Abstract
Terpenoid metabolites are important to the cellular function, structural integrity, and pathogenesis of the human-specific pathogen Mycobacterium tuberculosis (Mtb). Genetic and biochemical investigations have indicated a role for the diterpenoid isotuberculosinol (isoTb) early in the infection process. There are only two genes (Rv3377c and Rv3378c) required for production of isoTb, yet these are found in what appears to be a five-gene terpenoid/isoprenoid biosynthetic operon. Of the three remaining genes (Rv3379c, Rv3382c, and Rv3383c), previous work has indicated that Rv3379c is an inactive pseudo-gene. Here we demonstrate that Rv3382c and Rv3383c encode biochemically redundant machinery for isoprenoid metabolism, encoding a functional 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB) for isoprenoid precursor production and a geranylgeranyl diphosphate (GGPP) synthase, respectively, for which the Mtb genome contains other functional isozymes (Rv1110 and Rv0562, respectively). These results complete the characterization of the isoTb biosynthetic operon, as well as further elucidating isoprenoid metabolism in Mtb. In addition, we have investigated the evolutionary origin of this operon, revealing Mtb-specific conservation of the diterpene synthase genes responsible for isoTb biosynthesis, which supports our previously advanced hypothesis that isoTb acts as a human-specific pathogenic metabolite and is consistent with the human host specificity of Mtb. Intriguingly, our results revealed that many mycobacteria contain orthologs for both Rv3383c and Rv0562, suggesting a potentially important role for these functionally redundant GGPP synthases in the evolution of terpenoid/isoprenoid metabolism in the mycobacteria.
Keywords: isoprenoid biosynthesis, molecular evolution, virulence, terpenoids
INTRODUCTION
Each year, Mycobacterium tuberculosis (Mtb) accounts for ~1.7 million deaths as the main causative agent of the human disease Tuberculosis (Rohde et al., 2007), and the ability of this deadly pathogen to infiltrate macrophage cells from the mammalian immune system remains an active area of investigation (Russell, 2007). While the complete pathogenic mechanism remains unknown, the bacteria appear to launch a multi-factorial attack (Russell et al., 2010), with many bacterial genes involved in pathogenesis (Joshi et al., 2006). Notably, a mutational screen for genes involved in the early infection process highlighted a role for Rv3377c and Rv3378c, loss of which reduced the ability of Mtb to block acidification of the engulfing phagosome compartment, leading to suppressed bacterial proliferation in macrophage cell cultures (Pethe et al., 2004). These genes have since been characterized as encoding a pair of sequentially acting diterpene synthases responsible for the production of isotuberculosinol (isoTb; Nakano et al., 2005a,b; Mann et al., 2009b; Maugel et al., 2010; Prach et al., 2010; Spangler et al., 2010; Hoshino et al., 2011; Mann et al., 2011b; Nakano et al., 2011), a diterpenoid capable of arresting phagosomal maturation in vitro (Mann et al., 2009b). Thus, isoTb appears to play an immunomodulatory role in the Mtb infection process (Mann and Peters, 2012).
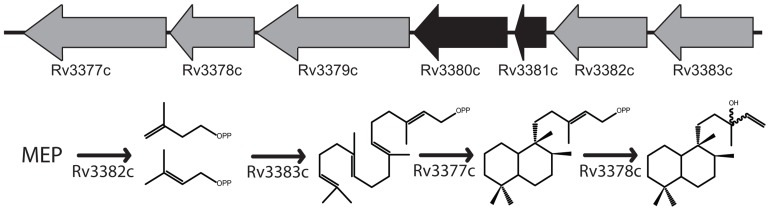
Rv3377c and Rv3378c fall within a larger operon that appears to have been acquired via horizontal gene transfer before the divergence of Mtb and Mycobacterium bovis (Becq et al., 2007). The operon includes Rv3377c–Rv3383c, with the intervening Rv3380c and Rv3381c representing apparent transposable elements inserted into a preexisting operon (Camus et al., 2002), such that only five genes seem to be relevant. By homology, these all appear to encode enzymes that could be involved in various stages of isoTb production (Figure 1). However, only Rv3377c and Rv3378c were isolated in the genetic screen targeting the early infection process, despite the saturating nature of that study (Pethe et al., 2004). The remaining genes do not appear to be required for any essential cellular function, as indicated by previously reported genetic screens (Sassetti et al., 2001, 2003; Sassetti and Rubin, 2003; Joshi et al., 2006), leaving uncertain their function, if any, in mycobacterial terpenoid biosynthesis. It has already been reported that Rv3379c is an inactive pseudo-gene (Bailey et al., 2002), leaving the function of the remaining genes in question.
FIGURE 1.
Proposed isoTb biosynthetic operon. The five-gene operon including the two previously characterized diterpene synthases required for isoTb biosynthesis and the proposed roles of the remaining uncharacterized genes in the biosynthetic pathway. Genes depicted in the order suggested by their numbering rather than orientation of open reading frame.
Mycobacterium tuberculosis uses the methyl-erythritol-5-phosphate (MEP) pathway for isoprenoid precursor biosynthesis (Rohmer et al., 1993). The resulting 5-carbon isoprenyl isomers, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), are then coupled together to form longer acyclic isoprenyl diphosphates such as the 10-carbon geranyl diphosphate (GPP), 15-carbon farnesyl diphosphate (FPP), and 20-carbon diterpenoid precursor geranylgeranyl diphosphate (GGPP). These are then used in cell wall assembly (e.g., the further elongated decaprenyl diphosphate), synthesis of other essential biomolecules (e.g., menaquinone), as well as isoTb biosynthesis. In the isoTb operon, Rv3379c is homologous to the deoxy-xylulose-5-phosphate synthases that initiate the MEP pathway, and is annotated as dxs2, although it has been reported to be an inactive pseudo-gene (Bailey et al., 2002). Rv3382c encodes a putative 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, the LytB that catalyzes the final step of the MEP pathway, producing IPP and DMAPP (Rohdich et al., 2002), but this has not previously been investigated. Rv3383c encodes a member of the isoprenyl diphosphate synthase (IDS) family, which generally catalyze isoprenyl diphosphate chain elongation. Accordingly, the IDS encoded by Rv3383c is likely to produce the GGPP required for isoTb biosynthesis, but this also has not yet been investigated. Here, we not only verify the biochemical function of both Rv3382c and Rv3383c, but also the presence of robust isozymes catalyzing the same reactions encoded elsewhere in the Mtb genome, suggesting functional redundancy consistent with the reported genetic screens. In addition, bioinformatics analysis of the available mycobacterial genome sequences indicates asymmetric conservation of isoTb biosynthetic machinery in Mtb relative to the greater Mycobacterium tuberculosis complex (MTC), consistent with a previously hypothesized role for isoTb in the highly infectious nature of Mtb in humans, along with the surprising finding of wider conservation of dual GGPP synthases in a sub-group of mycobacteria.
MATERIALS AND METHODS
GENERAL
All trans (E) isomers of GPP, FPP, and GGPP were purchased from Isoprenoids, LC (Tampa, FL, USA). Unless otherwise noted, all other chemicals were purchased from Fisher Scientific (St. Louis, MO, USA) and all molecular biology reagents were purchased from Invitrogen (Carlsbad, CA, USA). Gas chromatography-flame ionization (GC-FID) detection was carried out using an Agilent 6890 GC-FID (Agilent Technologies, Santa Clara, CA, USA) using a previously described method (Schmidt and Gershenzon, 2008).
BIOINFORMATICS
Genes of interest were identified via basic local sequence alignment (BLAST) against the M. tuberculosis H37Rv genome (Cole et al., 1998). Phylogenetic analysis of GGPP synthases (GGPPS) was accomplished via UPGMA modeling of amino acid sequences (Sneath and Sokal, 1973), using CLC Sequence Viewer 6.0 (CLC Bio, Cambridge, MA, USA). Complete operon analysis was performed against publically available sequences of Mtb diversity strains (Hershberg et al., 2008), M. canettii (Hershberg et al., 2008), M. marinum (Stinear et al., 2008), and M. ulcerans (Stinear et al., 2007), as well as previously published sequences of M. bovis (Garnier et al., 2003), and M. bovis BCG (Brosch et al., 2007), and the pre-publication reads of M. africanum (available via the Sanger Institute). Genomic regions were aligned using BLAST analysis and synteny mapping via the Tuberculosis Database (Reddy et al., 2009).
CLONING
Genes of interest were cloned by PCR amplification from Mtb CDC1551 genomic DNA and confirmed via complete sequencing. These were transferred into pENTR/SD/D-TOPO (Gateway, Invitrogen) by directional recombination. Rv2173 and Rv0562 were modified to change the first codon to ATG to enable expression in Escherichia coli, for which purpose all genes were recombined into pDEST17 expression vectors for production with an N-terminal 6xHis tag.
LytB COMPLEMENTATION
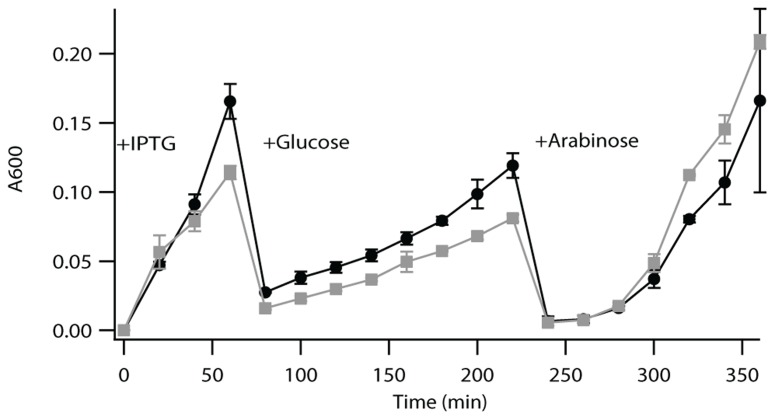
LytB complementation was initially assessed by colony counting assays, carried out in three individual experiments, each in duplicate. 50 ng of plasmid (pDEST14) containing Rv3382c or Rv1110 were each electroporated into 100 μL electro-competent E. coli cells containing LytB under arabinose promoter control (McAteer et al., 2001) and were plated on NZY medium in the absence of arabinose, which allows only for growth of LytB complemented cells. Electroporated E. coli cells plated in the presence and absence of 0.2% arabinose were used as controls. More quantitative analysis of LytB activity was accomplished by comparison of relative growth rates, carried out in triplicate. Colonies from Plasmid (+) plates were grown to saturation in LB medium overnight. Cultures were diluted 1/100 into fresh LB medium containing 0.5 mM isopropylthiogalactoside (IPTG) and grown for 1 h, then diluted 1/50 into media containing 50 μg/mL carbenicillin and 0.2% (v/v) glucose to inhibit the arabinose promoter and thus completely inhibit any expression of E. coli LytB. After 220 min, cultures were diluted 1/50 into medium containing 0.2% (v/v) arabinose, thus activating the chromosomal LytB and allowing for comparative analysis of the relative health of both complemented strains (i.e., to demonstrate that expression of Rv3382c did not exert any toxicity relative to expression of Rv1110 instead). Culture growth was monitored every 20 min by measuring OD600.
PUTATIVE GGPP SYNTHASE EXPRESSION
Putative GGPP synthases in pDEST17 were transformed into the C41 strain of E. coli (Lucigen, Middleton, WI, USA), and plated on NZY agar with carbenicillin (50 μg/mL) selection. Colonies were inoculated in 50 mL of NZY medium and grown to an optical density at 600 nm (OD600) of 0.6 at 37°C before being transferred to 16°C for 1 h prior to induction with 0.5 mM IPTG, with subsequent 14–16 h incubation. Cells were harvested via centrifugation and lysed in 1 mL lysis buffer (10 mM Tris-Cl, pH 6.8, 10% glycerol, 1 mM dithiothreitol) via sonication. These lysates were clarified via centrifugation (10 min × 10,000g) and the resulting cell free extracts assayed for GGPP synthase activity. As E. coli does not harbor contaminating GGPP synthases (Burke and Croteau, 2002), any resulting production of GGPP was attributed to expression of the recombinant gene.
GGPP SYNTHASE ASSAY
Assay buffer (50 mM sodium phosphate (pH 7.0), 10% glycerol, 5 mM MgCl2, 1 mM dithiothreitol) was mixed with 20 μM IPP and the equivalent concentration of indicated allylic isoprenyl diphosphate. To each assay, 100 μL of purified protein (~1 mg/mL) expressed as a 6×His fusion in pDEST17 was added, totaling 1 mL total volume. Assays were conducted at 37°C for indicated time and halted via incubation at 100°C for 5 min. After cooling, 1.5 U calf intestinal alkaline phosphatase (New England Biolabs, Ipswich, MA, USA) was added according to manufacturer protocol. The phosphatase reaction was incubated overnight at 37°C and extracted thrice with 1 mL hexanes. Samples were concentrated and analyzed via GC-FID and compared to similarly, dephosphorylated authentic GGPP.
KINETIC ANALYSIS OF Rv3383c AND Rv0562
The 6×His tagged versions of these enzymes were purified via Ni-NTA Superflow resin (Novagen, Merck, Germany) following the manufacturer directions. Protein was analyzed via denaturing gel electrophoresis (SDS-PAGE), demonstrating >90% purity. Enzymes were assayed as described against a substrate gradient (0–40 μM) with IPP and DMAPP/FPP for 5 min with 100 nM enzyme. Post-extraction, cembrene was added to a final concentration of 5 μM as an internal standard for quantification. Product concentration was calculated by integration of the product peak area and comparison to internal standard peak area. Michaelis–Menten curves were fit to the observed data (KaleidaGraph 4.0, Synergy Software, Reading, PA, USA), resulting in R2 values ≥0.95.
RESULTS
POTENTIAL ISOZYMES FOR BOTH Rv3382c AND Rv3383c
To begin investigating the hypothesized biochemical redundancy, the uncharacterized Rv3382c and Rv3383c were analyzed and compared to the rest of the Mtb genome. Rv3382c is 990 bp long and encodes a protein of 329 amino acids (AA) that has been annotated as lytB1, while Rv3383c is 1053 bp long and encodes a 350 AA protein annotated as idsB (Camus et al., 2002). Both genes are non-essential (Sassetti et al., 2003), and neither is associated with pathogenicity in published genetic screens (Sassetti and Rubin, 2003; Pethe et al., 2004; Fortune et al., 2005; Joshi et al., 2006). Protein–protein BLAST analysis of Rv3382c and Rv3383c against the H37Rv genome revealed putative paralogs for both. Specifically, Rv3382c shares 50% AA identity with Rv1110, which is similarly annotated as lytB2 (Camus et al., 2002). There are four IDS paralogs to Rv3383c in the Mtb genome: Rv3398c, a previously identified FPP synthase annotated as idsA1 (Dhiman et al., 2004), which shares 35% AA identity with Rv3383c; Rv2173, annotated as idsA2, which shares 29% AA identity; Rv0562, annotated asgrcC1, which shares 30% AA identity; and Rv0989c, that we have recently demonstrated encodes a functional GPP synthase likely involved in decaprenyl diphosphate biosynthesis (Mann et al., 2011a), annotated as grcC2, which shares 28% AA identity.
FUNCTIONAL ANALYSIS OF Rv3382c AND Rv1110
LytB is a [4Fe–4S] protein that has been shown to be difficult to work with in vitro, due to complications from both oxygen corruption and substrate availability (Adam et al., 2002; Wolff et al., 2003). Accordingly, such enzymatic activity is most often demonstrated by a complementation approach (McAteer et al., 2001; Rohdich et al., 2002; Hsieh and Goodman, 2005). Briefly, this utilizes a previously developed strain of E. coli in which the endogenous LytB gene (EG11081) has been put under control of an arabinose promoter, such that bacterial growth requires either supplementation with arabinose or complementation with a functional LytB (McAteer et al., 2001). Both Rv3382c and Rv1110 were able to complement the growth of this strain of E. coli in the absence of arabinose, demonstrating functional LytB activity. Relative growth rate analysis indicates that complementation with Rv3382c results in somewhat less vigorous growth than does complementation with Rv1110, which does not appear to be due to a toxicity effect (Figure 2). This result then suggests that Rv3382c exhibits somewhat less robust LytB activity than Rv1110, which might be due to Rv3382c being 8 AA shorter at the N-terminus. However, this region does not contain any residues known to be involved in enzymatic activity, and the exact mechanism underlying reduced function of the LytB encoded by Rv3382c is unclear.
FIGURE 2.
Complementation assay for LytB activity. Growth curve of lytB- E. coli complemented with Rv1110 (circles) or Rv3382c (squares). These bacteria were grown to saturation, then diluted 1/100 into fresh media containing 0.5 mM IPTG. After 60 min, cultures were diluted 1/50 in media containing selective antibiotic and 0.02% glucose (to suppress endogenous LytB expression). After 220 min, cultures were diluted 1/100 in media containing selective antibiotic, but 0.2% arabinose instead of glucose, which activated expression of the endogenous LytB, to demonstrate that the Rv3382c gene itself did not have any deleterious effects (i.e., as both recombinant strains then exhibited equivalent growth).
ANALYSIS OF PUTATIVE GGPP SYNTHASES
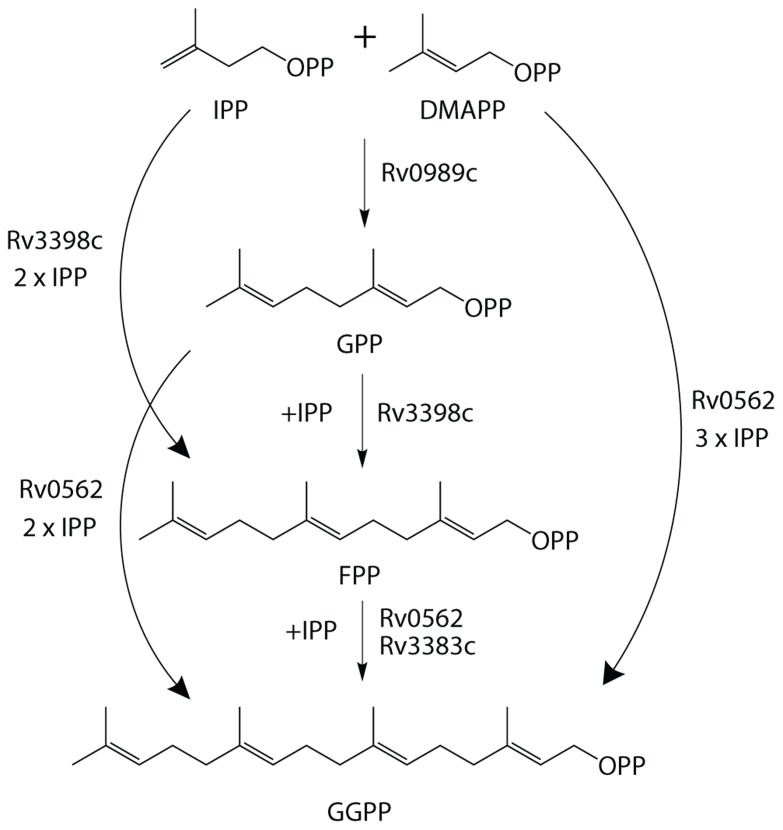
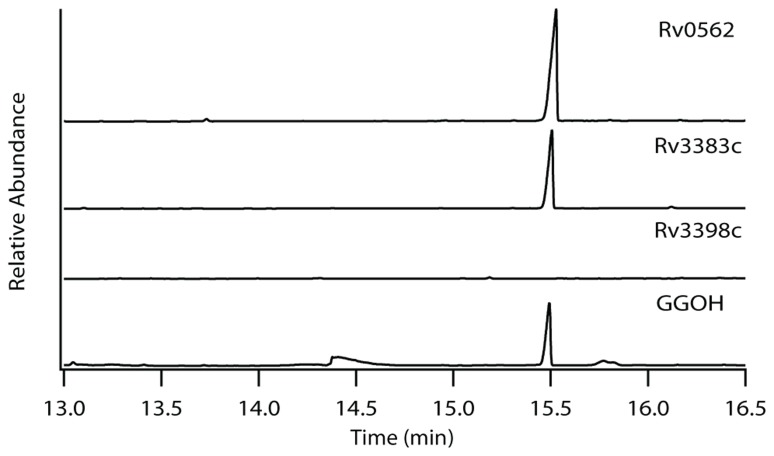
The production of GGPP is catalyzed by the condensation of IPP with allylic isoprenyl diphosphate acceptors, such as DMAPP (which then requires the addition of three molecules of IPP), GPP (e.g., that produced by Rv0989c, which then requires the addition of two molecules of IPP), or FPP (e.g., that produced by Rv3398c, which then only requires the addition of a single molecule of IPP; Figure 3). Thus, the putative GGPP synthases identified above were screened for such activity by assaying purified recombinant proteins expressed in E. coli, in the presence of IPP plus DMAPP, GPP, or FPP (Table 1). Both Rv0562 and Rv3383c were able to add IPP to FPP, producing GGPP (Figure 4). In addition, Rv0562 is able to produce GGPP using either DMAPP, GPP, or FPP as the allylic precursor, while Rv3383c can synthesize GGPP only when fed FPP (Table 1). By contrast, Rv2173 only exhibited intermittent GGPP synthase activity in these assays, indicating that its primary product may not be GGPP. Further studies are ongoing with this enzyme to clarify its function.
FIGURE 3.
Substrate plasticity of GGPPS. GGPPS enzymes are sometimes able to utilize a variety of substrates to synthesize the 20-carbon product, GGPP. Outlined are the various biosynthetic pathways for GGPP production in Mtb.
Table 1.
Kinetic analysis of confirmed GGPP synthases.
| Enzyme | IPP | DMAPP | GPP | FPP | |||
|---|---|---|---|---|---|---|---|
| KM (μM) | kcat (min-1) | KM (μM) | kcat (min-1) | KM (μM) | kcat (min-1) | KM (μM) | |
| Rv0562 | 1.7 ± 0.7 | 0.17 ± 0.02 | 5 ± 2 | 0.3 ± 0.2 | 2.3 ± 0.8 | 0.7 ± 0.1 | 11 ± 3 |
| Rv3383c | 11 ± 2 | NR | NR | NR | NR | 1.0 ± 0.5 | 19 ± 5 |
NR, no reaction.
FIGURE 4.
Production of GGPP by Rv3383c and Rv0562. GC-FID chromatogram of GGPP-derived GGOH production in Rv0562 and Rv3383c, but not the previously confirmed (E,E)-FPP synthase Rv3398c, as compared to dephosphorylated authentic standard.
EVOLUTION OF THE isoTb OPERON WITHIN THE MYCOBACTERIA
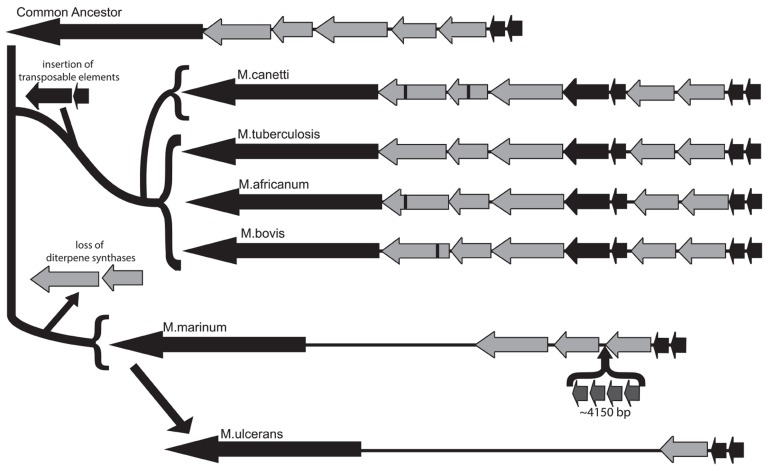
The commonly accepted evolutionary scheme for pathogenic mycobacteria indicates that the MTC emerged from a common ancestor, which was most recently shared with M. canettii, to create a pool of human and animal pathogens, including M. bovis and M. africanum, along with Mtb (Brosch et al., 2002). Early analysis of the Mtb genome isolated a putative five-gene operon involved in isoprenoid biosynthesis at coordinates 3790.85–3799.64 kb and containing loci Rv3377–Rv3383c. This operon was identified as potential horizontal gene transfer products from an exogenous source due to aberrations in GC content within this region (Becq et al., 2007). Rv3377c and Rv3378c are diterpene synthases that produce the immunomodulatory diterpenoid isoTb via cyclization of the diterpenoid precursor GGPP (Nakano et al., 2005a,b; Mann et al., 2009b; Maugel et al., 2010; Prach et al., 2010; Spangler et al., 2010; Hoshino et al., 2011; Mann et al., 2011b; Nakano et al., 2011). In our characterization of the diterpene cyclase encoded by Rv3377c, we noted that this gene was conserved in all four of the Mtb strains whose genomes were available at that time, while the M. bovis homolog contained an inactivating frameshift mutation that was conserved in the two sequenced strains, along with another that we investigated (Mann et al., 2009a). Based on the wider host range, but reduced infectivity in humans, of M. bovis, we then speculated that isoTb might play a role in the highly infectious nature of Mtb in humans. Given the current availability of genome sequences from a number of diverse Mtb strains (Comas et al., 2010), as well as several members of the MTC and other mycobacteria (Reddy et al., 2009), these were analyzed for the presence of all or part of the isoTb operon. Corresponding sequences could be found not only in the MTC and M. canettii, but also the much more distantly related M. marinum, although it is not present in the even more distantly related M. leprae, suggesting that the isoTb operon was acquired prior to separation of the lineages leading to M. marinum and the MTC/M. canettii. We further characterized the existing polymorphisms of not only the diterpene synthases [i.e., Rv3377c and Rv3378c, as previously reported (Mann and Peters, 2012)], but also the entire isoTb operon in these mycobacteria. Notably, very few mutations are found within the unique diterpene synthase encoding genes (i.e., Rv3377c and/or Rv3378c) in the sequenced strains of Mtb (Comas et al., 2010), with none in the highly infectious Beijing subgroup. By contrast, the other species within the MTC, as well as M. canettii, all have accrued frameshift and/or non-synonymous mutations in Rv3377c or Rv3378c. The range of observed mutations in these diterpene synthases is consistent with the current view of mycobacteria evolution, with only a single non-synonymous mutation found in the more recently derived M. africanum. By contrast, the more distantly related M. canettii contains both non-synonymous and frameshift mutations, wherein the frameshift mutation presumably precludes synthesis of isoTb, much as we previously demonstrated for M. bovis (Mann et al., 2009a). In addition, the much earlier diverging M. marinum does not contain the diterpene synthase genes at all, although it does have the other genes from this operon. Specifically, M. marinum has retained orthologs to the GGPP synthase encoded by Rv3383c, the LytB encoded by Rv3382c, and the pseudo-DXS encoded by Rv3379c, although there also has been an apparent transposon-mediated insertion within the operon. We suggest that this reflects loss of the diterpene synthases, rather than their later addition in the M. canettii/MTC lineage, as it is unclear why the other genes, which together produce the diterpene precursor GGPP, would have been assembled together in the absence of the downstream diterpene synthases (and prior to the acquisition of the carotenoid biosynthetic machinery, i.e., as seen in M. marinum). In the genome of M. ulcerans, which is derived from M. marinum (Stinear et al., 2007), the operon has been further reduced, with only the Rv3383c ortholog remaining (Figure 5). While this observation is consistent with the previously noted severe genome reduction of M. ulcerans (Stinear et al., 2000), M. marinum is believed to have undergone parallel evolution from a common ancestor with Mtb, but is generally considered to have retained and incorporated more genes, as expected for a more general pathogen, relative to the human host restricted Mtb (Stinear et al., 2007). Accordingly, isoTb production seems to be most highly conserved in Mtb, despite its comparatively specialized genome, which is consistent with our previous speculation positing a role for isoTb in the highly infectious nature of this human-specific pathogen (Mann and Peters, 2012).
FIGURE 5.
Conservation of the isoTb operon in the MTC and closely related mycobacteria. Analysis of the isoTb operon in the context of wider mycobacterial evolution indicates the operon was likely acquired in the most recent common ancestor to the MTC and M. marinum. The functional genes in the operon (light gray arrows) were retained during the speciation events leading to M. canetti, Mtb, M. africanum, and M. bovis, following which the operon independently evolved in each species, as evidenced by unique insertion–deletion and/or non-synonymous mutations in the diterpene synthases (black bars). Further, following separation of M. marinum and the MTC, the diterpene synthases were lost from M. marinum and several genes inserted, thus disrupting the operon. During later reductive speciation leading to M. ulcerans, two of the remaining genes were lost, leaving only the homolog to Rv3383c and flanking segments of the genome. Notably, retention of fully functional diterpene synthase encoding genes (Rv3377c and Rv3378c) is only found in the human-specific pathogen Mtb.
CONSERVATION OF GGPP SYNTHASES WITHIN THE MYCOBACTERIA
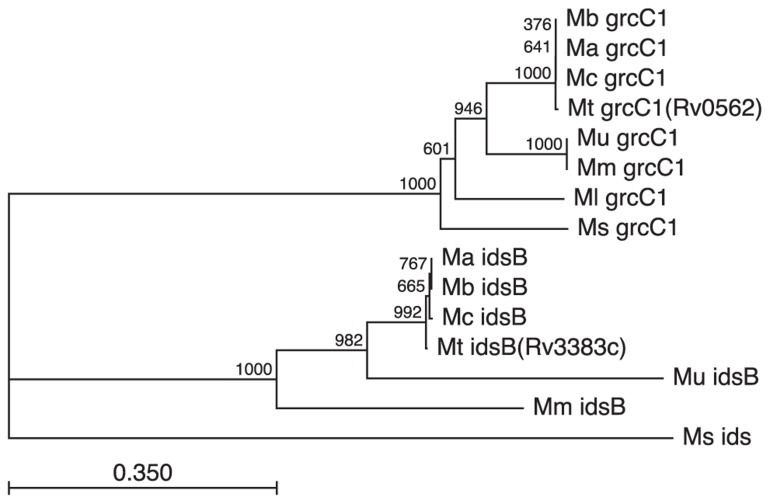
Although the ability to produce isoTb has not been found and seems unlikely to occur outside of Mtb, it is notable that the GGPP producing IDS found in Mtb are subject to stronger conservation (Figure 6). Strikingly, the GGPP synthase encoding Rv3383c/idsB from the isoTb operon was retained even through the reductive evolution process leading to M. ulcerans where the remainder of the operon is lost (Figure 5). While the presence of a GGPP synthase is likely due to its role in production of carriers involved in cell wall synthesis, the retention of multiple GGPP synthases in these mycobacteria is striking, albeit somewhat puzzling. Rv0562/grcC1 is likely the ancestral GGPP synthase and is widely found in the family Mycobacterium, e.g., an ortholog is present in the fast-growing, non-pathogenic saprophyte M. smegmatis. However, those mycobacteria derived from the MTC and M. marinum ancestral strain also have retained the additional GGPP synthase encoded by Rv3383c/idsB, even upon loss of the other genes in the isoTb operon (e.g., in M. ulcerans). In the case of M. marinum, this might be due to its production of GGPP-derived carotenoids (Ichiyama et al., 1988), although M. marinum also has acquired multiple other IDS homologs. In any case, this rationale does not hold true for other mycobacteria, leaving unclear what selective pressure led to the wider conservation of Rv3383/idsB noted here. Interestingly, this specific conservation of dual GGPP synthase homologs across these diverse species of mycobacteria suggests a more significant role for GGPP than previously presumed, consistent with the previous observation of large pools of GGPP-derived geranylgeraniol in Mtb cellular extracts (Crick et al., 2000; Mann et al., 2011b). However, it should be noted that the increased sequence divergence of the Rv3383c/idsB orthologs in M. marinum and M. ulcerans also might reflect functional diversification.
FIGURE 6.
Conservation of GGPP synthases in the MTC and closely related mycobacteria. GGPP synthases appear to have assumed additional wider importance in mycobacteria, as evidenced by the retention of Rv3383c/idsB despite loss of the remaining isoTb operon. Retention of orthologs for both Rv3383c/idsB and Rv0562/grcC1, based on both sequence similarity and genome positioning, is found in all sequenced members of the Mycobacterium family derived from the last common ancestor of the MTC and M. marinum. Species designated by genus and species initials (i.e., Mt, M. tuberculosis; Mb, M. bovis; Ma, M. africanum; Mc, M. canettii; Mm, M. marinum; Mu, M. ulcerans; Ml, M. leprae; Ms, M. smegmatis) and genes as annotated in the Mtb genome (i.e., either grcC1 or idsB), with the next closest ids homolog from M. smegmatis shown as the outgroup sequence). Not that while many of the listed species do contain other IDS encoding genes, these are relatively distantly related to those shown here.
DISCUSSION
Mycobacterium tuberculosis is a persistent and deadly human pathogen whose ability to infiltrate and spread within the human immune system has led to intense interest in all aspects of its life cycle (Russell et al., 2010). Isoprenoid metabolites play a variety of roles in Mtb and other bacteria, including electron transfer components and cell wall biosynthesis (Daum et al., 2009). At least in the case of Mtb, this appears to extend to its pathogenic lifestyle (Dhiman et al., 2004; Kaur et al., 2004; Holsclaw et al., 2008; Mann et al., 2009b). In particular, the bioactive diterpenoid isoTb seems to contribute to early processes in Mtb infections, based on both its biological activity (Mann et al., 2009b), and the reduced proliferation of Mtb unable to produce isoTb (Pethe et al., 2004). Moreover, specific conservation of the requisite diterpene synthases Rv3377c and Rv3378c in Mtb relative to the rest of the species in the MTC (Figure 5), has led to the suggestion that isoTb may play a role in the highly infectious nature of Mtb in humans (Mann and Peters, 2012), making this a potentially human-specific pathogenic natural product.
The two diterpene synthases required for isoTb biosynthesis were isolated in a screen for genes contributing to the early infection process. However, while the identified Rv3377c and Rv3378c are found in a larger five-gene operon, the remaining three genes were not isolated in this genetic screen, despite its saturating nature (Pethe et al., 2004). Presumably, these three genes originally functioned to increase flux into and through the MEP isoprenoid precursor pathway (i.e., Rv3379c/dxs2 and Rv3382c/lytB1), as well as direct it toward diterpenoid biosynthesis via production of GGPP (i.e., Rv3383c/idsB). This would enable production of isoTb by the accompanying diterpene synthases (i.e., Rv3377c and Rv3378c). While the isoTb operon appears to have been acquired via horizontal gene transfer prior to the speciation of Mtb and M. bovis (Becq et al., 2007), its presence in M. canettii demonstrates even earlier acquisition. Moreover, the presence of several of these genes in M. marinum suggests that the operon was present in the last common ancestor of the MTC/M. canettii and M. marinum. In any case, Rv3379–Rv3383c appear to be functionally redundant as none of these genes is essential in Mtb (Sassetti et al., 2001, 2003; Sassetti and Rubin, 2003; Joshi et al., 2006), nor do they appear to be absolutely required for isoTb production (Pethe et al., 2004). Consistent with this hypothesis it was previously reported that Rv3379c is an inactive pseudo-gene (Bailey et al., 2002).
It had already been predicted that the Mtb genome encodes two functional LytB for the MEP isoprenoid precursor pathway (Camus et al., 2002), and our studies confirm that both Rv1110/lytB2 and Rv3382c/lytB1 exhibit such activity (Figure 2). Given the non-essential nature of either of these genes indicated by previously reported genetic screens (Sassetti et al., 2001, 2003; Sassetti and Rubin, 2003; Joshi et al., 2006), our results indicate that Rv1110 and Rv3382c are functionally redundant. The essential nature of isoprenoid metabolism, along with the sole use of the distinct MEP pathway for precursor supply in bacteria indicates that the enzymes mediating this process might provide good targets for development of antibacterial agents (Singh et al., 2007; Eoh et al., 2009; Obiol-Pardo et al., 2011). However, it should be noted that, despite their analogous activity, the LytB paralogs characterized here share only 50% AA identity, and such divergence suggests that it may be difficult to simultaneously inhibit both gene products, i.e., to pharmaceutically block this step of the MEP pathway in Mtb.
Our results further demonstrate that Mtb contains two GGPP synthases, encoded by Rv3383c/idsB and Rv0562/grcC1 (Table 1 and Figure 3). Given the non-essential nature of Rv3383c indicated by previously reported genetic screens (Sassetti et al., 2001, 2003; Sassetti and Rubin, 2003; Joshi et al., 2006), our results suggest that Rv3383c is functionally redundant. By contrast, Rv0562 has been found to be essential in Mtb (Sassetti et al., 2003). Its ancestral function presumably was to initiate the production of long chain isoprenylated carriers required for cell wall assembly, as this is derived from GGPP in mycobacteria other than Mtb (Besra et al., 1994), and Rv0562 is extremely well conserved in the mycobacterial family (Figure 6). However, Mtb uses (Z,E)-FPP instead of GGPP as the precursor for cell wall isoprenylated carrier biosynthesis (Schulbach et al., 2000). Thus, particularly since Rv0562 is located just downstream of genes encoding other menaquinone biosynthetic machinery in the Mtb genome (Camus et al., 2002), we hypothesize that it may instead be required for generation of GGPP for production of this essential electron carrier, presumably as precursor to the known longer chain isoprenyl sidechain (Holsclaw et al., 2008). As an alternative and/or supplemental rationale for the essential nature of Rv0562, our results indicate that this gene encodes a GGPP synthase with more robust and enzymatic activity than that encoded by Rv3383c (i.e., that encoded by Rv0562 can produce GGPP from any length allylic precursor, rather than being limited to FPP as is the IDS encoded by Rv3383c).
In conclusion, our results clarify not only the role of all the genes in the operon associated with isoTb biosynthesis, but also previous metabolic, biochemical, and genetic observations regarding isoprenoid metabolism in Mtb, as described above. Nevertheless, there are questions that arise from the results reported here, such as the role of the remaining uncharacterized IDS in the MTC, and the puzzling observation of widespread conservation of Rv3383c/idsB, presumably encoding a second GGPP synthase, in mycobacteria, which we hope to address in future studies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Michael Phillips (Centre for Agricultural Genomics, Barcelona, Spain) for the kind gift of the conditional LytB strain of E. coli, and gratefully acknowledge support for this project from the NIH (GM076324 to Reuben J. Peters).
REFERENCES
- Adam P., Hecht S., Eisenreich W., Kaiser J., Grawert T., Arigoni D., et al. (2002). Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc. Natl. Acad. Sci. U.S.A. 99 12108–12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. M., Mahapatra S., Brennan P. J., Crick D. C. (2002). Identification, cloning, purification, and enzymatic characterization of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Glycobiology 12 813–820 [DOI] [PubMed] [Google Scholar]
- Becq J., Gutierrez M. C., Rosas-Magallanes V., Rauzier J., Gicquel B., Neyrolles O., et al. (2007). Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol. 24 1861–1871 [DOI] [PubMed] [Google Scholar]
- Besra G. S., Sievert T., Lee R. E., Slayden R. A., Brennan P. J., Takayama K. (1994). Identification of the apparent carrier in mycolic acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 91 12735–12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R., Gordon S. V., Garnier T., Eiglmeier K., Frigui W., Valenti P., et al. (2007). Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. U.S.A. 104 5596–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R., Gordon S. V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., et al. (2002). A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U.S.A. 99 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke C., Croteau R. (2002). Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J. Biol. Chem. 277 3141–3149 [DOI] [PubMed] [Google Scholar]
- Camus J. C., Pryor M. J., Medigue C., Cole S. T. (2002). Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148 2967–2973 [DOI] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 537–544 [DOI] [PubMed] [Google Scholar]
- Comas I., Chakravartti J., Small P. M., Galagan J., Niemann S., Kremer K., et al. (2010). Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick D. C., Schulbach M. C., Zink E. E., Macchia M., Barontini S., Besra G. S., et al. (2000). Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 182 5771–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum M., Herrmann S., Wilkinson B., Bechthold A. (2009). Genes and enzymes involved in bacterial isoprenoid biosynthesis. Curr. Opin. Chem. Biol. 13 180–188 [DOI] [PubMed] [Google Scholar]
- Dhiman R. K., Schulbach M. C., Mahapatra S., Baulard A. R., Vissa V., Brennan P. J., et al. (2004). Identification of a novel class of omega,E,E-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J. Lipid Res. 45 1140–1147 [DOI] [PubMed] [Google Scholar]
- Eoh H., Brennan P. J., Crick D. C. (2009). The Mycobacterium tuberculosis MEP (2C-methyl-d-erythritol 4-phosphate) pathway as a new drug target. Tuberculosis (Edinb.) 89 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune S. M., Jaeger A., Sarracino D. A., Chase M. R., Sassetti C. M., Sherman D. R., et al. (2005). Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U.S.A. 102 10676–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T., Eiglmeier K., Camus J. C., Medina N., Mansoor H., Pryor M., et al. (2003). The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U.S.A. 100 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Lipatov M., Small P. M., Sheffer H., Niemann S., Homolka S., et al. (2008). High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6 e311 10.1371/journal.pbio.0060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsclaw C. M., Sogi K. M., Gilmore S. A., Schelle M. W., Leavell M. D., Bertozzi C. R., et al. (2008). Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem. Biol. 3 619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Nakano C., Ootsuka T., Shinohara Y., Hara T. (2011). Substrate specificity of Rv3378c, an enzyme from Mycobacterium tuberculosis, and the inhibitory activity of the bicyclic diterpenoids against macrophage phagocytosis. Org. Biomol. Chem. 9 2156–2165 [DOI] [PubMed] [Google Scholar]
- Hsieh M. H., Goodman H. M. (2005). The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol. 138 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama S., Shimokata K., Tsukamura M. (1988). Relationship between mycobacterial species and their carotenoid pigments. Microbiol. Immunol. 32 473–479 [DOI] [PubMed] [Google Scholar]
- Joshi S. M., Pandey A. K., Capite N., Fortune S. M., Rubin E. J., Sassetti C. M. (2006). Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl. Acad. Sci. U.S.A. 103 11760–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur D., Brennan P. J., Crick D. C. (2004). Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J. Bacteriol. 186 7564–7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Peters R. J. (2012). Isotuberculosinol: the unusual case of an immunomodulatory diterpenoid from Mycobacterium tuberculosis. Med. Chem. Commun. 3 899–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Prisic S., Hu H., Xu M., Coates R. M., Peters R. J. (2009a). Characterization and inhibition of a class II diterpene cyclase from Mycobacterium tuberculosis: implications for tuberculosis. J. Biol. Chem. 284 23574–23579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Thomas J. A., Peters R. J. (2011a). Rv0989c encodes a novel (E)-geranyl diphosphate synthase facilitating decaprenyl diphosphate biosynthesis in Mycobacterium tuberculosis. FEBS Lett. 585 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Vanderven B. C., Peters R. J. (2011b). Magnesium depletion triggers production of an immune modulating diterpenoid in Mycobacterium tuberculosis. Mol. Microbiol. 79 1594–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Xu M., Chen X., Fulton D. B., Russell D. G., Peters R. J. (2009b). Edaxadiene: a new bioactive diterpene from Mycobacterium tuberculosis. J. Am. Chem. Soc. 131 17526–17527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugel N., Mann F. M., Hillwig M. L., Peters R. J., Snider B. B. (2010). Synthesis of (+/-)-nosyberkol (isotuberculosinol, revised structure of edaxadiene) and (+/-)-tuberculosinol. Org. Lett. 12 2626–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer S., Coulson A., Mclennan N., Masters M. (2001). The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J. Bacteriol. 183 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano C., Okamura T., Sato T., Dairi T., Hoshino T. (2005a). Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem. Commun. 2005 1016–1018 [DOI] [PubMed] [Google Scholar]
- Nakano C., Ootsuka T., Takayama K., Mitsui T., Sato T., Hoshino T. (2011). Characterization of the Rv3378c gene product, a new diterpene synthase for producing tuberculosinol and (13R, S)-isotuberculosinol (nosyberkol), from the Mycobacterium tuberculosis H37Rv genome. Biosci. Biotechnol. Biochem. 75 75–81 [DOI] [PubMed] [Google Scholar]
- Nakano C., Sato T., Hoshino T. (2005b). Structure of diterpene produced by Rv3378c gene product from Mycobacterium tuberculosis. Koryo Terupen oyobi Seiyu Kagaku ni kansuru Toronkai Koen Yoshishu 49 247–249 [Google Scholar]
- Obiol-Pardo C., Rubio-Martinez J., Imperial S. (2011). The methylerythritol phosphate (MEP) pathway for isoprenoid biosynthesis as a target for the development of new drugs against tuberculosis. Curr. Med. Chem. 18 1325–1338 [DOI] [PubMed] [Google Scholar]
- Pethe K., Swenson D. L., Alonso S., Anderson J., Wang C., Russell D. G. (2004). Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc. Natl. Acad. Sci. U.S.A. 101 13642–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prach L., Kirby J., Keasling J. D., Alber T. (2010). Diterpene production in Mycobacterium tuberculosis. FEBS J. 277 3588–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T. B., Riley R., Wymore F., Montgomery P., Decaprio D., Engels R., et al. (2009). TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 37 D499–D508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K., Yates R. M., Purdy G. E., Russell D. G. (2007). Mycobacterium tuberculosis and the environment with the phagosome. Immunol. Rev. 291 37–54 [DOI] [PubMed] [Google Scholar]
- Rohdich F., Hecht S., Gartner K., Adam P., Krieger C., Amslinger S., et al. (2002). Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. U.S.A. 99 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M., Knani M., Simonin P., Sutter B., Sahm H. (1993). Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295(Pt 2) 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. G. (2007). Who puts the tubercle in tuberculosis? Nat. Rev. Microbiol. 5 39–47 [DOI] [PubMed] [Google Scholar]
- Russell D. G., Barry C. E., III, Flynn J. L. (2010). Tuberculosis: what we don’t know can, and does, hurt us. Science 328 852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H., Rubin E. J. (2001). Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 98 12712–12717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H., Rubin E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48 77–84 [DOI] [PubMed] [Google Scholar]
- Sassetti C. M., Rubin E. J. (2003). Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 100 12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Gershenzon J. (2008). Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69 49–57 [DOI] [PubMed] [Google Scholar]
- Schulbach M. C., Brennan P. J., Crick D. C. (2000). Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J. Biol. Chem. 275 22876–22881 [DOI] [PubMed] [Google Scholar]
- Singh N., Cheve G., Avery M. A., Mccurdy C. R. (2007). Targeting the methyl erythritol phosphate (MEP) pathway for novel antimalarial, antibacterial and herbicidal drug discovery: inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) enzyme. Curr. Pharm. Des. 13 1161–1177 [DOI] [PubMed] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical Taxonomy: The Principles and Practice of Numerical Classification. San Francisco: W. H. Freeman [Google Scholar]
- Spangler J. E., Carson C. A., Sorensen E. J. (2010). Synthesis enables a structural revision of the Mycobacterium tuberculosis-produced diterpene, edaxadiene. Chem. Sci. 1 202–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Jenkin G. A., Johnson P. D., Davies J. K. (2000). Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182 6322–6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., et al. (2008). Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Seemann T., Pidot S., Frigui W., Reysset G., Garnier T., et al. (2007). Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17 192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff M., Seemann M., Tse Sum Bui B., Frapart Y., Tritsch D., Garcia Estrabot A., et al. (2003). Isoprenoid biosynthesis via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe–4S] protein. FEBS Lett. 541 115–120 [DOI] [PubMed] [Google Scholar]