | 109.00 |
| C3—C2—C7B | 120.5 (7) | H3A—C8A—H4A | 109.00 |
| N4—C3—C2 | 120.7 (2) | C7A—C8A—H3A | 109.00 |
| N4—C3—C31 | 116.1 (2) | C7A—C8A—H4A | 109.00 |
| C2—C3—C31 | 123.1 (2) | C7A—C8A—H5A | 109.00 |
| N4—C5—C6 | 120.09 (19) | H4A—C8A—H5A | 109.00 |
| N4—C5—C51 | 115.46 (18) | C7B—C8B—H8B | 109.00 |
| C6—C5—C51 | 124.42 (18) | C7B—C8B—H10B | 109.00 |
| N1—C6—C5 | 119.9 (2) | H8B—C8B—H9B | 109.00 |
| N1—C6—C61 | 114.7 (2) | C7B—C8B—H9B | 109.00 |
| C5—C6—C61 | 125.43 (18) | H9B—C8B—H10B | 109.00 |
| C2—C7A—C8A | 108.0 (8) | H8B—C8B—H10B | 110.00 |
| C2—C7B—C8B | 111.3 (10) | C33—C32—H32 | 120.00 |
| C3—C31—C32 | 121.7 (2) | C31—C32—H32 | 120.00 |
| C3—C31—C36 | 119.54 (19) | C32—C33—H33 | 120.00 |
| C32—C31—C36 | 118.7 (2) | C34—C33—H33 | 120.00 |
| C31—C32—C33 | 120.6 (3) | C35—C34—H34 | 120.00 |
| C32—C33—C34 | 119.9 (3) | C33—C34—H34 | 120.00 |
| C33—C34—C35 | 120.3 (3) | C36—C35—H35 | 120.00 |
| C34—C35—C36 | 119.8 (3) | C34—C35—H35 | 120.00 |
| C31—C36—C35 | 120.7 (2) | C31—C36—H36 | 120.00 |
| C5—C51—C52 | 119.45 (18) | C35—C36—H36 | 120.00 |
| C52—C51—C56 | 118.07 (19) | C53—C52—H52 | 119.00 |
| C5—C51—C56 | 122.43 (18) | C51—C52—H52 | 119.00 |
| C51—C52—C53 | 121.0 (2) | C54—C53—H53 | 120.00 |
| C52—C53—C54 | 120.1 (3) | C52—C53—H53 | 120.00 |
| C53—C54—C55 | 119.6 (2) | C53—C54—H54 | 120.00 |
| C54—C55—C56 | 120.5 (2) | C55—C54—H54 | 120.00 |
| C51—C56—C55 | 120.7 (2) | C56—C55—H55 | 120.00 |
| C6—C61—C62 | 122.11 (19) | C54—C55—H55 | 120.00 |
| C62—C61—C66 | 119.2 (2) | C51—C56—H56 | 120.00 |
| C6—C61—C66 | 118.6 (2) | C55—C56—H56 | 120.00 |
| C61—C62—C63 | 120.3 (2) | C61—C62—H62 | 120.00 |
| C62—C63—C64 | 119.9 (3) | C63—C62—H62 | 120.00 |
| C63—C64—C65 | 120.1 (3) | C62—C63—H63 | 120.00 |
| C64—C65—C66 | 120.2 (3) | C64—C63—H63 | 120.00 |
| C61—C66—C65 | 120.3 (3) | C65—C64—H64 | 120.00 |
| C2—C7A—H1A | 110.00 | C63—C64—H64 | 120.00 |
| C2—C7A—H2A | 110.00 | C66—C65—H65 | 120.00 |
| C8A—C7A—H1A | 110.00 | C64—C65—H65 | 120.00 |
| C8A—C7A—H2A | 110.00 | C61—C66—H66 | 120.00 |
| H1A—C7A—H2A | 108.00 | C65—C66—H66 | 120.00 |
| C6—N1—C2—C3 | 4.6 (4) | N1—C6—C61—C66 | −46.4 (3) |
| C6—N1—C2—C7A | −164.7 (5) | C5—C6—C61—C62 | −48.8 (3) |
| C2—N1—C6—C5 | 6.2 (3) | C5—C6—C61—C66 | 134.3 (2) |
| C2—N1—C6—C61 | −173.1 (2) | C3—C31—C32—C33 | 178.1 (3) |
| C5—N4—C3—C2 | 6.1 (4) | C36—C31—C32—C33 | −1.4 (4) |
| C5—N4—C3—C31 | −176.5 (2) | C3—C31—C36—C35 | −177.1 (2) |
| C3—N4—C5—C6 | 4.8 (3) | C32—C31—C36—C35 | 2.5 (3) |
| C3—N4—C5—C51 | −173.2 (2) | C31—C32—C33—C34 | −0.2 (4) |
| N1—C2—C3—N4 | −11.2 (4) | C32—C33—C34—C35 | 0.8 (5) |
| N1—C2—C3—C31 | 171.6 (2) | C33—C34—C35—C36 | 0.2 (4) |
| C7A—C2—C3—N4 | 156.3 (6) | C34—C35—C36—C31 | −1.9 (4) |
| C7A—C2—C3—C31 | −20.9 (7) | C5—C51—C52—C53 | 179.4 (2) |
| N1—C2—C7A—C8A | −70.8 (7) | C56—C51—C52—C53 | 1.9 (3) |
| C3—C2—C7A—C8A | 120.9 (7) | C5—C51—C56—C55 | −179.0 (2) |
| N4—C3—C31—C32 | 134.5 (2) | C52—C51—C56—C55 | −1.6 (3) |
| N4—C3—C31—C36 | −45.9 (3) | C51—C52—C53—C54 | −1.2 (4) |
| C2—C3—C31—C32 | −48.2 (4) | C52—C53—C54—C55 | 0.1 (4) |
| C2—C3—C31—C36 | 131.4 (3) | C53—C54—C55—C56 | 0.2 (4) |
| N4—C5—C6—N1 | −11.3 (3) | C54—C55—C56—C51 | 0.6 (4) |
| N4—C5—C6—C61 | 167.98 (19) | C6—C61—C62—C63 | −177.1 (2) |
| C51—C5—C6—N1 | 166.6 (2) | C66—C61—C62—C63 | −0.3 (3) |
| C51—C5—C6—C61 | −14.1 (3) | C6—C61—C66—C65 | 178.3 (2) |
| N4—C5—C51—C52 | −27.8 (3) | C62—C61—C66—C65 | 1.3 (4) |
| N4—C5—C51—C56 | 149.6 (2) | C61—C62—C63—C64 | −0.6 (3) |
| C6—C5—C51—C52 | 154.2 (2) | C62—C63—C64—C65 | 0.5 (3) |
| C6—C5—C51—C56 | −28.4 (3) | C63—C64—C65—C66 | 0.6 (4) |
| N1—C6—C61—C62 | 130.5 (2) | C64—C65—C66—C61 | −1.5 (4) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK5151).
References
- Agilent (2012). CrysAlis PRO Agilent Technologies, Yarnton, Oxfordshire, England.
- Anuradha, N., Thiruvalluvar, A., Pandiarajan, K., Chitra, S. & Butcher, R. J. (2009). Acta Cryst. E65, o106. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812039827/tk5151sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812039827/tk5151Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812039827/tk5151Isup3.cdx
Supplementary material file. DOI: 10.1107/S1600536812039827/tk5151Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report