Abstract
Lens epithelium-derived growth factor (LEDGF), a ubiquitously expressed nuclear protein, acts by interacting with DNA and protein and is involved in widely varying cellular functions. Despite its importance, the mechanism(s) that regulate naturally occurring LEDGF activity are unidentified. Here we report that LEDGF is constitutively Sumoylated, and that the dynamical regulatory mechanism(s), Sumoylation and deSumoylation act as a molecular switch in modulating DNA binding and transcriptional activity of LEDGF with the functional consequences. Using bioinformatics analysis coupled with in vitro and in vivo Sumoylation assays, we found that lysine (K) 364 of LEDGF was Sumoylated, repressing its transcriptional activity. Conversely, mutation of K364 to arginine (R) or deSumoylation by Senp-1, a nuclear deSumoylase, enhanced the transactivation capacity of LEDGF and its cellular abundance. The enhancements were directly correlated with an increase in LEDGF’s DNA binding activity and small heat shock protein (Hsps) transcription, while the process was reversed in cells overexpressing Sumo1. Interestingly, cells expressing Sumoylation-deficient pEGFP-K364R protein showed increased cellular survival compared with the wild-type LEDGF protein. The findings provide insights into regulation and regulatory functions of LEDGF in Sumoylation-dependent transcriptional control that may be essential for modifying the physiology of cells to maintain cellular homeostasis. These studies also provide new evidence of the important role of post-translational modification in controlling LEDGF function.
Keywords: LEDGF, Senp-1, Sumo1, Sp1, Hsp27
Introduction
Lens epithelium derived growth factor (LEDGF) is an ubiquitous and stress-inducible transcriptional nuclear protein that has important functions in regulating cell proliferation, differentiation, and apoptosis [1–3]. In addition, the involvement of aberrant expression of LEDGF with cancer or progression of cancer has been documented [4, 5]. The critical importance of LEDGF in cell biology was shown when genetic ablation studies revealed that deficiency of LEDGF may lead to embryonic lethality or perinatal death, while some survivors have many abnormalities [6]. LEDGF is identical to p75 and was originally identified as a transcriptional coactivator as well as a transactivator of stress response genes, thereby providing cytoprotection against various stressors [3, 7]. LEDGF belongs to the hepatoma-derived growth factor family of proteins [8]. Its gene yields two proteins by alternate splicing: the coactivator p52 (exons 1–9 plus part of intron 9) and LEDGF (exons 1–15) [9, 10]. Both are karyophilic proteins, but differ in their nuclear localization patterns. Structurally, LEDGF contains several functional domains, which are involved in protein-protein and protein-DNA interactions and are responsible for various activities of LEDGF [9, 11, 12]. Studies have shown that LEDGF is tightly bound to chromatin during all phases of cell cycle, and this binding is mediated by the functional interaction of PWWP domain, Nuclear Localization Signal (NLS) and two AT hook motifs located in N-terminal region. A stretch of 58 amino acids, PWWP domain bound to stress response elements (STRE; A/TGGGGA/T) and C-terminal LEDGF with two helix-turn-helix (HTH)-like domains, has been found to bind to a heat shock element (HSE; nGAAn) of stress-related genes [13]. A transactivation assay using Hsp27 promoter revealed that both HTH domains contribute in a cooperative manner to the transactivation potential of LEDGF. N-terminal domain is involved in stabilizing the LEDGF-DNA binding complex. These studies underscore the importance of LEDGF in controlling many cellular functions that may depend upon cellular microenvironment and thus modifying the physiology of cells to maintain cellular homeostasis.
Recently, the role of LEDGF in modulating diverse cellular functions has been documented. It is involved in human immunodeficiency virus type-1 (HIV-1) integration and prevention of proteosomal degradation of HIV-1 integrase [14]. LEDGF/p75, but not p52, was identified as the prominent interaction partner of HIV-1 integrase [15, 16]. This interaction of HIV-1 integrase with the C-terminal integrase-binding domain of LEDGF/p75 is crucial for HIV-1 replication [14]. The protein is also involved in autoimmune disorders and cancer, through the formation of chimeric protein with NUP98 [5, 17]. A high expression level has been reported in prostate tumors [5]. We have shown that expression of LEDGF is developmentally regulated, and the level of expression influences cellular fate [18, 19]. LEDGF plays a role in lens epithelial to fiber cell terminal differentiation, and more recently the presence of this molecule in discrete regions and cell types within the fetal and adult brain suggest that it is involved in neuro-epithelium stem cell differentiation and neurogenesis [20]. All these functions collectively underscore the wide range of roles played by this molecule, ranging from cellular protection to association with cellular abnormalities. The mechanism by which LEDGF is involved in various cellular events and the specific ways in which it plays its diversified roles need to be investigated. We predict that the functions of LEDGF may be associated with its modifications such as phosphorylation and/or Sumoylation, and these modifications may be responsible for its diversified cellular functions. Recently it has been documented that LEDGF is a target of Sumo, and Sumoylation of LEDGF leads to repression of small heat shock protein transcription [21]. Even so, it remains unclear how naturally occurring LEDGF functions within the transcriptional regulatory programs that govern and regulate transactivation of stress response genes. Several possibilities exist: LEDGF may repress transcription by interacting with corepressors following Sumoylation, or LEDGF Sumoylation may decrease its DNA binding activity and definably contribute to the dynamic regulation of transcription that occurs in response to signals of cellular status. However, the mechanism involved and the molecular components that mediate such dynamic function of LEDGF or at the same-time transcriptional status of LEDGF have not been identified, nor have the functional consequences of LEDGF Sumoylation and deSumoylation on cell fate.
The small ubiquitin-like modifier (Sumo) has been shown to regulate cellular processes by controlling the localization, function and expression, and stability of large numbers of cellular proteins [22]. The complexity of LEDGF gene regulation and function was explained recently by ectopically expressing LEDGF posttranslational modification, Sumoylation. However, still not known is the fate of naturally occurring LEDGF Sumoylation and how Sumoylated LEDGF affects its own function(s) in normal physiological conditions as well as in cells facing stress or during aging [23]. Sumoylation is reversible, and the removal of Sumo, deSumoylation, is catalyzed by Sumo-specific proteases. Lysines (K) are major sites of protein modification [24]. Sumo modification of transcriptional protein is an important mechanism for achieving dynamic regulation of gene expression. However, most Sumoylated proteins have been shown to repress gene transcription [25–29]. In a reversible post-transcriptional modification, Sumo(s) are covalently linked to lysine residues of the target proteins [30]. Sumo is an 11.5 kDa ubiquitin-related protein and close relative to ubiquitin [31, 32]. Although Sumoylation is enzymatically similar to ubiquitinization, the two require different sets of enzymes. The Sumo-activating enzymes SAE1/SAE2 activate Sumo in an ATP-dependent manner. Activated Sumo is then transferred to the Sumo-conjugating enzyme Ubc9, which mediates conjugation of Sumo to an exposed lysine residue in the target protein. Sumoylation is enhanced by Sumo ligase, a diverse group of proteins that stabilize and direct the interaction between Ubc9 and its Sumoylation targets. Upon conjugation, Sumo can be efficiently removed from its targets by Sumo proteases, resulting in very low steady-state levels of the Sumo-modified forms for most Sumo targets [33, 34]. A long list of transcriptional factors includes heat shock proteins HSF1 and HSF2 that are modified by Sumo1. However, in contrast to HSF2, the HSF1 protein is not constitutively modified by Sumo1 and instead is only modified after cells are exposed to stress conditions [35, 36].
Sumoylation can be readily reversed by a family of Sumo-specific proteases (Senps). Senp-1 is a nuclear protease that appears to deconjugate a large number of Sumoylated proteins [37]. Senp-2 is a nuclear-envelope-associated protease that has activity similar to Senp-1 [38, 39]. Two additional Sumo-specific proteases, Senp-3/SMT3IP1 and Senp-6/SUSP1, have also been reported [40, 41]. Even though the ability of Senps to reverse Sumoylation is well established, the specificity of each Senp and the difference in each regulatory pathway mediated by these Senps has yet to be defined. We found that LEDGF transcription is markedly enhanced by Senp-1. Sumoylation has been linked to transcription repression in an increasing number of Sumoylated transcription factors or cofactors [42]. Several lines of evidence indicate the ability of LEDGF to bind to many proteins [11, 12, 43] including Sumo1 [21]. It seems Sumo motif (K364) present within integrase binding domain (with highest probability score) in the C-terminal of LEDGF protein is crucial for its constitutive Sumoylation and function(s) as identified by two different computational prediction programs, SUMOplot analysis program (Abgent) and PIC-Based Sumo Site Prediction Server. At present, however, the influence of endogenous LEDGF Sumoylation/deSumoylation upon LEDGF activity is in infancy. It is also unclear if Senp-1 or Sumo alters DNA binding activity and function in cells under normal physiological conditions or cells facing stresses. Also unknown is whether Sumoylaion or deSumoylation influences LEDGF’s own expression levels, as these factors may be involved in downstream signaling by modulation of transcription.
Using several biochemical approaches, here we revealed that LEDGF is Sumoylated in vitro as well as in vivo, and K364 present in the C-terminal domain (within IBD) is critical for constitutive Sumoylation of LEDGF and its regulatory function. We also showed that transcriptional activity of LEDGF was altered as a consequence of Sumo1 conjugation, and conversely, deSumoylation by Senp-1 or disruption of Sumo1 Motif; lysine (K) residue to arginine (R) impaired the Sumoylation process and promoted the transactivation capacity of LEDGF (pEGFP-K364R), which is accompanied by increased DNA binding and activation of stress response heat shock proteins and cell growth. Our data reveal a regulatory mechanism for naturally occurring LEDGF mediated by transcriptional modulation of its own and target genes via a dynamic process of Sumoylation and deSumoylation, which may have implications in cellular survival when faced with stress and human disorders.
Results
LEDGF is present in Sumoylated form and colocalizes with Sumo1 in human LECs
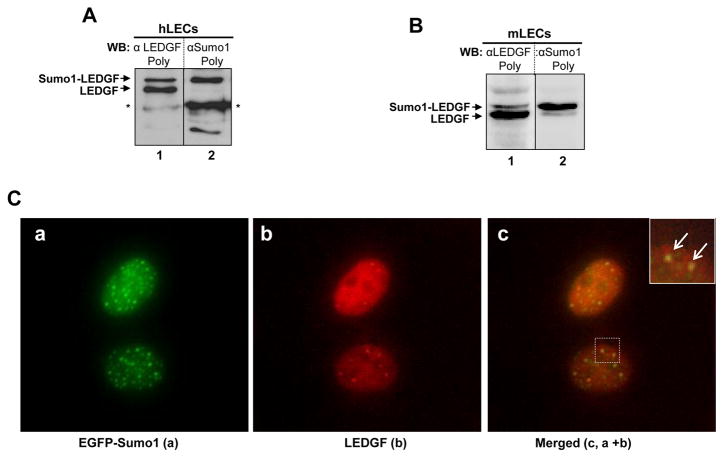
LEDGF is both a coactivator and transactivator, and most of the transcriptional proteins have been shown to be Sumoylated [44]. Considering the broad range of substrates in which the Sumoylation pathway is involved, it would not be surprising if LEDGF was to be a target of Sumo modification. Recently It has been shown that exogenous LEDGF is polySumoylated [21]. Our purpose was to observe if endogenous LEDGF is sumoylated. Western analysis of LEDGF expression reveals two forms (duplet) that are detected only by polyclonal antibodies (Lifespan Biosciences or Santa Cruz Biotech). To test whether either of these forms is yielded by Sumo1 conjugation of LEDGF, we examined the possible presence of Sumo1-modified LEDGF forms by using a membrane that had been previously probed with anti-LEDGF polyclonal antibody, and restriping and reprobing it with Sumo1 antibody (Santa Cruz Biotech). Of the two LEDGF-antibody-reacting bands (Fig. 1A; lane 1, protein bands (upper) and (lower), only the upper band of approximately ~87 kDa was recognized by Sumo1 antibody (lane 2, upper band, a). Additionally, we examined mouse (m) LECs (Fig. 1B) if the LEDGF is in Sumoylated form as in hLECs. Interestingly, similar results (two bands) were obtained with these cells, suggesting that LEDGF of 75kDa was modified with Sumo1 [45, 46] , giving rise to a band of ~87kDa (Fig. 1B). Moreover, mobility of the shifted band was dependent upon the percentage of SDS-gel (7.5%–12%) used to resolve the protein.
Fig.1.
LEDGF was present in both Sumoylated and unSumoylated form in lens epithelial cells. Nuclear extracts were prepared from human (h) and mouse (m) LECs, resolved on 7.5% SDS-PAGE (A and B), and processed for Western analysis. Membranes were striped/restriped and immunostained with LEDGF polyclonal (A, lane 1, duplet band, upper and lower) or Sumo1 antibody (A, lane 2). *denotes nonspecific band. (B) Representative Western analysis using mLECs nuclear extract with LEDGF (B, lane 1) or Sumo1 antibody (B, lane 2). (C) Immunofluorescence images showing colocalization of Sumo1 and LEDGF in nucleus. Cells were transiently transfected with pEGFP-Sumo1 and immune-stained with antibodies specific to LEDGF and Sumo1; a) pEGFP-Sumo1 (green), b) LEDGF (red), and c) merged images (c, a plus b; yellow/orange [islet; arrow indicates enlarged images of green and red merged; yellow/orange dot]). (D) Intrinsic LEDGF protein is a substrate for Sumo1 in vivo. hLECs were over expressed with pEGFP-Sumo1. Cells transfected with EGFP-empty vector served as control (lanes 1, 3 and 5). Nuclear extract were prepared 48h of post-transfection and subjected to immunoprecipitation (IP) using LEDGF monoclonal or mouse IgG (control) antibody. Input and IP samples were resolved on 4–20% SDS-PAGE and immunoblotted with LEDGF polyclonal (D, lanes 1, 2), Sumo1 polyclonal (D, lanes 3, 4), or GFP polyclonal (D, lanes 5, 6) antibody and visualize as described in ‘Materials and Methods’ section. a; samples pulled with LEDGF antibody, b; control IgG pulled samples. c; input samples subjected to IP experiments. IP experiments revealed presence of three bands with LEDGF antibody; ~75kDa (unSumoylated endogenous LEDGF), ~87kDa (endogenous Sumoylated LEDGF) and ~115kDa (endogenous LEDGF Sumoylated by exogenous Sumo1), indicating that LEDGF may contains a single site for Sumo1 protein. *denotes nonspecific band. (E) hLECs were over expressed with EGFP-Sumo1. 48h post transfection, nuclear extract was prepared and immunoprecipitation was performed using Sumo1 polyclonal antibody. Input and IP samples were resolved on 4–20% SDS-PAGE and immunoblotted with LEDGF polyclonal (E, lanes 1,2) or Sumo1 polyclonal (E, lanes 3, 4) antibody. Approximately 87kDa (endogenous Sumoylated LEDGF) and ~115kDa (endogenous LEDGF Sumoylated by exogenous EGFP-Sumo1) protein bands could be detected.
Furthermore, LEDGF is predominately localized in nucleus [10, 13, 47–49], and Sumo1 is known to be localized and exerts its genetically defined activity in nucleus [50]. As our Western blot data indicated that a fraction of naturally occurring LEDGF was constitutively Sumoylated, we next examined whether LEDGF and Sumo1 are colocalized. We performed immunocytochemistry using Sumo1 or LEDGF antibody, respectively. Immunofluorescence analysis determined that both molecules colocalized in nucleus. Merged images of Sumo1 (Green) and LEDGF (Red) that yielded yellow color or granules revealed further that a portion of total LEDGF protein interacted with Sumo1 (Fig. 1C), suggesting that only a certain amount of LEDGF is Sumoylated.
We wished to know more about if endogenous LEDGF is, indeed, Sumoylated, we performed immunoprecipitation assay using antibody specific to LEDGF or IgG antibody (control) or Sumo1 antibody. To this end, hLECs were transiently transfected with EGFP-Sumo1. 48h later nuclear extracts were processed for immunoprecipitation and immunoblotted as decribed in ‘Materials and Methods’ section. Cells transfected with EGFP-empty vector were taken as control (lanes 1, 3 and 5). Results revealed the presence of three bands with anti-LEDGF polyclonal antibody (Fig. 1D; a, lane 2: unSumoylated endogenous LEDGF, ~75kDa; endogenous Sumoylated LEDGF, ~87kDa and endogenous LEDGF Sumoylated by exogenous EGFP-Sumo1, ~115kDa). The same membrane was probed with anti-Sumo1 that gave rise two bands (Fig. 1D; a, lane 4: endogenously Sumoylated LEDGF, ~87kDa and endogenous LEDGF Sumoylated with exogenous EGFP-Sumo1, ~115kDa), while only one band could be detected when the same membrane probed with anti-GFP antibody (Fig. 1D; a, lane 6: endogenous LEDGF Sumoylated with EGFP-Sumo1). To validate further Sumoylation of endogenous LEDGF, nuclear extracts were immunoprcipitated with anti-Sumo1 antibody and immunoblotted with anti-LEDGF antibody (Fig. 1E; lane 1) or anti-Sumo1 antibody. Cells transfected with EGFP-empty vector were taken as control (lanes 1 and 3). The data revealed the presence of two bands; Sumoylated LEDGF (~87kDa) and endogenous LEDGF Sumoylated with exogenous EGFP-Sumo1 (~115kDa). In contrast, nuclear extract immnoprecipitated with IgG antibody (control) did not produce any detectable specific band with either of specific antibodies, validating specificity of experiments (Fig. 1D; b). Collectively, these results indicate that a fraction of LEDGF is present in mono-Sumoylated form. Taken together, endongenous LEDGF was Sumoylated with endogenous Sumo1 as well as exogenously expressed EGFP-Sumo1, suggesting further that LEDGF is substrate for Sumo1.
LEDGF is Sumoylated both in vitro and in vivo, and conserved lysine (K) 364 is subject to Sumoylation
Ubc9 is an E2 conjugation enzyme for protein Sumoylation, and interaction with UBC9 is a feature of many proteins subsequently modified by Sumo1. To test whether this occurs in LEDGF–Sumo1 conjugation, we performed in vitro Sumo1 conjugation assay. We prepared constructs expressing GST-LEDGF and GST-Sumo1 as described in the ‘Materials and Methods’ section, and verified expression of these two through Western blot (data not shown). Purified GST-LEDGF protein was incubated with purified Sumo1, activating enzyme E1 (SAE1/2) and conjugating enzyme E2 (Ubc9) in a buffer containing an ATP-generating system. As shown in Fig. 2A, in the presence of enzymes, a higher molecular weight band was recognized by antibodies specific to GST (left panel, lane 1), Sumo1 (middle panel, lane 1), or LEDGF (right panel, lane 1) in Western blot, confirming the identity of a slowly migrating Sumoylated LEDGF band. The data indicate that LEDGF is substrate for Sumo1, and Ubc9 is essential as an interacting partner for the Sumoylation process. In contrast, purified GST protein did not show interactions, so we could not detect such a band (data not shown). HSF1 is known to be Sumoylated. In parallel experiment we also used HSF1 Sumoylation experiment as control to validate the experiments (Fig. 2B).
Fig. 2.
LEDGF underwent Sumo1 modification in vitro. (A) The in vitro Sumoylation assay was done according to the manufacturer’s protocol. Briefly, a combination of E1 enzyme, E2 (Ubc9) enzyme, Sumo1 protein and GST-LEDGF was mixed with in 20 μl reaction mixture containing Sumoylation buffer. Following incubation at 30°C for 3h, reaction product was incubated with 2X SDS-gel loading buffer and processed for Western blot analysis using anti–GST, Sumo1, and LEDGF antibodies. (B) GST-HSF1 was used as positive control to verify the identity of experiments [94].
Next, we determined the critical region or Sumo1 conjugation site(s) of LEDGF protein using SUMOplot prediction, a Web-based tool for predicting consensus Sumoylation (Abgent). Sequence analysis predicted six Sumo1 conjugation sites (Fig. 3A); however, the highest score was predicted with lysine (K) 364 (LKID) present at C-terminal of LEDGF. To ascertain accurate Sumoylation site(s) of LEDGF, we generated various deletion constructs of GST-LEDGF expression plasmids containing predicted sequences and we also utilized previously prepared LEDGF constructs in expressing prokaryotic vectors. In vitro Sumoylation assay disclosed that C-terminal LEDGF ranging from 170 to 530 amino acids (aa) is, indeed, Sumoylated (Fig. 3B, lane 3), while N-terminal (1–250aa) was not, emphasizing that C-terminal LEDGF bore Sumo1 conjugation motif.
Fig. 3.
Sumo1 conjugation motif resided in the carboxyl terminus of LEDGF and Sumoylated. (A) Schematic representation of full-length LEDGF protein showing the putative Sumo1 conjugation motif(s) as predicted by SUMOplot (Abgent). The position of modifiable lysine (K) site(s) in each Sumo1 site is indicated by number. GST-linked full-length LEDGF -or NH2 - or COOH- LEDGF used in assay are shown as Full, N and C, respectively. The amino acid sequence of LEDGF was analyzed to identify possible Sumoylation sites. Full and deleted NH2-and COOH-LEDGF constructs (N, 1-250aa or C, 170-530aa) were generated using PCR with LEDGF sense and antisense primers and cloned into pGEX-2T vector. Recombinant protein was purified and used for assay. (B) C-terminal LEDGF was targeted by Sumo1 conjugation, while NH2 terminus was not. To analyze which region of LEDGF was Sumoylated, N-terminal (N, 1-250aa) and C-terminal (C, 170-530aa) deletion constructs of LEDGF were subjected to in vitro Sumoylation assay. Western blot analysis was conducted with LEDGF antibody. Sumoylated C-terminal LEDGF band with retarded mobility (B, lane 3); NH2–terminal, lanes, 1 and 2. (C) Western analysis image showing the expression of purified recombinant NH2-and COOH-LEDGF protein immunostained with anti-GST antibody.
Furthermore, a closer analysis of the predicted sequence revealed that C-terminal of LEDGF contained only one Sumo1 conjugation motif with the highest score (SUMOplot, Score < 90) while other two had lowest scores. Additionally, in vitro Sumoylation assay and SUMOplot prediction supported the fact that K364 (LKID) can be an actual site for LEDGF Sumoylation, which is also evolutionarily well conserved (Fig. 4). Based on these facts, to determine if lysine within the integrase binding domain (IBD) [21] is required for the Sumoylation of LEDGF, we mutated lysine (K) to arginine (R), generating LEDGF mutant (K364R) and performed in vivo Sumoylation experiment as described in ‘Materials and Methods’ section. Coexpression of EGFP-Sumo1 and lysine mutated forms of EGFP-LEDGF (K364R) showed that EGFP-LEDGF (K364R) was not Sumoylated, and thus could not be detected by anti-Sumo1 antibody (Fig. 5A, right panel, lane 3). Conversely, wild type EGFP-LEDGF or endogenous LEDGF was Sumoylated, and therefore migrated more slowly on gel, yielding band: EGFP-LEDGF plus EGFP-Sumo1 (Fig. 5A, ~143kDa, lane 2), endogenous LEDGF plus EGFP-Sumo1 (Fig. 5A, ~115kDa, lane 1, 2, 3), endogenous LEDGF plus endogenous Sumo1 (~87kDa, lane 1, 2, 3). Next, the same blotted membrane was probed with anti-LEDGF (Fig. 5B) or anti-GFP (Fig.5C) antibody to examine if the Sumo1 recognized band is indeed endogenous /exogenous LEDGF. Results revealed that anti-LEDGF (Fig. 5B) or anti-GFP (Fig. 5C) antibody detected the same protein bands as recognized by anti-Sumo1 antibody (Fig. 5A). Thus our data revealed that disruption of K364R attenuated LEDGF Sumoylation (Fig. 5A lane 3), and data points that K364 was an active and effective Sumoylation site in LEDGF, although residual Sumoylation may have occurred at the lysine residues outside IBD domain of LEDGF, but we were unable to detect them in our system [21].
Fig. 4.
Sumo1 motif (K364) within the IBD domain of COOH-LEDGF was evolutionarily well conserved. Bioinformatics analysis and SUMOplot, a web-based program, were used to evaluate Sumo1 conjugation motifs present in LEDGF protein. Sequence alignment of human, bovine, mouse, chicken and xenopus LEDGF protein was done to identify evolutionarily conserved Sumoylation sites (ClustalW). Important lysine residues are indicated in red bold (Score; <90).
Fig. 5.
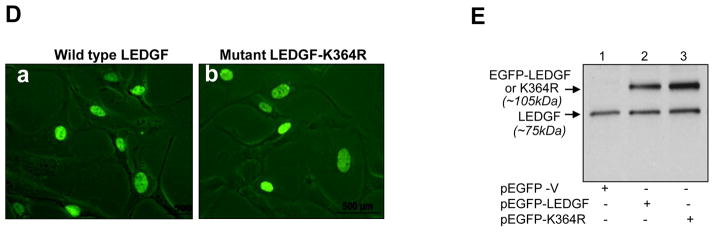
LEDGF was modified by Sumo1 in vivo, and conserved lysine (K) 364 in the IBD domain of COOH-terminal is a Sumo1 conjugation motif. Cells were co-expressed with pEGFP-Sumo1 and pEGFP-LEDGF or pEGFP-K364R along with pCMV-Ubc9. Cellular extracts were processed for immunoprecipitation (IP) using anti-LEDGF monoclonal antibody. (A) The membrane was immunoblotted with anti-Sumo1 antibody: endogenous Sumoylated LEDGF (~87kDa), endogenous LEDGF Sumoylated with exogenous EGFP-Sumo1 (~115kDa; EGFP-Sumo1+LEDGF) and exogenous LEDGF Sumoylated with exogenous EGFP-Sumo1 (~143kDa; EGFP-LEDGF+EGFP-Sumo1). (B) The same membrane was immunoblotted with anti-LEDGF antibody: unSumoylated endogenous LEDGF (~75kDa), unSumoylated EGFP-LEDGF or K364R (~105 kDa), endogenous Sumoylated LEDGF (~87kDa), and endogenous LEDGF Sumoylated with exogenous EGFP-Sumo1 (~115kDa; *EGFP-Sumo1+LEDGF) and exogenous EGFP-LEDGF plus exogenous EGFP-Sumo1 (~143kDa; **EGFP-LEDGF+EGFP-Sumo1). (C) The same membrane was immunoblotted with anti-GFP antibody: EGFP-LEDGF or K364R (~105 kDa), Endogenous LEDGF Sumoylated with exogenous EGFP-Sumo1 (~115kDa; *EGFP-Sumo1+LEDGF) and exogenous EGFP-LEDGF Sumoylated with EGFP-Sumo1 (~143kDa; **EGFP-LEDGF+EGFP-Sumo1). Sumoylated form of Mutant LEDGF (K364R) could not be detected; indicating Lysine (K) at 364 is the major Sumoylation site in LEDGF protein.
(D) Immunofluorescence images showing localization of LEDGF and its mutant form, EGFP-K364R. Cells were transfected with wild type pEGFP-LEDGF (left panel) or mutant LEDGF pEGFP-K364R (right panel) and fluorescence images of live cells were recorded after 24 h transfection under inverted fluorescence microscope (Nikon Eclipse Ti-U). E, Western analysis of cellular extract obtained from LECs transfected with pEGFP-LEDGF or pEGFP-K364R following equalization with GFP (O.D. at Ex485/Em530).
We next tested if Sumoylation/deSumoylation alters LEDGF localization, a nuclear protein, we transfected cells with pEGFP-K364R (mutant LEDGF) and wild type pEGFP-LEDGF. Disruption of Sumo1 motif did not affect LEDGF localization pattern, and predominantly localized in nucleus as wild type LEDGF (Fig. 5D). Next we tested identity and expression levels of LEDGF and mutant LEDGF EGFP-K364R by Western analysis (Fig. 5E). We found that protein bands of mutant LEDGF EGFP-K364R and wild type EGFP-LEDGF (Fig. 5E, lane 2 vs 3, upper band) were of correct molecular weight size (~105kDa), suggesting that protein integrity is maintained. In addition, consistent with findings of recent published works showing that protein Sumoylation may lead to protein stabilization or vice versa [21], Our result also revealed that EGFP-LEDGF (K364R) is more stable than wildtype-EGFP-LEDGF. However, our results are consistent with earlier reports that Sumoylation destabilizes LEDGF proteins and decreases its half-life [21].
A dynamical process, Sumoylation/deSumoylation affects transactivation capacity of LEDGF and regulates transcription
Given the roles of Sumoylation and deSumoylation of proteins in modulation of transcriptional activity, and in vitro and in vivo Sumoylation of LEDGF (current data, Figs. 1–5) as well report of others [21], we first examined whether Sumo1/Senp-1 alters the endogenous expression of LEDGF target gene, Hsp27. Cellular extracts isolated from Sumo1 or Senp-1 transfected cells were analyzed by Western blot using antibodies specific to Sumo1, Senp-1 or LEDGF (monoclonal antibody), or Hsp27. We found that Sumo1 downregulated the expression of LEDGF (Fig. 6A, b) and Hsp27 (Fig. 6A, c), and that the decrease of Hsp27 was dependent upon the concentration of Sumo1 (Fig. 6A, a), while Senp-1 increased the expression of LEDGF (Fig. 6B, b) and Hsp27 (Fig. 6B, c) proteins in dose-dependent fashion. Our data indicated that the decreased expression of Hsp27 in cells was associated with the increased Sumoylation of LEDGF.
Fig. 6.
(A) Sumo1 and Senp-1-dependent regulation of endogenous LEDGF activity in regulating Hsp27 protein expression. Cells were transfected with different concentrations of pEGFP-Sumo1 (0.5, 1.0 and 2.0 μg). Cell extracts isolated after 48h of transfection were resolved on 10% SDS-PAGE, and Western analysis was conducted using specific antibodies as indicated. The same membrane was utilized to visualize the relative expression levels. (B) Cells were transfected with pFlag-Senp-1 at different concentrations as indicated. Western analysis was carried out and membranes were striped/restriped and immune-stained with Senp-1 or Hsp27 antibodies. β-actin antibody was used as an internal control and to normalize expression. (* p<0.05; **p<0.001)
(C) Disruption of LEDGF Sumoylation motif, K364 (K to R) promoted its transcriptional capacity. Cells were transfected either with pEGFP-vector, pEGFP-LEDGF or pEGFP-K364R and αB crystallin (left part), or Hsp27 (right part) promoters linked to CAT reporter vector. After 72h cell lysates were analyzed for CAT activity. (D) LEDGF siRNA assay showing the involvement of Sumo1 and Senp-1 in modulating transcriptional activity of LEDGF. Cells were transiently cotransfected with pCAT-Hsp27 reporter plasmid and pEGFP-Sumo1 (2 μg) or pFlag-Senp-1 (0.15 μg) or with or without siRNA specific to LEDGF or with empty vector as indicated. CAT activity was monitored. Results are represented as a histogram, and promoter activity was compared between siRNA-LEDGF transfected and untransfected cells (D; left half, black bar and right half, black bar). The data represent the mean ± SD from three independent experiments. (* p<0.05; **p<0.001) statistically significant difference. (E) Silencing of LEDGF by specific siRNA was confirmed through western analysis (upper panel). The membrane was striped and stained with β-actin antibody to normalize expression.
Next, we performed a series of cotransfection transactivation assays using mutants of LEDGF pEGFP-K364R at Sumo1 conjugation site (Fig. 6C), pEGFP-Sumo1 or pFlag-Senp-1 (Fig. 6D). The specificity of LEDGF-dependent activity was evaluated by using siRNA specific to LEDGF (Fig. 6E). First we compared the transactivation ability of wild-type (WT)-LEDGF with that of Sumoylation-deficient mutant (LEDGF) pEGFP-K364R to activate the small heat shock protein genes, αB-crystallin and Hsp27. LEDGF binds to heat shock element (HSE, nGAAn) in Hsp27 and αB-crystallin promoters and to up-regulate their transcription [2, 3, 51]. LECs were cotransfected either with pEGFP-vector, pEGFP-LEDGF (WT) or pEGFP-K364R (mutant) plasmid along with reporter plasmid Hsp27 or αB-crystallin promoters linked to CAT vector, and cell extracts were analyzed with CAT-ELISA. Both forms of LEDGF activated CAT gene expression above the basal level (Fig. 6C, light gray bar). However, the transactivation potential of pEGFP-K364R in Hsp27 or αB-crystallin promoter activity was significantly greater than that of pEGFP-LEDGF (Fig. 6C, black bar). Because single mutation disrupts Sumoylation of LEDGF, our data indicated that covalent modification of LEDGF by Sumo1 transrepressed LEDGF’s transactivation capacity. Collectively, results revealed an increase in the transactivation potential of LEDGF when lysine (K) 364 was changed to arginine (R).
Sumoylation and deSumoylation are reversible processes [52], and the removal of Sumo1, deSumoylation, is catalyzed by Sumo-specific proteases. However, Senp-1 is most abundantly engaged in deSumoylation of proteins [37]. However, it was not clear whether cellular expression levels of Senp-1 or Sumo1 influenced LEDGF transcriptional activity. Thus, to identify the critical link among Sumoylation (EGFP-Sumo1), deSumoylation (Senp-1) and LEDGF (by applying siRNA of LEDGF) in vivo, we utilized transactivation assays by cotransfecting cells with the plasmid(s) of above-mentioned reagents and reporter plasmids, pCAT-Hsp27. Promoter activity was dramatically increased (Fig. 6D, left part, black bar), and the activity was dependent specifically upon LEDGF as evidenced by LEDGF-specific interference (Fig. 6D, right part, black bar). In contrast, reduced activity was observed in cells cotransfected with pEGFP-Sumo1 (Fig. 6D, light gray bars vs gray bars). Also, western analysis revealed that LEDGF siRNA used for the assay effectively reduced the expression of LEDGF protein, (Fig. 6E, lane 2) and that reduced promoter activity was correlated with reduced expression of LEDGF protein.
These data implied that Sumoylation and deSumoylation of LEDGF affects its transcriptional efficacy. Moreover, we also recognized that Sumo1 modification of LEDGF may change its conformation, making LEDGF less accessible to nucleus. This possibility was ruled out when transfection assay revealed nuclear localization of mutant LEDGF- K364R, and the localization pattern was indistinguishable from that of LEDGF (Fig. 5D). Collectively the findings provide evidence that Sumoylation/deSumoylation effectively influenced the regulatory activity of LEDGF, in turn affecting expression of its target genes.
Sumoylation diminished the DNA binding activity of LEDGF, suggesting that reduced transactivation activity was dependent upon LEDGF-DNA interaction
The transactivation experiments did not reflect whether modulation in transactivation activity of LEDGF was associated with its DNA binding. Recent evidence revealed that Sumo1 conjugation may alter DNA-binding activity of proteins [53, 54]. To determine whether Sumoylation affected the DNA binding activity of LEDGF, we carried out gel-shift mobility assay. Nuclear extracts from cells transfected with pFlag-Senp-1, pEGFP-Sumo1 or empty vector were incubated with radio-labeled probes containing HSE (heat shock element) or STRE (stress response element) and processed for gel-shift assay to examine the influence of Sumoylation of LEDGF on its DNA binding activity (Fig. 7A). The binding of LEDGF in nuclear extract from Sumo1-transfected cells to probe was reduced significantly (Fig. 7A, lane 2) compared to vector-transfected cells (lane 1). Conversely, LEDGF in nuclear extracts of Senp-1-transfected cells bound with greater affinity to the same probe and formed complex, Cm1 (Fig. 7A, lane 3). The data demonstrated that Senp-1 induced increased LEDGF interaction with its binding sites. Previously we had shown that LEDGF binds to HSE (nGAAn) and STRE and activates transcription of stress associated genes [3, 18, 49].
Fig. 7.
(A) Senp-1 increased the DNA-binding activity of LEDGF. Nuclear extracts isolated from cells transfected with pFlag-Senp-1, pEGFP-Sumo1 or plasmid vector were incubated with 32p-labeled wild type probe (left panel) or its mutant (right panel) and then processed for gel-shift mobility assay. The effects of Sumo1 (lane 2) and Senp-1 (lane 1) on LEDGF DNA binding compared to control (lane 1) are shown. Cm1 indicates DNA and LEDGF complex. (B) Right graph indicates binding intensity (in pixels).
Next, we examined whether the increased binding of LEDGF was associated with greater/lesser abundance in cells or with its modulated affinity to DNA binding due to Sumoylation. Initially, levels of LEDGF in cells transfected with Senp-1, Sumo1 or empty vector were subjected to Western analysis using anti-LEDGF monoclonal antibody. Cell extracts from pFlag-Senp-1-transfected cells contained increased levels of LEDGF compared to cells transfected with Sumo1 or vector (Fig. 6A and B). This indicated that Senp-1-mediated abundance of deSumoylated LEDGF may be at least one cause of increased DNA binding as evidenced by Fig. 6B, b. Sumoylation and deSumoylation have been shown to influence protein stability and integrity [55, 56], and deSumoylated LEDGF protein is known to have greater stability as it is less susceptible to degradation than Sumoylated LEDGF [21]. Moreover, LEDGF is an inducible and constitutively transcriptionally active gene, and Sumo acts as a controller for constitutive transcription and during activation of inducible genes [57]. The presence of Sumo1 at transcriptionally active gene LEDGF suggests that Sumo1 plays a role in regulating LEDGF transcription [42, 58–60].
Interestingly, while evaluating the expression level of LEDGF in cells overexpressing Senp-1, we observed that LEDGF’s expression increased with an increase in Senp-1 concentration (Fig. 6). We speculated existence of three possibilities: (i) DeSumoylation of LEDGF increased its half-life by slowing its degradation processing by ubiquitization pathway(s) opposed to Sumo1 as Sumo1 conjugation of LEDGF decreased LEDGF half-life or stability [21]. (ii) Senp-1 enhanced LEDGF transcription by enhancing LEDGF’s transactivator, thereby increasing expression of LEDGF in cells. (iii) Senp-1 enhanced abundance of LEDGF expression via both mechanisms, however Sumoylation increased LEDGF degradation pathways is known [21]. Therefore we proceeded to explore whether mRNA of LEDGF was increased in cells harboring higher level of Senp-1.
Sumo-protease, Senp-1 enhanced expression of LEDGF mRNA by promoting LEDGF gene transcription in LECs
Endopetidase Senp-1 has been shown to remove Sumo from conjugated protein substrate including transcriptional proteins and to modulate their transcriptional activity, which involves Sumo conjugation in repression of gene transcription [37]. However, in the current study (Fig. 6), we observed that the level of LEDGF protein was elevated in cells following increased expression of Senp-1, and the increased expression of LEDGF was one factor in increased Hsps expression. Reasoning that expression levels of Senp-1 may increase LEDGF expression by promoting LEDGF gene transcription, we determined first whether Senp-1 expression could regulate endogeneous expression of LEDGF mRNA in LECs. By real-time PCR we found that cells overexpressing Senp-1 displayed greater abundance of LEDGF mRNA (Fig. 8A, black bar) than cells with vector or Sumo1 plasmids. Sumo1 transfected cells displayed reduced expression of LEDGF mRNA, indicating that Sumo1 repressed the transcription of LEDGF as opposed to Senp-1 (Fig. 8A, open bar vs gray bar vs black bar). Because modulation of LEDGF expression occurred at the mRNA level, we postulated that Sumoylation/deSumoylation may occur in the transcriptional protein responsible for transcriptional activation of LEDGF. To find out this, we generated truncated constructs of LEDGF promoter linked to CAT vector as described earlier [61]. Cells were cotransfected with a series of 5'-deletion mutant constructs of LEDGF gene promoter linked to CAT containing common 3' end or their wild type and pFlag-Senp-1 as shown in Fig. 8B and reported earlier [61]. Cellular extracts of transfectant(s) were analyzed for promoter activity by monitoring CAT reporter protein as described in ‘Experimental Procedure’ section. Data analysis showed that Senp-1 dramatically increased LEDGF gene promoter activity (black bars vs. gray bar), and stimulation in promoter activity was Senp-1 dependent (Fig. 8B, black bar). Significantly increased promoter activity was observed with reporter plasmid containing −170/+35 and −127/+35 bp promoter regions, while lowest promoter activity was shown by deletion mutant plasmid with −28/+35, suggesting that transcriptional protein binding response element(s), which are plausibly involved and activated by Senp-1 in transactivating LEDGF, can be present between −170 and −28nts (Fig. 8B).
Fig. 8.
(A) LECs overexpressing Senp-1 displayed elevated expression of LEDGF mRNA than cells overexpressing Sumo1. Cells were transfected either with vector plasmid (open bar), pEGFP-Sumo1 (gray bar) or pFlag-Senp-1 (black bar). Total RNA extracted from transfectant was submitted to real-time PCR using primers specific to LEDGF. Values were normalized with β–actin and presented (* p<0.05; **p<0.001).
(B) Senp-1-dependent transactivation of deletion mutants of 5′-proximal regulatory region of LEDGF gene promoter. (a) Schematic representation of partial constructs of LEDGF gene promoter linked to CAT vector [61]. Various deletion mutants of LEDGF gene promoter linked to reporter plasmid CAT were engineered as described in the text [61]. Extracts isolated from cells cotransfected with deletion mutants, plasmid constructs and pFlag-Senp-1 or pFlag-Senp-1 mutant as indicated were tested for CAT activity. The transactivation activities of truncated constructs in the presence of pFlag-Senp-1 mutant (B, black bar) or pFlag-Senp-1 (B, gray bar) are shown. Transfection efficiencies were normalized using pSEAP basic vector. The data represent the means ± SD from three independent experiments (* p<0.05; **p<0.001).
(C) Senp-1 dramatically enhanced LEDGF transcription in concentration-dependent fashion. hLECs were co-transfected with LEDGF-CAT plasmid (−170/+35; based on results obtained from Experiment 8B) with increasing concentration of pFlag-Senp-1 (gray bars; 0.05, 0.15, 0.5, 2 and 2μg) or its mutant plasmids (black bar). After 72h of transfection, extracted cell lysates from these transfectants were analyzed for CAT activity. CAT activity is shown as a histogram (pFlag-Senp-1 mut, black bar; pFlag-Senp-1, gray bar) (**p<0.001).
(D) LEDGF transcription was downregulated in cells overexpressing Sumo-1 in comparison to Senp-1. hLECs were co-transfected with LEDGF-promoter linked to CAT vector (−170/+35) along with either pEGFP-Sumo1 or pFlag-Senp-1. Cell lysates isolated after 72h of transfection were examined. The effects of pEGFP-Sumo-1 (black bar) or pFlag-Senp-1 (light gray bar) on CAT activity are shown. Empty CAT-vector served as a control (open bar). The transfection efficiencies were normalized using pSEAP basic vector. The data represent the means ± SD from three independent experiments (**p<0.001).
LEDGF gene promoter region spanning from −170 to −28 contained critical responsive element(s) for transcription factor sensitive to Senp-1 and/or Sumo1 modification
To determine whether the transcription factor binding region of 5'-promoter between -170/-28nt is, indeed, responsive to Senp-1 and/or Sumo1-dependent regulation, we cotransfected cells with reporter plasmid containing LEDGF promoter spanning −170 to +35 linked to CAT and pFlag-Senp-1 (0.05, 0.15, 0.5, 2 μg), or Sumo1, or control vector(s). Titration of Senp-1 showed a dose-dependent enhanced transcription of LEDGF (Fig. 8C). Transcriptional activity of LEDGF was at its peak (~40 fold) at 2μg of Senp-1 concentration (Fig. 8, gray bar). Even at a very low concentration (150ng), Senp-1 induced LEDGF’s transcription significantly compared to control (black bars). This indicates transcription of LEDGF transcription by Senp-1, and showed that transcription factors (activator/enhancer) were a target of Sumo1 modification.
Sumo proteins are known to influence the activities of other proteins by direct conjugation. We found that Senp-1 upregulated LEDGF transcription, so it is possible that Sumo conjugation to some factor(s) is the mechanism underlying the action of Sumo1 on LEDGF transcription. To test whether Sumo1 repressed the transcription of LEDGF, we transfected cells with pEGFP-Sumo1, pFlag-Senp-1 empty vector along with reporter plasmid, pCAT-LEDGF (−170/+35) and performed transactivation assays. Enforced expression of Sumo1 led to a decrease in LEDGF’s promoter activity (Fig. 8D, black bar vs gray bar). On the other hand, Senp-1 increased the promoter activity significantly (Fig. 8D, light gray bar vs dark gray or black bar) as observed in previously. Collectively, our data demonstrated that LEDGF expression was regulated by dynamical processes of Sumoylation and deSumoylation. We posit that the transcriptional regulator of LEDGF is a target of such a process, and that process controls LEDGF transcription in accord with cellular needs and background.
Senp-1 potentiated Sp1 activation of LEDGF transcription
To further investigate the potential cis-regulatory elements of LEDGF promoter involved in transcriptional control of the expression of LEDGF, we analyzed 5'-LEDGF promoter sequences (within −170 to -28) [61] using a Web-based computer program for predicting putative transcription factor binding sites (MatInspector, Genomatix). Our unpublished findings coupled with the current analysis of LEDGF promoter convinced us that three putative Sp1 sites in LEDGF promoter positioned at -50/−43 (Sp1-3), -109/-102 (Sp1-2), and -146/-139 (Sp1-1) were responsible for transcription. The most importantly, it has been shown recently by other research group that Sp1 is Sumoylated by Sumo1, and that Sumoylation attenuates Sp1-dependent transcription [62]. Conversely, Senp-1 was found to stabilize Sp1 activity. Under this scenario, we predicted that modulation in transcription of LEDGF by Senp-1 and Sumo1 should be associated with regulation of Sp1 by these molecules. To test whether regulation of LEDGF is indeed under the control of the regulatory activity of Sp1, we performed cotransfection transactivation experiments using wild type LEDGF promoter (−170/+35) or its mutant at all Sp1 sites linked to reporter gene CAT along with pFlag-Senp-1 plasmid of variable concentration (Fig. 9). Interestingly, cellular extract from Senp-1-transfected cells showed higher LEDGF promoter activity, and promoter activity increased with an increase of Senp-1 concentration (Fig. 9, black bar). However, mutant promoter showed some activity to Senp-1, but was significantly lower than that of wild-type promoter (right part, light gray bar vs black bar). We think that promoter activity in response to Senp-1 should be associated with other cofactors/factor which may be target for Sumoylation/deSumoylation process. As whole our results revealed that expression and function of LEDGF is regulated by at posttranslational as well as transcriptional levels.
Fig. 9.
Senp-1 promoted LEDGF transcription through specificity protein, Sp1. Upper panel, schematic illustration of wild type LEDGF gene promoter construct (−170/+35) containing three Sp1 binding elements (WT) and its mutant mutated at all three Sp1 sites (disrupted using site-directed mutagenesis) linked to CAT vector. Lower panel, cells were transfected with either wild type (left part) or mutant (right part) LEDGF promoter constructs. Both groups were cotransfected with pFlag-Senp-1 (0.0 μg/ml, light gray bars; 0.50 μg/ml, black bars and 0.05 μg/ml, dark gray bars). The transfection efficiencies were normalized using pSEAP basic vector. The data represent the means ± SD from three independent experiments (* p<0.05; **p<0.001).
Sumoylation and desumoylation of LEDGF affected growth and survival of cells at 37°C as well as during heat stress
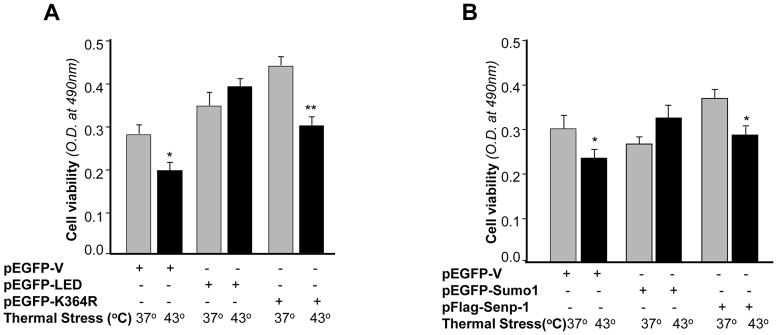
Sumoylation and deSumoylation are dynamic processes, and either activation or inhibition of those processes would have a profound effect on subsequent cellular events. However, LEDGF expression and stability in response to stressors is imperative for maintaining its ability to coordinate the stress response and protect cells, and it acts by upregulating stress response genes [3, 23, 49, 61, 63, 64]. Under conditions of stress, the current study showed that increased expression and stability of LEDGF appeared to be attributable, in part, to its transcriptional and post-transcriptional control by Sumoylation/deSumoylation. We next examined whether LEDGF’s protective activity against environmental stressors was altered. Cells transfected with wild type LEDGF, mutant LEDGF pEGFP-K364R or pEGFP-vector were submitted to heat stress [49, 65]. Interestingly, cells overexpressing mutant LEDGF pEGFP-K364R at Sumo1 conjugation site showed better growth than wild type LEDGF under normal physiological conditions (Fig. 10, 37°C; gray bars). In contrast, the cells with mutant LEDGF pEGFP-K364R did not have resistance against heat stress compared to wild type LEDGF transfected cells (Fig. 10, 43°C, black bars). However, we could not be able to explain how the protective mechanism of LEDGF and mutant LEDGF-K364R became different in cells facing stress and cells at normal physiological condition. Next, our interest was to examine the effect of Senp-1 or Sumo1 expression on cell survival. Cells were overexpressed with either pEGFP-Sumo1 or pFlag-Senp-1, and were subjected to heat stress. Increased cell growth was seen in cells overexpressing Senp-1 compared to cells overexpressing Sumo1, under normal physiological conditions (Fig. 10B, gray bars). But the phenomenon was reversed when cells were subjected to heat stress (Fig. 10B, 43°C, black bars), and survival of cells overexpressing wild type LEDGF was increased. Results of both experiments (Fig. 10A and B) were similar, and we believe that Fig. 10A defines specific and selective functions of Sumo1 or Senp-1 in context to LEDGF, while the results in Fig. 10B may indicate broader effects of Sumoylation and deSumoylation, and may also include regulatory activity of Sp1 and Sp1-mediated regulation of LEDGF transcription. As a whole, these data suggest that Sumoylation and deSumoylation play a critical role in cellular integrity and expression of LEDGF, one which defines the function of LEDGF depending upon cellular microenvironment and cellular needs.
Fig. 10.
(A) Disruption of LEDGF Sumoylation motif, K364 (K to R) promoted hLECs growth and survival at normal physiological temperature, but failed to do so during heat stress. Cells were transfected with pEGFP-vector, pEGFP-LEDGF or pEGFP-K364R plasmid. After 48 h, MTS assay was performed (A, gray bars). The data represent the mean ± SD from three independent experiments. In a parallel experiment, cells transfected with the above constructs were subjected to heat stress (43°C for 1h), and viability was monitored with MTS assay (A, black bars) following a recovery period. Transfection efficiency was normalized with values of EGFP. The data represent the means ± SD from three independent experiments (* p<0.05; **p<0.001).
(B) Influence of Sumo1 on hLECs growth and survival at normal physiological temperature or under heat stress. Cells were transfected with pEGFP-vector, pEGFP-Sumo1 or pFlag-Senp-1 as indicated. Cells were submitted for MTS assay after 48h of transfection with (black bars) or without (gray bars) heat stress. The data represent the mean ± SD from three independent experiments (* p<0.05).
Discussion
LEDGF belongs to the hepatoma-derived growth factor (HDGF) family of proteins that consists of a well-conserved N-Terminal amino acid sequence called the HATH (homologous to amino terminus of HDGF) region [10, 66, 67]. It is a stress-inducible and acts transcriptional survival factor and coactivator [3, 23, 61]. LEDGF plays a pivotal role in cellular survival, and is known to be a preventive factor in eye disorders [7, 23, 63, 68]. Recently, elevated expression of LEDGF has been reported in cancer cells [5] and it has been implicated in carcinogenesis and cancer progression. The involvement of LEDGF has been reported in HIV-1 integration [14], autoimmune disorders [5, 69–72], and cancer. It acts by forming a chimeric protein with NUP 98 [73]. In addition, LEDGF plays a role in lens epithelial-to-fiber cell terminal differentiation [19]. These studies underscore the wide range of roles played by LEDGF, ranging from cellular protection to promotion of cellular abnormalities. However, the mechanisms by which it is involved in various cellular events and how it specifically plays its various roles remain the subject of active investigation. In the present study, we attempted to identify certain regulatory roles, and to show that at least some functions of LEDGF are associated with its posttranslational modification Sumoylation/deSumoylation, which regulates LEDGF transactivation capacity (Fig. 6). The same reversible modifier, Sumoylation, regulates LEDGF expression by regulating its transactivator, specificity protein Sp1 (Fig. 9). Furthermore, biological functions of molecules are primarily dependent on their cellular localization, expression levels and posttranslational modification. Posttranslational modifications such as phosphorylation, acetylation, ubiquitization and/or Sumoylation can alter the activity of proteins by modifying their DNA binding affinity, transactivation capacity [24, 74–76]. Recently Sumoylation has been implicated in diverse regulatory functions including subcellular compartmentalization, protein stability, chromatin structure regulation, transcription factor activity, DNA binding, and protein complex assembly [42, 44]. A growing list of transcription factors and co-regulators has been shown to be modified by Sumo, indicating that Sumo modification is important in the regulation of gene transcription [77, 78]. The process involved in the Sumoylation pathway is analogous to ubiquitization and requires a specific E1 activating enzyme (SAE1/SAE2), a Sumo-specific E2-conjugating enzyme (Ubc9), and an E3 ligating enzyme [78, 79]. The Sumoylation target is a lysine that occurs in the consensus motif; ¥ KXE, where ¥ is a hydrophobic amino acid and X is any residue. Our study with deletion mutants and point mutation disclosed that LKID positioned at 364aa of the LEDGF was a Sumo1 modification motif. We identified K364as the primary site and found that it lies inside the IBD domain [80–82]. Furthermore, in the current study, we found that an amount of naturally occurring LEDGF is constitutively Sumoylated by Sumo1 to lysine (K) 364 within IBD domain, which is an evolutionarily well conserved region (Fig. 4) [83]. By using SUMOplot (a web-based program to predict Sumo1 conjugation in protein, Abgent) coupled with in vitro and in vivo Sumoylation assays, we found that LEDGF is Sumoylated by Sumo1 and Senp-1 as the Sumo proteases for LEDGF deSumoylation. Interestingly, protein expression analysis showed that sufficient endogenous LEDGF exists in Sumoylated form in the nuclear extracts of hLECs and mLECs (Fig. 1). It was shown that LEDGF is polySumoylated [21] ; however, surprisingly, we found that endogenous LEDGF is monoSumoylated (Fig. 1, 2 and 3). This discrepancy may be associated with cell types, cell background, or cellular microenvironment or due to ectopic/forced expression of molecules.
The most important finding of our study is that the expression and transcriptional activity of LEDGF is regulated by the dynamical process of Sumoylation and deSumoylation. Sumo1 conjugation with LEDGF was further evidenced by immunoprecipitation experiments in which proteins were first immunoprecipitated with anti-Sumo1 antibody and analyzed in immunoblot with LEDGF antibody (Fig. 5). These data indicate that endogenous LEDGF with a molecular weight of ~87kDa was conjugated with Sumo1. However, mobility of Sumoylated LEDGF was retarded, reflecting an ~12–15 kDa band shift, which demonstrates that naturally occurring cellular LEDGF modification may involve only a single Sumo1 molecule in LECs under normal physiological conditions (Fig. 1). Moreover, we could not rule out polySumoylation of LEDGF in other cell types or when cells are ectopically overexpressed with interacting molecules, Sumo1 and LEDGF. The Sumoylation of endogenous LEDGF may require conformational changes that are not readily available to EGFP-linked Sumo1 in presence of endogenous Sumo1. Possibly, low levels of other enzymes involved in Sumoylation processing did not cooperate with exogenously expressed Sumo1 in the recruitment of proteins that are Sumoylated before they associate with gene [84] . Moreover, posttranslational modification by Sumoylation has been reported for a variety of proteins, including several transcriptional regulators, such as p53, c-Jun, c-Myb, AP-2, androgen receptor, promyelocytic leukemia protein (PML), and IκBα [85–87]. Covalent attachment of the Sumo1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity [85]. Sumo1 modification has been found to have diverse substrate-specific functions. It has been shown to act antagonistically to ubiquitization by enhancing protein stability, as exemplified by Sumoylation of the NF-κB inhibitor IκBα [88]. Sumo1 modification of IκBα inhibits NF-κB activation. Our findings indicate that Sumoylation, in general, has a suppressive effect on LEDGF transcription, at least in LECs, a mutant LEDGF (K364R) cannot be Sumoylated, and overexpression of this mutant protein increases the transcriptional activity of LEDGF. Also, overexpression of Senp-1 enhances LEDGF’s transcriptional activity. Senp-1 is a nuclear protease that appears to de-conjugate a large number of Sumoylated proteins [39]. It has been identified in organisms ranging from yeast to mammals. Although several Senps have been reported in mammals, Senp-1 is the most often studied and is known to regulate the activity of many transcriptional factors [86]. We found that Senp-1 profoundly enhanced LEDGF-dependent transcription, which is very important in light of our earlier research showing LEDGF to be implicated in tumor progression and highly expressed in cancer cells (unpublished data). Senp-1 is also highly expressed in prostate cancer cells, and silencing of Senp-1 gene attenuates progression and growth of such cells [89]. We expect that silencing of Senp-1 will also reduce LEDGF expression. Elevated LEDGF/p75 expression has also been found in human breast and bladder carcinomas, and its ectopic overexpression increases the tumorigenic potential of human cancer cells in murine models [4].
Sumo1 modification may alter other properties of its targets and consequently their activities. p53, it affects the transactivation capacity of p53 as well as its intracellular localization [90, 91]. A family of cystein proteases (Senps) specifically hydrolyzes Sumo isopeptide bonds. For example, mammals have at least nine Senps, localized in different subcellular compartments, such as at the PML nuclear bodies (Senp-1), the cytoplasm (Senp-6), the nucleolus (Senp-3), and the nuclear pore (Senp-2). In our studies, the enzymatic activity of Senp-1 was used to independently confirm the effect of Sumoylation on the transactivating capacity of LEDGF. We found that Sumoylation/deSumoylation is at least one mechanism of controlling LEDGF activity, and it may be associated with cellular background. In particular, Sumoylation of LEDGF led to transrepression of stress response genes such as small heat shock protein genes (Fig. 6). Our experiments showed that cells transfected with mutant EGFP-LEDGF (K364R) had higher activation of small heat shock protein genes. In addition, we found that Senp-1 upregulated hsps transcription and endogenous expression levels more than did Sumo1 (Fig. 6). It was interesting to observe the finding depicted in Fig. 6A and B, showing that an increase in Sumo1 concentration was associated with a decrease in LEDGF protein level (Fig. 6A, panel a and b ) and with a decline in the expression of Hsp27 protein (Fig. 6A, panel c). In contrast, an increase expression of Senp-1 was associated with an increase of LEDGF and Hsp27 protein level (Fig. 6B).
Furhther more, our data demonstrate that LEDGF and mutant LEDGF pEGFP-K364R localization patterns were indistinguishable from one another, indicating that Sumo1 conjugation or deSumoylation of LEDGF did not alter LEDGF’s subcellular localization, making LEDGF available for interaction with DNA binding. Furthermore, with Western analysis, we examined whether mutant LEDGF pEGFP-K364R is more prevalent than LEDGF in cells (Fig. 5), arguing that deSumoylated LEDGF is stable and not a target of a quick ubiquitization pathway as is Sumoylated LEDGF. While these results were derived from exogenously expressed LEDGF tagged to EGFP, similar results were reported earlier [21]. Moreover, stabilization and regulation of LEDGF response in favor of cellular survival during environmental stress is essential. In general, regulation of biological processes can be achieved through controlling transcription and translational level. In the current study, we found that LEDGF activity was under the control of both levels. Moreover, LEDGF activated stress response genes by binding STRE and HSE. However, Sumo1 modification of HSF2 has been reported to have greater DNA binding activity. Goodson and associates have proposed that this increase of DNA binding activity is associated with conformational changes [92]. Furthermore, Our experiments also revealed that Senp-1 enhanced the DNA binding activity of LEDGF in nuclear extract of cells in comparison to controls, while it was decreased in cells treated with Sumo1, emphasizing that change in DNA binding activity of LEDGF can be related to either conformational changes or less abundance of LEDGF protein due to repression of LEDGF expression or stability due to Sumoylation (Fig. 7A). Sumo1 conjugation of LEDGF has been reported to decrease half-life of LEDGF [21]. Our data, however, revealing an increased level of LEDGF protein in Senp-1 treated cells (Figs. 5 and 6B), raises the possibility that this increase of LEDGF protein may be associated with deSumoylation of LEDGF. Several reports have shown that Sumoylation both stabilizes and destabilizes the proteins and determines their fate and integrity [62]. It is also possible that Sumoylation of LEDGF may alter its localization patterns. In this case Sumoylated LEDGF is not available to bind the DNA. But our study ruled out this possibility, as localization patterns of LEDGF and mutant LEDGF pEGFP-K364R were indistinguishable and both predominantly localized in nucleus (Fig. 5D). However, the level of LEDGF Sumoylation and its potential for repression can be regulated by the opposing activities of Sumoylases and deSumo proteases. Thus balance in expression and activity of the two are essential to determine the cellular signaling that determines the fate of cells.
Furthermore, our work showed that the protein level of LEDGF was increased with an increase of Senp-1, while levels were decreased in cells overexpressing Sumo1. Two possibilities seemed to exist: (i) LEDGF accumulated in the presence of Senp-1 (increase of half-life) due to inhibition of degradation by deSumoylation of LEDGF [21, 52], or (ii) Senp-1-dependent upregulation of LEDGF transactivation counteracted transrepression by Sumo1. Real-time PCR data revealed that the latter was the case; expression of LEDGF mRNA was significantly increased in cells overexpressing Senp-1 (Fig. 8A) compared to cells overexpressing Sumo1 or control vector. Senp-1-induced increased expression of LEDGF mRNA indicated that LEDGF may be transcriptionally regulated. Our cotransfection experiments involving promoter activity assay revealed, indeed, that Senp-1 upregulated LEDGF transcription, and promoter activity was dramatically increased in a dose-dependent fashion (Fig. 8B). The evidence of Senp-1’s role in controlling other transcription factors is indeed compelling. It functions in two ways, by up- or down-regulating transcriptional activity and by modulating DNA binding activity, which is exemplified by GATA-1, HSF1 or NF-kB to name a few [93–95]. However, LEDGF activity was not fully eliminated by Sumoylation or deSumoylation. We reasoned that LEDGF in cells is not absolutely Sumoylated or deSumoylated, as both are dynamic phenomena. We surmise that the balance is established to maintain optimum cellular signaling beneficial to cells and cell background. Certainly, further investigation is required as to how this balance of Sumoylation/deSumoylation occurs in cells. However, Sumoylation is a very dynamic process involving on one hand the conjugation components and on other hand the deconjugation machinery. Thus, short period of cycles of conjugation and deconjugation may state the existence of usually very low steady state of target gene modification within cellular microenvironment [96]. However, this may be true for transcription factors such as c-Jun and c-fos are deSumoylated within minutes. In contrast, RanGAP-1, which is particularly stably and constitutively Sumoylated and is almost (possibly) fully deconjugated, arguing each protein has differential sensitivity and selectivity. Our results also showed that a portion of LEDGF is constitutively Sumoylated and may have differential affinity to Sumo1 conjugation, and this affinity may be changed according to cellular background.
Our study is the first demonstration of the functional significance of LEDGF deSumoylation. While Senp-1 enhanced the binding of LEDGF to its binding element, this may be attributable to a higher abundance of unSumoylated protein due to higher transcriptional expression of LEDGF and its deSumoylation by Senp-1 (Figs. 6 and 8). However, no effect of LEDGF Sumoylation on its chromatin binding activity has been shown. The difference may lie in conformational changes in exogenously expressed LEDGF and Sumo1 or different cell types used for study or cellular microenvironment. Several reports have emphasized that Sumo or Senp activities are dependent on cell type and cell backround or cellular microenvironment [97]. Furthermore, we found that LEDGF promoter ranging from -170/+35 bore Sp1 binding sites and predicted that transrepression of LEDGF or its transactivation can be associated with Sp1. Our data revealed that Sp1 indeed regulated LEDGF transcription. Senp-1 upregulation of Sp1 may be associated with its higher abundance in cells. Sumo1 conjugation is known to decrease Sp1 transcriptional activity [62]. Our results clearly demonstrate that Sumo1 downregulated LEDGF transcription, and this repression of LEDGF gene may be due to destabilization and degradation of Sp1 by Sumo1 [62, 98]. A previous report stated that constitutively Sumo-modified Sp1 was a poorer transactivator than Sp1 [62]. Modification of Sp1 by Sumo1 was found to affect the stability of Sp1, thereby inhibiting Sp1-dependent transcription. Another report indicated that Sumoylation inhibits the cleavage of the negative N-terminal regulatory domain of Sp1 and p21WAF1/CIP1 transcriptional activity [62, 98]. Only a small fraction of total Sp1 is Sumo1-modified, and this difference in activity suggests that Sumoylation plays an important role in Sp1-dependent transcription. When a LEDGF transactivation assay was performed after overexpressing hLECs with Sumo1 protein, we noticed a reduced CAT values. In a similar assay after overexpressing Sumo hydrolase, Senp-1, the CAT value was much higher. Our finding demonstrated that mutant LEDGF pEGFP-K364R (where Sumo1 conjugation sites were disrupted) enhanced cell growth and cell survival. Results with cells that received Senp-1 were similar, suggesting that deSumoylation is functionally important for cells in normal physiological conditions. While Sumoylation in normal conditions may maintain cellular homeostasis by restricting cell growth, and such cells gain resistance against stressors, Sumo- transfected cells were more resistant to heat stress and survived better than Senp-1 transfected cells. However, further work is needed to determine the underlying mechanism by which Sumo1 conjugation of LEDGF provides cytoprotection against stress. We are exploring whether Sumoylated LEDGF interacts with other proteins that stablize the protective function of LEDGF during stress.
Finally, we found that LEDGF activity was controlled by dual mechanisms that include transcription and posttranslational modification of LEDGF, and these processes are under the control of the reversible Sumoylation process. Based on our results, we propose LEDGF Sumoylation and deSumoylation as a molecular switch that determines cellular integrity/fate by regulating LEDGF-dependent cellular signaling within the cellular microenvironment. If this process goes awry, the aberrant expression of LEDGF leads to cellular abnormalities. In conclusion, our data show that naturally occurring LEDGF is constitutively Sumoylated in a process involving Ubc9 (E2), that Senp-1 is one peptidase involved in LEDGF deSumoylation, and that this dynamic process controls LEDGF activity by regulating its expression and protein modification.
Materials and Methods
Cell Culture
Human lens epithelial cells (hLECs) (a gift of Dr. V. N. Reddy, Eye Research Institute, Oakland University, Rochester, MI, U.S.A) [99] and mouse LECs (mLECs) were maintained routinely in our laboratory following the method as described elsewhere [99]. Briefly, cells were cultured in a 75-mm tissue culture flask in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 15% or 10% heat-inactivated fetal bovine serum (FBS), 100 μg/ml streptomycin and penicillin in a 5% CO2 environment at 37°C following the standard methods. Cells were harvested and cultured in 96, 24, 48 or 6 well plates and 100 mm petri dishes according to the requirement of the experiment.
Generation of prokaryotic expression vectors
Subcloning techniques described by Sambrook et al [100] were used throughout these experiments. A fusion protein between LEDGF and partial LEDGF with glutathione-S-transferase (GST), generated by inserting the entire coding sequence of the LEDGF cDNA into the BamHI and EcoRI sites of a pGEX-2T vector (Pharmacia Biotech, Piscataway, NJ), was used to transform Escherichia coli (BL21) [101]. Deletion constructs of GST-LEDGF, N-terminal (1–250 aa) and C-terminal (170–530 aa) were also prepared. The expression of the GST-LEDGF fusion protein was induced with isopropyl-D-thiogalactopyranoside (IPTG). Proteins were purified with glutathione-Sepharose 4B beads (Pharmacia Biotech) following the manufacturer's protocol. Similarly, a construct containing a green fluorescent protein (GFP) and LEDGF cDNA was also generated with the ‘living color system’ (Clontech, Palo Alto, CA) using the plasmid vector pEGFP-C1 (Clontech) for eukaryotic expression [9]. Also, the full length of Sumo1 was subcloned into pGEX-5X-3 (Pharmacia Biotech) vector for prokaryotic expression by creating BamH1 and Xho1 sites in the forward and reverse primers, respectively (Forward primer 5’-CAGGATCCTCATGTCTGACCAGGAGGCA-3 ’ and Reverse Primer 5 ’-CCGTCTCGAGCACCACATTACAAAAGAAC-3’. Additionally, GST-HSF1 construct was made by subcloning full length of HSF1coding region into pGEX-5X-3 vector using specific primers and sequenced. Sequence was confirmed by automated DNA sequencing and purified plasmid was transformed into E. coli BL21 and protein was purified.
Construction of pEGFP-Sumo1
For the eukaryotic expression, full-length of Sumo1 cDNA was sub cloned into pEGFP-C1 vector. The coding region of Sumo1 was amplified by PCR from human lens cDNA library using forward (5'-CCGTCGACATGTCTGACCAGGAG-3') and reverse primer (5'-TCGGATCCGTTTTGAACACCACA-3') with restriction enzyme sites, SalI and BamHI. The PCR product was digested and ligated into pEGFP vector. pFlag-Senp-1 was a generous gift from Dr. Yeh, University of Texas MD Anderson Cancer Center, Texas. All the Transfection experiments were carried out either with Superfactamine Reagent (Invitrogen) or using Neon Transfection system (Invitrogen).
Construction of LEDGF gene promoter linked to CAT
5' flanking region of human LEDGF gene was isolated and sequenced as reported previously [61]. A construct of –5139 bp was prepared by ligating it to basic pCAT vector (Promega). Similarly, constructs of different sizes were prepared with were made prepared with appropriate sense primers bearing SacI or MluI and antisense with NheI and ligated into pCAT-basic vector as described earlier [61]. The plasmid was amplified and used for CAT assay.
Construction of Hsp27 and alpha B-crystallin-CAT
pCAT-Hsp27 and pCAT-αB-crystallin constructs were engineered as described previously [49]. Briefly, the 5'-flanking region of the human Hsp27 gene was isolated with a genomic PCR kit (Clontech) using specific primers. A forward primer containing a SacI site (5'-GCGTCGAGCTCTCGAATTCATTTGCTT-3') and reverse primer with a XhoI site (5'-GCTCTCGAGGTCTGCTCAGAAAAGTGC-3') were used to generate the fragment, which was cloned between the EcoRI sites of the TA vector (Invitrogen Corp., Carlsbad, CA, USA). Similarly, a fragment comprising the 5'-flanking region of the human αB-crystallin promoter (a gift from Dr. Piatigorsky, NEI, NIH) was prepared using specific primers. A forward primer with a SacI site (5'-CTCTCTTCCAAGAGCTCACAAAG-3') and reverse primer containing a XhoI site (5'-ATGGTGGCTACTCGAGAGTGA-3') were used to generate the above fragment, which was cloned between the EcoRI sites of the TA vector. The Hsp27 and αB-crystallin/TA constructs were digested with SacI and XhoI and promoter fragments were ligated to pCAT-Basic vector (Promega), using the appropriate restriction enzymes.
Preparation of small interfering RNAs
The LEDGF-specific small interfering (si)RNA expression plasmid was designed according to the method described earlier [64]. The sequence was selected from location 1340–1360 (5'-AAAGACAGCATGAGGAAGCGA-3'). The sense and antisense oligonucleotides with the internal loop were synthesized by Invitrogen. These were annealed and ligated into the BamHI and HindIII sites of pSilencer 4.1-CMV hygro (Ambion). pSilencer 4.1- pCMVhygro expressing a scrambled siRNA (Ambion) was used as a control.
Site Directed Mutagenesis (SDM)
GST-K364R and pEGFP-K364R constructs carrying a substitution at lysine 364 for arginine was prepared using the Quickchange Site-Directed Mutagenesis kit (Stratagene) following the company's protocol. Briefly, amino acid exchanges were generated by point mutations in the pEGFP-LEDGF using the following complimentary primers (K364R) Forward: 5’-ACATGCTGAGATTAAGAATTCACTCAGAATTGATAATCTTGA-3’ Reverse: 5’-TCAAGATTATCAATTCTGAGTGAATTCTTAATCTCAGCATGT-3’ Following site-directed mutagenesis, XL1-Blue super-competent cells (Stratagene Inc, CA) were subsequently transformed with mutated cDNA, and clones were grown on Luria-Bertani/Kanamycin plates. The plasmid was amplified, and the mutation was confirmed by sequencing. The mutated plasmid was transfected and intracellular translocation was determined with fluorescent microscopy.
Cotransfection and Promoter activity assay
The CAT assay was performed using a CAT-ELISA kit (Roche Diagnostics). Cells were transfected/co-transfected with various reporter constructs (pCAT-LEDGF, pCAT-Hsp27 and pCAT-αB crystallin) and/or pEGFP-Sumo1, pFlag-Senp-1, pEGFP-LEDGF or pEGFP-K364R expression vectors. After 48 or 72 h of incubation, cells were harvested, and extracts were prepared and protein was normalized. CAT-ELISA was performed to monitor CAT activity, following the manufacturer's protocol. Absorbance was measured at 405 nm using a microtiter plate ELISA reader. Trans-activation activities were adjusted for transfection efficiencies using GFP values.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as described earlier [101]. Oligos containing HSE or STRE elements were commercially synthesized, annealed, and end-labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England Biolabs, Inc). The binding reaction was performed in 20 μl of binding buffer containing 20 mM Tris-HCl (pH 8.0), 75 mM KCl, 5% glycerol, 50 μg/ml bovine serum albumin, 0.025% Nonidet P-40, 1 mM EDTA, 5 mM DTT, and 1 μg of poly (dI/dC). Five fmol (1000 cpm) of the end-labeled probe were incubated on ice with GST-LEDGF or GST-K364R fusion protein. Samples were then loaded on 5% polyacrylamide gel in 0.5X TBE buffer for 2h at 10 V/cm. The gel was dried and autoradiographed.
SUMOplot or Sumo site analysis and in vitro Sumoylation assay
To predict and identify Simulation site(s) in LEDGF protein two different computational prediction programs, SUMOplot analysis program (Abgent) and PIC-Based Sumo Site Prediction Server were used. In vitro Sumo modification reaction was performed in accordance with manufacture’s protocol (LAE Biotech # K007 kit). Bacterial expressed GST, GST-LEDGF, GST-K364R and GST-HSF1 were purified using glutathione-Sepharose 4B (Pharmacia) as per manufacture's specifications and then used as substrates in the in vitro reaction. 500ng GST-LEDGF, GST-K364R or GST-HSF1, 150ng SAE I/SAE II, 1μg Ubc9, and 1μg Sumo1 were incubated in the reaction buffer containing 20mM Hepes (pH 7.5), 5mM MgCl2, and 2mM ATP in a total volume of 20μl for 1h at 37°C. Reactions were stopped by addition of 2X SDS-PAGE sample buffer and subjected to SDS-PAGE followed by Western analysis. Blotted membrane was immunostained with primary antibodies (anti-GST, anti-Sumo1 or anti-LEDGF). Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies. Specific protein bands were visualized by incubating the membrane with luminal reagent (Santa Cruz Biotechnology) and recorded with FUJIFILM-LAS-4000 luminescent image analyzer (FUJIFILM Medical system).
In vivo Sumoylation assay
hLECs were co-transfected with either pEGFP-LEDGF or pEGFP-K364R and pEGFP-Sumo1. After 48h, cytoplasmic and nuclear extracts were prepared as described earlier [49, 101]. Nuclear extract was incubated with 3–4 μg anti-LEDGF monoclonal antibody (BD Biosciences (cat. no. 611714 ) in binding buffer provided in IP kit (Pierce), and kept at 4°C for 2h followed by addition of 40μl of Protein A–Sepharose pre-absorbed with BSA and rotated overnight at 4°C. The immunoprecipitates were collected by centrifugation and washed several times prior to boiling in SDS-sample buffer. Precipitates were resolved on polyacrylamide gels (4–20% SDS gel) and analyzed through Western blotting using anti-LEDGF monoclonal or anti-Sumo1 or anti-GFP antibodies.
Western blot analysis
Whole cell extracts or nuclear extracts were prepared as described earlier [49, 101] and protein blot analysis was conducted. Equal amounts of protein samples were loaded onto a SDS-10% or 7.5% or 4–20% (w/v) PAGE, blotted onto a PVDF membrane, and then immunostained with primary antibodies at appropriate dilutions. LEDGF monoclonal (BD Biosciences; cat. no. 611714) and polyclonal (Lifspan Bioscience; cat. no. LS-C31241 and SantaCruz Biotech; cat. no. sc-33371 ) antibodies were used. GFP (Cat. no. sc-8334), GST (Cat. no. sc-53909), Sumo1 (Cat. no. sc-9060), Senp-1 (Cat. no. sc-46634) and Hsp27 (Cat. no. sc-1048) antibodies were purchased from SantaCruz Biotechnology. The membranes were then incubated with horseradish peroxidase conjugated with secondary antibodies. Specific protein bands were visualized by incubating the membrane with luminal reagent (Santa Cruz Biotechnology) and recorded with FUJIFILM-LAS-4000 luminescent image analyzer (FUJIFILM Medical system Inc, USA). To ascertain comparative expression and equal loading of the protein samples, the membrane stained earlier was stripped and re-probed with β-actin antibody (Abcam).
Real-time PCR or Quantitative PCR
Total RNA was isolated using the single-step guanidine thiocyanate/ phenol/chloroform extraction method (Trizol Reagent; Invitrogen) and converted to cDNA using Superscript II RNAase H-Reverse Transcriptase. Quantitative real-time PCR was performed with SYBR Green Master Mix (Roche Diagnostic Corporation, Indianapolis, IN) in a Roche R LC480 Sequence detector system (Roche Diagnostic Corporation). PCR conditions consisted of 10-min hot start at 95°C, followed by 45 cycles of 10s at 95°C, 30s at 60°C, and 10s at 72°C. Primer sequence as follows: LEDGF, 5'-CAGCAACAGCATCTGTTAATCTAAA-3' and Reverse primer: 5'-GGGCTGTTTTACCATTTTGG-3'; β-actin, 5’-CCAACCGCGAGAAGATGA-3’ and 5’-CCAGAGGCGTACAGGGATAG-3’. Expression levels of target genes were normalized to the levels of β-actin as an endogenous control in each group.
Cell survival assay (MTS assay)
A colorimetric MTS assay (Promega) was performed as described earlier [49, 101] . This assay of cellular proliferation uses 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2 to 4-sulfophenyl)-2H-tetrazolium salt (MTS; Promega, Madison, MI, USA). Upon addition to medium containing viable cells, MTS is reduced to a water-soluble formazan salt. The OD490 nm value was measured after 4h with an ELISA reader.
Statistical method
Data are presented as Mean ± S.D. of the indicated number of experiments. Data were analyzed by Student's t-test when appropriate. A p value of < 0.05 and <0.001 was defined as indicating a statistically significant difference.
Acknowledgments
Grants provided by the National Eye Institute, NIH (EY-13394 and EY017613) (to DPS) and Research for Preventing Blindness (R.P.B) are gratefully acknowledged. Grant support by American Health Assistance Foundation (AHAF) (to NF) is gratefully acknowledged.
The abbreviations
- LEDGF
lens epithelium-derived growth factor
- Sumo1
small ubiquitin-like modifier-1
- Senp-1
sumo-specific proteases-1
- GFP
green florescence
- GST
glutathione-s-transferase
References
- 1.Nguyen TA, Boyle DL, Wagner LM, Shinohara T, Takemoto DJ. LEDGF activation of PKC gamma and gap junction disassembly in lens epithelial cells. Exp Eye Res. 2003;76:565–572. doi: 10.1016/s0014-4835(03)00049-6. S0014483503000496 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Singh D, Kimura A, Chylack L, Shinohara T. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene. 2000;242:265–273. doi: 10.1016/s0378-1119(99)00506-5. [DOI] [PubMed] [Google Scholar]
- 3.Shinohara T, Singh DP, Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog Retin Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. S1350946202000071 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Daugaard M, Kirkegaard-Sørensen T, Ostenfeld MS, Aaboe M, Høyer-Hansen M, Ørntoft TF, Rohde M, Jäättelä M. Lens Epithelium-Derived Growth Factor Is an Hsp70-2 Regulated Guardian of Lysosomal Stability in Human Cancer. Cancer Res. 2007;67:2559–2567. doi: 10.1158/0008-5472.can-06-4121. [DOI] [PubMed] [Google Scholar]
- 5.Daniels T, Zhang J, Gutierrez I, Elliot ML, Yamada B, Heeb MJ, Sheets SM, Wu X, Casiano CA. Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. doi: 10.1002/pros.20112. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland HG, Newton K, Brownstein DG, Holmes MC, Kress C, Semple CA, Bickmore WA. Disruption of Ledgf/Psip1 Results in Perinatal Mortality and Homeotic Skeletal Transformations. Mol Cell Biol. 2006;26:7201–7210. doi: 10.1128/mcb.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui H, Lin L-R, Singh DP, Shinohara T, Reddy VN. Lens Epithelium-Derived Growth Factor: Increased Survival and Decreased DNA Breakage of Human RPE Cells Induced by Oxidative Stress. Invest Ophthalmol Vis Sci. 2001;42:2935–2941. [PubMed] [Google Scholar]
- 8.Dietz F, Franken S, Yoshida K, Nakamura H, Kappler J, Gieselmann V. The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem J. 2002;366:491–500. doi: 10.1042/BJ20011811. BJ20011811 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh DP, Kimura A, Chylack LT, Jr, Shinohara T. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene. 2000;242:265–273. doi: 10.1016/s0378-1119(99)00506-5. S0378-1119(99)00506-5 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Ge H, Si Y, Roeder R. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723– 6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stec I, Nagl S, van Ommen G, den Dunnen J. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1– 5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 12.Bartholomeeusen K, De Rijck J, Busschots K, Desender L, Gijsbers R, Emiliani S, Benarous R, Debyser Z, Christ F. Differential interaction of HIV-1 integrase and JPO2 with the C terminus of LEDGF/p75. J Mol Biol. 2007;372:407–421. doi: 10.1016/j.jmb.2007.06.090. S0022-2836(07)00917-5 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Singh DP, Kubo E, Takamura Y, Shinohara T, Kumar A, Chylack LT, Jr, Fatma N. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J Mol Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. S0022-2836(05)01301-X [pii] [DOI] [PubMed] [Google Scholar]
- 14.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. M209278200 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci U S A. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. 0914142107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busschots K, Voet A, De Maeyer M, Rain JC, Emiliani S, Benarous R, Desender L, Debyser Z, Christ F. Identification of the LEDGF/p75 binding site in HIV-1 integrase. J Mol Biol. 2007;365:1480–1492. doi: 10.1016/j.jmb.2006.10.094. S0022-2836(06)01521-X [pii] [DOI] [PubMed] [Google Scholar]
- 17.Hussey D, Moore S, Nicola M, Dobrovic A. Fusion of the NUP98 gene with the LEDGF/p52 gene defines a recurrent acute myeloid leukemia translocation. BMC Genet. 2001;2:20. doi: 10.1186/1471-2156-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fatma N, Singh DP, Shinohara T, Chylack LT. Transcriptional Regulation of the Antioxidant Protein 2 Gene, a Thiol-specific Antioxidant, by Lens Epithelium-derived Growth Factor to Protect Cells from Oxidative Stress. J Biol Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 19.Kubo E, Singh DP, Fatma N, Shinohara T, Zelenka P, Reddy VN, Chylack LT. Cellular distribution of lens epithelium-derived growth factor (LEDGF) in the rat eye: loss of LEDGF from nuclei of differentiating cells. Histochem Cell Biol. 2003;119:289–299. doi: 10.1007/s00418-003- 0518-3. [DOI] [PubMed] [Google Scholar]
- 20.Chylack LT, Jr, Fu L, Mancini R, Martin-Rehrmann MD, Saunders AJ, Konopka G, Tian D, Hedley-Whyte ET, Folkerth RD, Goldstein LE. Lens epithelium-derived growth factor (LEDGF/p75) expression in fetal and adult human brain. Exp Eye Res. 2004;79:941–948. doi: 10.1016/j.exer.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 21.Bueno MT, Garcia-Rivera JA, Kugelman JR, Morales E, Rosas-Acosta G, Llano M. SUMOylation of the lens epithelium-derived growth factor/p75 attenuates its transcriptional activity on the heat shock protein 27 promoter. J Mol Biol. 2010;399:221–239. doi: 10.1016/j.jmb.2010.03.063. S0022-2836(10)00338-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Singh DP, Ohguro N, Chylack LT, Jr, Shinohara T. Lens epithelium-derived growth factor: increased resistance to thermal and oxidative stresses. Invest Ophthalmol Vis Sci. 1999;40:1444–1451. [PubMed] [Google Scholar]
- 24.Freiman RN, Tjian R. Regulating the Regulators: Lysine Modifications Make Their Mark. Cell. 2003;112:11–17. doi: 10.1016/s0092-8674(02)01278-3. [DOI] [PubMed] [Google Scholar]
- 25.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. p300 Transcriptional Repression Is Mediated by SUMO Modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 26.Hsu Y-HR, Sarker KP, Pot I, Chan A, Netherton SJ, Bonni S. Sumoylated SnoN Represses Transcription in a Promoter-specific Manner. J Biol Chem. 2006;281:33008–33018. doi: 10.1074/jbc.M604380200. [DOI] [PubMed] [Google Scholar]
- 27.Stielow B, Sapetschnig A, Wink C, Kruger I, Suske G. SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 2008;9:899–906. doi: 10.1038/embor.2008.127. doi: http://www.nature.com/embor/journal/v9/n9/suppinfo/embor2008127_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perdomo J, Verger A, Turner J, Crossley M. Role for SUMO Modification in Facilitating Transcriptional Repression by BKLF. Mol Cell Biol. 2005;25:1549–1559. doi: 10.1128/mcb.25.4.1549-1559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Dyck F, Delvaux EL, Van de Ven WJ, Chavez MV. Repression of the Transactivating Capacity of the Oncoprotein PLAG1 by SUMOylation. J Biol Chem. 2004;279:36121–36131. doi: 10.1074/jbc.M401753200. M401753200 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. Global Shifts in Protein Sumoylation in Response to Electrophile and Oxidative Stress. Chem Res Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 31.Muller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 32.Bailey D, O'Hare P. Comparison of the SUMO1 and ubiquitin conjugation pathways during the inhibition of proteasome activity with evidence of SUMO1 recycling. Biochem J. 2005;392:271–281. doi: 10.1042/bj20050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. M104714200 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem. 2001;276:18513–18518. doi: 10.1074/jbc.M008066200. M008066200 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Bailey D, O'Hare P. Characterization of the Localization and Proteolytic Activity of the SUMO-specific Protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- 38.Hang J, Dasso M. Association of the Human SUMO-1 Protease SENP2 with the Nuclear Pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 39.Cheng J, Wang D, Wang Z, Yeh ETH. SENP1 Enhances Androgen Receptor-Dependent Transcription through Desumoylation of Histone Deacetylase 1. Mol Cell Biol. 2004;24:6021–6028. doi: 10.1128/mcb.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hattersley N, Shen L, Jaffray EG, Hay RT. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol Biol Cell. 2011;22:78–90. doi: 10.1091/mbc.E10-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishida T, Yamada Y. SMT3IP1, a nucleolar SUMO-specific protease, deconjugates SUMO-2 from nucleolar and cytoplasmic nucleophosmin. Biochem Biophys Res Commun. 2008;374:382–387. doi: 10.1016/j.bbrc.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 42.Grace G. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr Opin Genet Dev. 2003;13:108–113. doi: 10.1016/s0959-437x(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 43.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. jcs.02995 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. embor738 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 46.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 47.Tsutsui KM, Sano K, Hosoya O, Miyamoto T, Tsutsui K. Nuclear protein LEDGF/p75 recognizes supercoiled DNA by a novel DNA-binding domain. Nucleic Acids Res. 2011;39:5067–5081. doi: 10.1093/nar/gkr088. gkr088 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishizawa Y, Usukura J, Singh D, Chylack L, Shinohara T. Spatial and temporal dynamics of two alternatively spliced regulatory factors, lens epithelium-derived growth factor (ledgf/p75) and p52, in the nucleus. Cell Tissue Res. 2001;305:107–114. doi: 10.1007/s004410100398. [DOI] [PubMed] [Google Scholar]
- 49.Singh DP, Fatma N, Kimura A, Chylack LT, Shinohara T. LEDGF Binds to Heat Shock and Stress-Related Element to Activate the Expression of Stress-Related Genes. Biochem Biophys Res Commun. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 50.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. doi: http://www.nature.com/nrm/journal/v8/n12/suppinfo/nrm2293_S1.html. [DOI] [PubMed] [Google Scholar]
- 51.Fatma N, Kubo E, Chylack LT, Jr, Shinohara T, Akagi Y, Singh DP. LEDGF regulation of alcohol and aldehyde dehydrogenases in lens epithelial cells: stimulation of retinoic acid production and protection from ethanol toxicity. Am J Physiol Cell Physiol. 2004;287:C508–516. doi: 10.1152/ajpcell.00076.2004287/2/C508. [pii] [DOI] [PubMed] [Google Scholar]
- 52.Yeh ETH. SUMOylation and De-SUMOylation: Wrestling with Life's Processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tateishi Y, Ariyoshi M, Igarashi R, Hara H, Mizuguchi K, Seto A, Nakai A, Kokubo T, Tochio H, Shirakawa M. Molecular Basis for SUMOylation-dependent Regulation of DNA Binding Activity of Heat Shock Factor 2. J Biol Chem. 2009;284:2435–2447. doi: 10.1074/jbc.M806392200. [DOI] [PubMed] [Google Scholar]
- 54.Wu S-Y, Chiang C-M. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 2009;28:1246–1259. doi: 10.1038/emboj.2009.83. doi: http://www.nature.com/emboj/journal/v28/n9/suppinfo/emboj200983a_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. S1097-2765(05)01903-9 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010;24:1242–1252. doi: 10.1101/gad.1917910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seeler J-S, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 59.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 60.Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 61.Sharma P, Fatma N, Kubo E, Shinohara T, Chylack LT, Jr, Singh DP. Lens epithelium-derived growth factor relieves transforming growth factor-beta1-induced transcription repression of heat shock proteins in human lens epithelial cells. J Biol Chem. 2003;278:20037–20046. doi: 10.1074/jbc.M212016200. M212016200 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Spengler ML, Brattain MG. Sumoylation Inhibits Cleavage of Sp1 N-terminal Negative Regulatory Domain and Inhibits Sp1-dependent Transcription. J Biol Chem. 2006;281:5567–5574. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- 63.Machida S, Chaudhry P, Shinohara T, Singh DP, Reddy VN, Chylack LT, Sieving PA, Bush RA. Lens Epithelium-Derived Growth Factor Promotes Photoreceptor Survival in Light-Damaged and RCS Rats. Invest Ophthalmol Vis Sci. 2001;42:1087–1095. [PubMed] [Google Scholar]
- 64.Takamura Y, Fatma N, Kubo E, Singh DP. Regulation of heavy subunit chain of gamma-glutamylcysteine synthetase by tumor necrosis factor-alpha in lens epithelial cells: role of LEDGF/p75. Am J Physiol Cell Physiol. 2006;290:C554–566. doi: 10.1152/ajpcell.00398.2005. 290/2/C554 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Awasthi N, Wagner BJ. Upregulation of Heat Shock Protein Expression by Proteasome Inhibition: An Antiapoptotic Mechanism in the Lens. Invest Ophthalmol Vis Sci. 2005;46:2082–2091. doi: 10.1167/iovs.05-0002. [DOI] [PubMed] [Google Scholar]
- 66.Nakamura H, Izumoto Y, Kambe H, Kuroda T, Mori T, Kawamura K, Yamamoto H, Kishimoto T. Molecular cloning of complementary DNA for a novel human hepatoma-derived growth factor. Its homology with high mobility group-1 protein. J Biol Chem. 1994;269:25143–25149. [PubMed] [Google Scholar]
- 67.Izumoto Y, Kuroda T, Harada H, Kishimoto T, Nakamura H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem Biophys Res Commun. 1997;238:26–32. doi: 10.1006/bbrc.1997.7233. [DOI] [PubMed] [Google Scholar]
- 68.Nakamura M, Singh DP, Kubo E, Chylack LT, Shinohara T. LEDGF: Survival of Embryonic Chick Retinal Photoreceptor Cells. Invest Ophthalmol Vis Sci. 2000;41:1168–1175. [PubMed] [Google Scholar]
- 69.Ayaki M, Ohoguro N, Azuma N, Majima Y, Yata K, Ibaraki N, Singh DP, Ko V, Shinohara T. Detection of cytotoxic anti-LEDGF autoantibodies in atopic dermatitis. Autoimmunity. 2002;35:319–327. doi: 10.1080/0891693021000003198. [DOI] [PubMed] [Google Scholar]
- 70.Ayaki M, Sueno T, Singh DP, Chylack JLT, Shinohara T. Antibodies to Lens Epithelium-Derived Growth Factor (LEDGF) Kill Epithelial Cells of Whole Lenses in Organ Culture. Exp Eye Res. 1999;69:139–142. doi: 10.1006/exer.1999.0701. [DOI] [PubMed] [Google Scholar]
- 71.Ganapathy V, Daniels T, Casiano CA. LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis. Autoimmun Rev. 2003;2:290–297. doi: 10.1016/s1568-9972(03)00063-6. S1568997203000636 [pii] [DOI] [PubMed] [Google Scholar]
- 72.Okamoto M, Ogawa Y, Watanabe A, Sugiura K, Shimomura Y, Aoki N, Nagasaka T, Tomita Y, Muro Y. Autoantibodies to DFS70/LEDGF are increased in alopecia areata patients. J Autoimmun. 2004;23:257–266. doi: 10.1016/j.jaut.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Ahuja H, Hong J, Aplan P, Tcheurekdjian L, Forman S, Slovak M. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 74.Luscher B, Christenson E, Litchfield DW, Krebs EG, Eisenman RN. Myb DNA binding inhibited by phosphorylation at a site deleted during oncogenic activation. Nature. 1990;344:517–522. doi: 10.1038/344517a0. [DOI] [PubMed] [Google Scholar]
- 75.Aziz N, Miglarese MR, Hendrickson RC, Shabanowitz J, Sturgill TW, Hunt DF, Bender TP. Modulation of c-Myb-induced transcription activation by a phosphorylation site near the negative regulatory domain. Proc Natl Acad Sci U S A. 1995;92:6429–6433. doi: 10.1073/pnas.92.14.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holmstrom S, Van Antwerp ME, Iñiguez-Lluhí JA. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc Natl Acad Sci U S A. 2003;100:15758–15763. doi: 10.1073/pnas.2136933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, Meluh PB, Pandolfi PP, Zon LI. SUMO-1 Protease-1 Regulates Gene Transcription through PML. Mol Cell. 2002;10:843–855. doi: 10.1016/s1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 78.Tatham MH, Jaffray E, Vaughan OA, Desterro JMP, Botting CH, Naismith JH, Hay RT. Polymeric Chains of SUMO-2 and SUMO-3 Are Conjugated to Protein Substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 79.Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. doi: http://www.nature.com/emboj/journal/v24/n3/suppinfo/7600552a_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. Journal of virology. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nature structural & molecular biology. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 82.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. Journal of cell science. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 83.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. The Journal of biological chemistry. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 84.Wang L, Zhang J, Banerjee S, Barnes L, Sajja V, Liu Y, Guo B, Du Y, Agarwal MK, Wald DN, et al. Sumoylation of vimentin354 is associated with PIAS3 inhibition of glioma cell migration. Oncotarget. 2010;1:620–627. doi: 10.18632/oncotarget.196. 101101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bies J, Markus J, Wolff L. Covalent Attachment of the SUMO-1 Protein to the Negative Regulatory Domain of the c-Myb Transcription Factor Modifies Its Stability and Transactivation Capacity. J Biol Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- 86.Cheng J, Perkins ND, Yeh ETH. Differential Regulation of c-Jun-dependent Transcription by SUMO-specific Proteases. J Biol Chem. 2005;280:14492–14498. doi: 10.1074/jbc.M412185200. [DOI] [PubMed] [Google Scholar]
- 87.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. S1097-2765(00)80133-1 [pii] [DOI] [PubMed] [Google Scholar]
- 88.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. S1097-2765(00)80133-1 [pii] [DOI] [PubMed] [Google Scholar]
- 89.Kaikkonen S, Jaaskelainen T, Karvonen U, Rytinki MM, Makkonen H, Gioeli D, Paschal BM, Palvimo JJ. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol Endocrinol. 2009;23:292–307. doi: 10.1210/me.2008-0219. me.2008-0219 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Sal GD. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem. 2001;276:18513–18518. doi: 10.1074/jbc.M008066200. M008066200 [pii] [DOI] [PubMed] [Google Scholar]
- 93.Yu L, Ji W, Zhang H, Renda MJ, He Y, Lin S, Cheng E-c, Chen H, Krause DS, Min W. SENP1-mediated GATA1 deSUMOylation is critical for definitive erythropoiesis. J Exp Med. 2010;207:1183–1195. doi: 10.1084/jem.20092215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson M, Park-Sarge O-K, Sarge KD. Regulation of Heat Shock Transcription Factor 1 by Stress-induced SUMO-1 Modification. J Biol Chem. 2001;276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 95.Mabb A, Miyamoto S. SUMO and NF-κB ties. Cell Mol Life Sci. 2007;64:1979–1996. doi: 10.1007/s00018-007-7005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 97.Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson L-M, Parini P, Jänne OA, Gustafsson J-Å, et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRβ in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y-T, Chuang J-Y, Shen M-R, Yang W-B, Chang W-C, Hung J-J. Sumoylation of Specificity Protein 1 Augments Its Degradation by Changing the Localization and Increasing the Specificity Protein 1 Proteolytic Process. J Mol Biol. 2008;380:869–885. doi: 10.1016/j.jmb.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 99.Ibaraki N, Chen S-C, Lin L-R, Okamoto H, Pipas JM, Reddy VN. Human Lens Epithelial Cell Line. Exp Eye Res. 1998;67:577–585. doi: 10.1006/exer.1998.0551. [DOI] [PubMed] [Google Scholar]
- 100.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- 101.Singh DP, Kubo E, Takamura Y, Shinohara T, Kumar A, Chylack LT, Jr, Fatma N. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J Mol Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. S0022–2836(05)01301-X [pii] [DOI] [PubMed] [Google Scholar]